Abstract
Background
Mathematical models have proven helpful in analyzing the virological response to antiviral therapy in hepatitis C virus (HCV) infected subjects.
Objective
To summarize the uses and limitations of different models for analyzing HCV kinetic data under pegylated interferon therapy.
Methods
We formulate mathematical models and fit them by nonlinear least square regression to patient data in order to estimate model parameters. We compare the goodness of fit and parameter values estimated by different models statistically.
Results/Conclusion
The best model for parameter estimation depends on the availability and the quality of data as well as the therapy used. We also discuss the mathematical models that will be needed to analyze HCV kinetic data from clinical trials with new antiviral drugs.
Keywords: Direct antivirals, interferon, hepatitis C virus, mathematical modeling, pegylated interferon, NS5B polymerase inhibitor, NS3/4A protease inhibitor
1. Introduction
1.1 Hepatitis C virus infection
Hepatitis C virus (HCV) is a positive sense RNA virus in the Flaviviridae family. It infects and replicates in hepatocytes (liver cells), but extrahepatic infection in peripheral blood mononuclear cells may also occur 1-3. HCV is genetically variable and genotypes 1 to 7 have been assigned 4. Globally, approximately 170 million people are chronically infected with HCV and 3 to 4 million persons are newly infected each year 5. Genotype 4 is the most prevalent in Africa while genotype 1 is the most prevalent in the other continents 6. HCV is a major risk factor for developing cirrhosis and hepatocellular carcinoma (HCC, liver cancer), which are the major causes of liver transplantation. In the United States, 16,000 individuals are waiting for livers to become available for transplantation 7. Reducing the prevalence of HCV infection and progression to HCC are important health care issues.
1.2 Therapy for chronic HCV infection
Due to the lack of an HCV vaccine, antiviral therapy is currently the only available treatment. However, antiviral therapy is not effective in all patients. Sustained virological response (SVR), which is defined as virus being undetectable at the end of therapy and 6 month later, is achieved only in 50% of treated patients when pegylated interferon α-2a or 2b (PEG-IFN)8, 9 plus ribavirin, the current standard of care, is used.
Recombinant forms of naturally occurring interferon (IFN), IFN α-2a 10 and IFN α-2b 11, were commonly used to treat chronic HCV infected patients in the 1990s. Due to the fact that IFN is a protein that would be digested if taken orally, IFN is administered by injection. Both forms of IFN have a short in vivo half-life (5.1 hours for IFN α-2a, and 2-3 hours for IFN α-2b), and thus the recommended dosing interval for IFN was three times a week. In order to increase the dosing interval peginterferon α-2a (PEG-IFNα-2a) 9 and peginterferon α-2b (PEG-IFN α-2b) 8 were introduced and approved by the FDA. Both PEG-IFN α-2a and PEG-IFN α-2b are recombinant IFN-α with a polyethylene glycol (PEG) chain covalently attached 12-14. The two forms of PEG-IFN not only differ in the type of IFN used but also in the size of the PEG molecule attached to IFN. The molecular weight of the PEG molecule is 40 kD in PEG-IFN α-2a and 12 kD in PEG-IFN α-2b. The large PEG molecule reduces IFN’s rate of elimination from the body 14 (Table 1, Fig. 1) 15. Therefore, PEG-IFN is administered only once weekly, compared with the three times a week administration for standard IFN, which increases compliance and is easier for patients to tolerate. In addition, clinical trials have shown that there is an improvement in the SVR rate when PEG-IFN instead of standard IFN is used in combination with daily ribavirin for 12 months (~45 % vs. ~55 %) 16-18.
Table 1.
Pharmacokinetic parameters of IFN-based drugs.
| Product | Elimination half life (hours) |
Absorption half life (hours) |
tmax (hours) |
Elimination rate (day−1) |
Absorption rate (day−1) |
|---|---|---|---|---|---|
| Roferon A (IFN α-2a) | 5.1 (3.7-8.5) | N/S | 7.3 | 3.26 (4.50-1.96)* | 3.31 (2.32-5.12)+ |
| Intron A (IFN α-2b) | (2-3) | N/S | (3-12) | (5.55-8.32)* | (0.14 -11.1)+ |
| PEGASYS® (peginterferon α-2a) | 80 (50 -140) | N/S | (72 - 96) | 0.21 (0.33-0.12)* | (0.18-0.72)+ |
| PEG-Intron™ (Peginterferon α-2b) | 40 (22-60) | 4.6 | (15 -44) | 0.42 (0.76- 0.28)* | 3.61+ (0.38 - 4.86)* |
Half life (absorption and elimination) and tmax as reported in 8-11 for subcutaneous infusion with the exception of the elimination rate of IFN α-2a, which was given for intravenous infusion. Averages (and ranges between parentheses) are shown. N/S - not shown in 8-11.
Estimated parameters using a non-compartmental model 73.
Estimated from tmax, the time to the peak of concentration, given by Eq. (6).
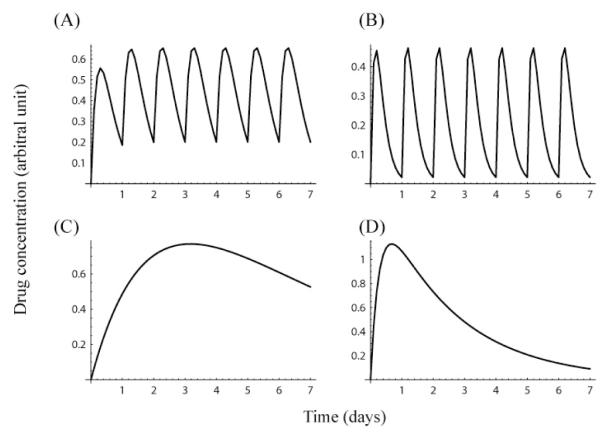
Figure 1.
Drug concentration in patient serum: IFN α-2a (A), IFN α-2b (B), PEG-IFN α-2a (C), and PEG-IFN α-2b (D). Simulation of drug concentrations in serum when IFN (A,B) is administered once daily, on days 0 through 6; and PEG-IFN (C,D) is administered once weekly at day 0 only. The curves represent the solution of the PK model, Eq. (5), using representative PK parameters for each drug. Parameters are: ke=3.26 day−1 and ka=3.31 day−1 (A), ke=7.0 day−1 and ka=5.0 day−1 (B), ke=0.21 day−1 and ka=0.45 day−1 (C), and ke=0.42 day−1 and ka=3.61 day−1 (D). For all drugs, we used an arbitrary value, 1.5 units, for FD/Vd.
To improve treatment outcome, there are many HCV drugs under development: both new IFN-based drugs 19, 20 and direct antivirals that inhibit HCV enzymes such as the HCV protease and HCV polymerase 21-26. These new drugs alone or in combination with PEG-IFN are expected to be the standard care for the treatment of chronic HCV infected patients in the next decade 21, 23.
2. Mathematical modeling: Basic to state-of-the-art
2.1 Why do we need mathematical modeling?
Mathematical modeling of viral infections under therapy has been successful in unraveling the dynamic aspect of chronic infection by providing estimates of key parameters. These estimates were obtained by analyzing the response of individual patients to antiviral therapy and then using nonlinear least squares regression to fit mathematical models to patient data. For example, using this approach to analyze human immunodeficiency virus (HIV) infection 27, 28, it was discovered that the dynamics of HIV infection were not slow even though HIV takes ~10 years, on average, from infection to cause full blown AIDS. Analysis of patient response to therapy showed that virus was both being produced and cleared at extremely rapid rates with close to the entire viral population being renewed every day of infection 29.
Analyses of HCV infection and treatment also showed that the viral half-life is short (~2-3 h) and the half-life of infected hepatocytes ranged from 2 to 70 days 30. Moreover, by modeling HCV RNA declines under treatment, it was proposed that the primary mode of action of interferon involved blocking viral production from infected cells rather than preventing cells from being infected 30. This approach was also instrumental in providing new insights into the mode of action of ribavirin 31 as well as shedding light on the differences in response of diverse patient populations32.
Another important clinical benefit of viral kinetics was the prediction of long term therapy outcome based on early decreases in viral load. For example, SVR is much more likely to be achieved in patients that have a rapid virological response (absence of detectable virus at 4 weeks after the initiation of therapy) or an early virological response (absence of detectable virus at week 12 or >2 log10 decline from baseline) than in patients with slower responses 33-38. Researchers also have found a correlation between achieving SVR and HCV RNA decline for shorter times, such as 1 day 39, 40, 1 week 41 or 2 weeks18, 42 after the initiation of therapy. As shown below, some viral kinetic parameters and pharmacokinetic/pharmacodynamic (PK/PD) parameters have been shown to be associated with SVR achievement.
2.2 Basic mathematical model of HCV dynamics under therapy
Under high-dose (5, 10, 15 million international unit) daily administration of IFN α-2b, the serum level of HCV RNA typically declines in a biphasic manner with a rapid first phase followed by a slower second phase 30. To describe the biphasic kinetics of decline, a basic mathematical model was designed (Fig. 2A). In this model, cells susceptible to infection, i.e., target cells, T, are infected by HCV, V, with rate constant β, resulting in infected cells, I. which produce new virions. The equations describing this system are 30, 41, 43-47.
| (1) |
where T and I are the number of target cells and infected cells, respectively, and V is the viral load.. The target cells are assumed to be generated at a constant rate s, and die at rate d per cell. Infected cells are lost at rate δ per cell, and free virions are produced at rate p per infected cell and are assumed to be cleared by a first order process with rate constant c. In principle, treatment with IFN α-2b could block de novo infection (reducing β) or interfere with virion production reducing p. Analyses of these possibilities showed that the primary mode of IFN action was to partially block virion production 30. This mode of action is incorporated into the model given by Eq. (1) by introducing the parameter ε(t), the effectiveness of IFN. The drug effectiveness, ε(t), is bounded between 0 and 1, where virion production is not suppressed if ε(t)=0 or is fully suppressed if ε(t)=1. To fit the kinetics of viral load decline under high dose daily IFN treatment, it was assumed that (i) the number of target cells, the number of infected cells and the HCV concentration in serum are in steady state before the initiation of therapy, (ii) the number of target cells remains constant for a short period after the initiation of therapy, and (iii) the drug effectiveness is initially zero but after some delay, t0, to account for both the pharmacokinetics of the drug and its need to bind cellular IFN receptors and stimulate expression of interferon regulated genes, the effectiveness increases to a constant level that is sustained until the end of therapy (Fig. 2B). That is, the drug effectiveness is essentially constant during treatment, ε(t)=ε once it becomes active at the site of infection (Fig. 2B). Because of this assumption we call this model a constant effectiveness (CE) model. With these assumptions, the CE model above can be solved explicitly and the solution is
| (2) |
where and A = (εc -λ2)/(λ1 - λ2).
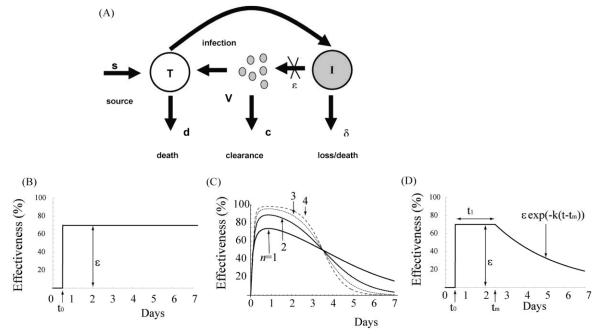
Figure 2.
(A) Schematic illustration of the model used to analyze HCV RNA kinetic data under therapy. Here T represents targets cells, I infected cells and V virus. Therapy is assumed to reduce the amount of virus produced per day by an infected cell from p to (1-ε(t))p. The effectiveness of therapy in blocking viral production, ε(t), is chosen differently in different models. (B) ε(t) for the CE model, (C) ε(t) for the PK/PD model, and (D) ε(t) for the DE model.
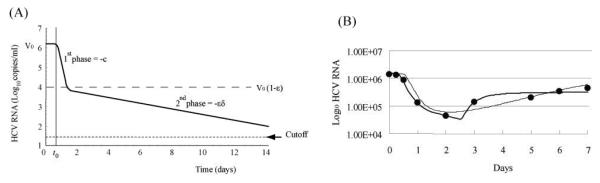
The slope of the first phase of HCV kinetic decline is dependent on the virion clearance rate, c; while the absolute level of decline is related to the effectiveness ε, such that ε=0.9 corresponds to ~1 log10 decline, ε=0.99 to ~2 log10 decline, etc. The second phase slope is approximated by the maximal drug effectiveness, ε, times the infected cell loss rate, δ 30 (Fig. 3A). In patients who have larger εδ, the virus become undetectable by the end of treatment. Thus, we can expect that the patients who have larger εδ may achieve an end of treatment response (ETR), defined as virus being undetectable at the end of therapy, and perhaps also an SVR.
Figure 3.
HCV RNA kinetics described by (A) the CE model, (B, thin line) the PK/PD model and (B, heavy line) the DE model. The CE model can not describe viral rebound. (B) also shows the best-fit of the PK/PD (thin line) and DE models (thick line) to HCV RNA data (filled circles) from a representative patient from 42 during the first week of PEG-IFN α-2b therapy.
The solution of the CE model, Eq. (2), has four parameters: t0, ε, c and δ, as well as V0, the initial viral load, which can be directly measured. Thus this equation, when it is fit to HCV kinetic data, allows one to estimate the viral kinetic and drug effectiveness parameters. This model has been successfully used to analyze a variety of HCV kinetic data 30, 43-45, and help make predictions or understand the response in different patient groups.
Despite its simplicity, the CE model may not always be the appropriate model to use to estimate HCV kinetic parameters or drug effectiveness. We have shown that this is the case under PEG-IFN α-2b therapy 42, 46-48. The concentration of PEG-IFN α-2b gradually increases and decreases over the week when it is administered once weekly (Fig. 1D, and Table 1). Thus, it is likely that the effectiveness of PEG-IFN α-2b also changes substantially over the week, violating the assumption of constant effectiveness. In fact, HCV kinetic data obtained during the first two weeks of PEG-IFN α-2b therapy shows that the HCV viral load oscillates 42, 49, 50, i.e., the viral load initially decreases but then partially rebounds before the next weekly drug dose. This fluctuation of HCV RNA in serum is synchronized with a fluctuation in the PEG-IFN α-2b concentration in serum 49, 50. These types of significant viral load fluctuations (>1 log10) are not observed under daily IFN α-2a 51 or IFN α-2b therapy 30. Importantly, the CE model under assumptions (i)-(iii) given above cannot describe a viral load rebound – its solution, Eq. (2), predicts that under therapy the viral load, V(t), will monotonically decrease from its initial value either reaching zero or a new constant value. If we fit the CE model to HCV kinetic data that contains a rebound and if blood is sampled frequently, the best-fit curve converges to a flat 2nd phase response in which the infected cell loss rate, δ̣, iṣ estimated as zero46. We have analyzed this issue exhaustively in computer simulation studies 47 46 and have shown that the parameter estimates obtained with the CE model under PEG IFNα–2b therapy are not reliable.
2.3 Pharmacokinetics of IFN and PEG-IFN
PEG IFN is given subcutaneously and is absorbed into the blood from the injection site. Assuming first order processes for absorption and elimination, the time evolution of the drug concentration in plasma is determined by the following equations 42, 48, 52, 53 31.
| (3) |
| (4) |
Here, Ma is the mass of the drug at the absorption site, M is the mass of drug in blood, and ka and ke are first order rate constants of absorption and elimination, respectively. Solving these equations with the initial conditions Ma(0) = FD and M(0) = 0, where D is the mass of the drug administered in a dose, with only a fraction, F, of the drug being bioavailable (0 ≤ F ≤ 1), gives the time evolution of the concentration of the drug in blood, C(t), following a single dose at t=0:
| (5) |
where we have assumed that PEG-IFN is distributed through a volume Vd. Equation (5) predicts that the maximum concentration, Cmax, is attained at t=tmax where tmax is given by
| (6) |
Both the absorption and elimination of PEG-IFN are slower than those of IFN, and the tmax of PEG-IFN is much larger than that of IFN (Table 1). Thus, the PEG-IFN concentration in serum changes gradually over the week when administered once weekly (Fig. 1). The drug elimination half-life and the time to peak drug concentration in serum are available 8-11. From this information, one can calculate the pharmacokinetic (PK) parameters (Table 1) that determine C(t).
2.4 Analyzing HCV kinetic data as well as PK data
Talal et al. 42 measured the HCV RNA levels as well as PEG-IFN α-2b concentration in serum frequently for the first two weeks of therapy of 24 patients coinfected with HCV and HIV. Patients received once weekly PEG-IFN α-2b therapy plus daily ribavirin for 48 weeks. We analyzed the HCV viral load data and pharmacokinetic (PK) data consisting of the drug concentration in serum using pharmacokinetic/pharmacodynamic (PK/PD) modeling. The PK/PD model assumed that the drug concentration in serum, C(t), is described by Eq. (5). For the pharmacodynamic model, we set ε(t) to 52, 54
| (7) |
Here τ is a delay between the time drug levels in serum are sensed by IFN receptors and the time it has an effect on reducing viral production, EC50 is the drug concentration at which PEG-IFN α-2b suppresses virion production by 50%, and the parameter n is the Hill coefficient (n ≥ 1), which controls how abruptly the effectiveness increases with higher drug concentrations. The drug effectiveness, ε(t) gradually increases and then decreases during the first week of therapy, as C(t) does the same for each patient individually (Fig. 2C).
Talal et al. 42 fitted Eq. (5) to each patient’s PK data to estimate ka, ke and FD/Vd. Then with these parameters fixed at their best-fit values, they fitted each patient’s HCV RNA kinetic data to estimate c, δ, V0̣, τ, EC50 and n. This model generated good fits to the viral load data, and demonstrated that taking the PK/PD aspects into consideration was crucial both for explaining the viral load rebound towards the end of the dosing interval and for obtaining sensible estimates for the viral kinetic parameters. Talal et al. 42 found that EC50 was significantly lower in sustained virological responders (patients who achieved SVR) than in non-responders (0.04 vs. 0.15 μg/L, p=.014). Additionally, the median therapeutic quotient (the ratio between the weekly average PEG-IFN concentration and EC50), and the PEG-IFN α-2b concentration at day 7 divided by EC50 were significantly higher in patients who achieved SVR than in patients who didn’t achieve SVR.
This PK/PD model can be extended to two weeks of therapy as shown in 42, or longer if frequent HCV RNA and PEG-IFN concentration data can be obtained over longer periods. To analyze PK data over multiple doses (e.g. two weeks of therapy), suitable modifications of the solution for C(t) were determined 55. One interesting aspect is that in some patients the values for drug absorption and elimination varied considerably from the first week to the second, indicating an effect of IFN exposure or other factors on its pharmacokinetics 42.
2.5 Simplified models of PK/PD for analyses of HCV viral load data
In many clinical studies, drug concentrations in serum are not measured. Thus, the modeling approach based on detailed PK/PD profiles is not useful. Shudo et al. 41 developed an alternative mathematical model where the drug’s effectiveness changes with time in a manner similar to that observed for PEG-IFN α-2b but which is analyzable in the absence of PK data. This model was called the decreasing effectiveness (DE) model. In the DE model, the viral kinetics for the first week of PEG-IFN α-2b therapy is governed by Eq. (1) but with ε(t) given by
| (8) |
where tm = t0 + t1 (Fig. 2D). In this model, after some delay, t0, the drug effectiveness increases instantaneously to a high level, ε, which is sustained for t1 days and then declines exponentially. This model ignores the PK details of PEG-IFN α-2b, and describes an approximation to the PD of the drug given by Eq. (8) (Fig. 2D). Thus, the DE model has fewer parameters than the full PK/PD model. With this simplification, we can estimate parameters without knowledge of drug concentration data. Shudo et al. 41 applied this model to 19 of the patient’s analyzed in Talal et al. 42. The fits were done for fixed delays t0 of 4 hours, 8 hours, and 12 hours, and the best delay was chosen based on the sum of squared residuals. The average drug effectiveness multiplied by the infected cell loss rate, which is the relevant parameter affecting the steepness of the second phase slope of viral load decline, was significantly larger in sustained virological responders than in non-responders (p=.03) 42.
2.6 Similarity and difference between PK/PD model and DE model
Both the PK/PD model and the DE model can describe the viral load rebound caused by fluctuations in drug effectiveness. However, the patterns of viral dynamics described in the two models are slightly different. For some patients (i.e. choices of parameters), the viral load rebound as described by the DE model occurred more abruptly than that described by the PK/PD model (Fig. 3B). The infected cell loss rate estimated by the DE model also tended to be higher than the one estimated by the PK/PD model (in 13/19 patients). By contrast, the average drug effectiveness over the first week of therapy estimated by the DE model and the PK/PD model were similar to each other. This suggests that even if PK data is unavailable, one can obtain an estimate of the average drug effectiveness that is close to the one estimated by HCV kinetic data plus PK data. Larger studies will be needed to establish this more definitively.
3. Selection of a model to analyze HCV RNA decline kinetics under therapy
3.1 HCV kinetic models under therapy
In addition to PEG-IFN α-2b treatment, there are other circumstances in which the CE model seems inappropriate. In clinical studies, the drug dose may be changed during therapy. For example, Bekkering et al.56 modeled a case in which 15 patients received 10 MU of IFN once daily for the first 3 days of therapy, followed by 3 MU IFN daily for 52 weeks. On average, viral load rebounded by 1.5 log10 HCV RNA copies/ml between days 3 and 4. Bekkering et al. 56 modified the CE model, and described the viral load rebound that was observed right after the dose change. Moreover, recently, a variety of other viral kinetic models have been developed, some including additional biological details, such as the proliferation of uninfected and infected cells 57, 58, possible enhancement of the infected cell loss rate during therapy due to an immune response 59, and extrahepatic infection 60. These detailed models can sometimes describe more complex viral load declines, such as a triphasic decline (i.e., a decline consisting of three phases, with a viral load plateau between two decays) 57-59. However, these models also introduce additional parameters, which in general, cannot be uniquely estimated from viral load data alone. Even if we find the “best parameter set” by nonlinear least square regression using the Levenberg–Marquardt algorithm 61, which is one of the most commonly used algorithms, the result we get might critically depend on the initial parameter guesses from which we started the algorithm, i.e., the algorithm may find a local optima not the global one. To avoid this problem, one can perform a global search, e.g. a grid search, in which the residuals are evaluated at each point on a grid spanning a biologically realistic range for each parameter. However, this method is computationally intensive. If one wants to evaluate the residuals at 100 values of each parameter using a mathematical model that has 4 parameters, 108 determinations are required. In addition, if the data is limited, a complex model with a larger number of parameters, such as the model of Dahari et al 58, 62 that incorporates liver-cell proliferation, which in full generality has 10 parameters, will never lead to reliable estimates for all of the parameters.
3.2 Usefulness of the CE model
Before the DE model was described, the CE model was fit to HCV kinetic data obtained under PEG-IFN α-2a therapy because PK data was unavailable 43-45 and in general PEG-IFN α-2a concentrations do not decline as rapidly between dosing interval as do PEG-IFN α-2b concentrations (Table 1). We recently fit both the CE model and the DE model to HCV kinetic data obtained during the first week of therapy of HIV/HCV coinfected patients who received PEG-IFN α-2a therapy from the study in 44. We compared the suitability of the two models using an F-test since the models are nested. We found the use of the CE model rather than the DE model was preferred for the analysis of this data (Shudo, unpublished). Because PEG-IFN α-2a is eliminated more slowly than PEG-IFNα-2b (Table 1 and Fig. 1C-D), the assumption of decreasing drug effectiveness in the DE model (Fig. 2C) adds additional parameters that are not justified statistically.
HCV kinetics under PEG-IFN α-2a therapy looks biphasic more often than under PEG-IFN α-2b therapy. We found that when using PEG-IFN α-2a, only 2 of 5 patients in one study 44 and 3 of 15 patients in another study (unpublished data) showed a rebound of HCV viral load during the first week of therapy. By contrast, in a study using PEG-IFN α-2b, a viral load rebound was observed in 13 of 19 patients 42.
As another example of the usefulness of the CE model, we fit a model that incorporates hepatocyte proliferation57, 58 as well as the CE model 30 to hepatitis B virus kinetic data that looks like it has a triphasic decline 57, 58. Still, an F-test supported the use of the CE model, which only generates a biphasic decline, rather than the extended model with liver proliferation 63, which can generate a triphasic decline.
4. Expert Opinion
We have reviewed a number of different models that describe the kinetics of HCV RNA decline under therapy: the CE model 30, a PK/PD model 42, 48 and the DE model 41. Deciding which model to use to analyze HCV RNA kinetic data is not a simple matter. First, if the drug being used is PEG-IFN α-2b given once weekly, drug concentration and presumably the drug’s effectiveness, ε, will vary between doses. In such cases, the best approach is to measure the drug concentration at each blood draw when HCV RNA is measured. If blood is drawn frequently, as in the study by Talal et al. 42, then a PK/PD model can be used to fit the data and estimate viral dynamic, pharmacokinetic and pharmacodynamic parameters. If drug concentration data is not available, then either the CE or DE model can be used. The solution to the CE model, given by, Eq. (2), will describe a biphasic HCV RNA decline but it is not able to describe a dataset in which HCV RNA rebounds. Thus, the data would need to be examined and if rebounds were prominent (e.g. >1 log10), Eq. (2) would not be an appropriate model. For such cases, if the viral load rebound is thought to be due to a decrease in drug concentration, as has been seen for PEG-IFN α-2b, then the DE model might be more suitable. The DE model has more parameters than the CE model and thus requires more HCV RNA measurements. Equation (1) with ε(t)=ε in which target cells are allowed to vary, can also describe a viral load rebound, but in this case the rebound is due to an increase in target cells in the presence of insufficient drug effectiveness 57, 58. Thus, if the rebound is taking many weeks and is not occurring towards the end of each weekly dosing interval, this model may be appropriate, but numerical solution of the differential equations (1) will be required rather than the use of Eq. (2).
One practical difficulty in using any of the three kinetic models is that frequent HCV RNA measurements need to be made if all the model parameters are to be estimated. For example, the rate of virion clearance, c, and the initial delay before HCV RNA begins to decline, t0, affect only the early viral dynamics. In order to estimate these parameters, frequent data needs to be collected during the first 1-2 days of therapy. Thus, in many studies these parameters are fixed to values found in studies with intense early sampling such as that of Neumann et al.30 The viral load at baseline before the first dose of drug is given, which we have called V0, can be directly measured or estimated from the data. We have found that in some patients in which frequent viral loads have been obtained early there are fluctuations in the HCV RNA level and it is better to fit V0 than to use a single viral load measurement.
At the moment, analysis of viral load data is still somewhat of an art and the choice of model and which parameters to fix and which to fit depend on the drug being used and the objective of the clinical study. HCV models are still evolving and some studies are aimed at improving the models and our understanding of HCV pathogenesis, while other studies are of a more clinical nature and simply want to compare the effects of drug A vs drug B.
A large number of new drugs are under development or are becoming available for the treatment of HCV infection 24, 26. As in the case of IFN and PEG-IFN, mathematical models should be useful in evaluating these new agents. Using mathematical models one can estimate in vivo parameters that are not directly measurable such as the drug’s effectiveness, ε, in blocking viral production. In addition, the modeling approaches described above also provide information about the effects of drug dose and dosing interval in inducing viral declines and allow one via extrapolation to estimate how long therapy will be needed to totally eliminate the virus and all infected cells.
4.1 Mathematical models to describe the HCV RNA declines observed with new agents
Albuferon® (Albinterferon) 19, 20, which is a new IFN based product, is a recombinant IFN α-2b molecule with albumin attached. Albumin weighs ~60 kD, which is larger than either of the two forms of polyethylene glycol used in PEG-IFN (40 kD: PEG-IFN α-2a; 12 kD: PEG-IFN α-2b). Because of this large albumin molecule, albinterferon is eliminated much more slowly (elimination half life ~ 144 hours; ke=0.12 day−1) 19 than PEG-IFN α-2a 9 (elimination half life ~80 hours; ke=0.21 day−1) or PEG-IFN α-2b (elimination half life ~40 hours; ke =0.42 day−1)8 (Table 1). Albinterferon, thus, has the advantage of being administered subcutaneously only once every 2 weeks 19. As in the case of once weekly administered PEG-IFN, modeling the effects of albinterferon on HCV RNA decline may require the use of PK/PD models or the DE model. Frequent viral kinetic data over the two week dosing interval will have to be analyzed to decide whether viral load rebounds are observed and whether drug pharmacokinetics needs to be taken into account. It will also be informative to study whether the early therapeutic quotient of this new formulation is predictive of SVR.
Another class of drugs in an advanced stage of development is the so called direct antivirals. These compounds interfere with specific steps of the HCV replication cycle. Inhibitors have been developed for both the NS3/4A serine protease and the NS5B RNA-dependent RNA polymerase of HCV. Remarkably potent in vivo antiviral activity has been demonstrated in early clinical trials for the protease inhibitors telaprevir (VX-950) 64 and boceprevir (SCH503034)25. Both nucleoside analog polymerase inhibitors, such as R1626 23, 65, and non-nucleoside NS5B polymerase inhibitors, such as GS919026, have also been developed. Although the actions of these drugs in reducing viral production can be modeled, as in standard models, through the appropriate estimation of a constant value for the drug effectiveness, ε, more specific models might be desired. For example, because these drugs are enzyme inhibitors, models based on competitive and non-competitive enzyme inhibitors could be explored in addition to the simple Hill function type pharmacodynamic relationship given by Eq. (7). In addition, the NS3/4A protease is not only involved in viral polyprotein processing, an essential step in viral replication, but it also cleaves two cellular proteins involved with IFN-signaling66. Therefore it is speculated that protease inhibitors might have a double effect and play a role in both inhibiting viral replication and restoring endogeneous synthesis of IFN 67. Lastly, because it is envisaged that some of these new drugs will ultimately be used in the context of combination therapy, PD models that incorporate the possibility of synergy between these inhibitors may lead to important clinical understanding of their interactions.
As we have studied for IFN based regimens, it will be interesting to determine from clinical trial data with these new direct antivirals, if viral kinetic and PK/PD parameters estimated early in therapy can distinguish between sustained virological responders and non-responders. To study this, we need to be careful in choosing appropriate mathematical models or, if necessary, to develop new models that take into account the drug dose, the dosing schedule, and PK parameters estimated in vitro or in early clinical studies.
The viral load declines more steeply under the effect of direct antivirals in combination with PEG-IFN than with direct antivirals alone 21-23, 25, 26. However, modeling combination therapy will be more difficult than modeling monotherapy. The drug effectiveness caused by direct antivirals administered every 8 hours or every 12 hours 22, 23, 25 may fluctuate more rapidly than the drug effectiveness caused by PEG-IFN administered every week. Further, synergistic interactions may occur that will need to be incorporated into pharmacodynamic relationships.
In addition, the emergence of drug resistant mutants is an important issue when patients are treated with direct antivirals 21, 22, because they impose a strong selective pressure. Serrazin et al. 24 observed that the viral load rebounded (breakthrough) a median of 2 log10 during 14 days of telaprevir monotherapy in 13/28 patients. In these 13 patients, the rebounding HCV virus contained drug resistant mutations. By contrast, viral load rebound was not observed in any of 12 patients during 28 days of therapy when a combination of telaprevir, PEG-IFN and ribavirin was used 22. Thus, mathematical modeling that takes into account the difference of direct antiviral effectiveness againstwild-type HCV and against drug-resistant HCV will also be required to analyze HCV kinetic data from patients treated with direct antivirals.
4.2 Mathematical model of intracellular dynamics
Is the antiviral effect enhanced additively or synergistically when direct antivirals and IFN are used at the same time? How much drug should be administered to get the best outcome? These questions are being addressed by clinical studies 19, however we may ask if modeling can make a contribution. To answer this question, one can introduce models of the intracellular dynamics of HCV RNA replication, as has been done for the replicon system 22, and then incorporate into these models the proposed effects of IFN and/or direct antivirals. The effect of IFN has been studied experimentally in HCV replicon systems 68-70, but its detailed mechanism of action is still not fully understood. Further, we do not know how direct antivirals, IFN and ribavirin interact with each other in hepatocytes when these drugs are used at the same time. For example, there are some suggestions that ribavirin might upregulate IFN stimulated genes in the presence of IFN thus leading to a synergy between these two drugs 71. In the future, detailed computer simulation models could be developed that include both viral replication and host cell signaling pathways so that the effects of IFN, ribavirin and of direct antivirals could be included. With such models one might be able to predict how much HCV RNA would be reduced by therapy with direct antivirals, IFN-based drugs, ribavirin and combinations of these drugs 72.
Acknowledgements
Portions of this work were done under the auspices of the U. S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grants R37 AI28433-18 and R01 RR06555-17 (ASP) and P20-RR18754 (RMR).
References
- 1.Barria MI, Vera-Otarola J, Leon U, et al. Influence of extrahepatic viral infection on the natural history of hepatitis C. Ann Hepatol. 2008 Apr-Jun;7(2):136–43. [PubMed] [Google Scholar]
- 2.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996 Feb 1;97(3):845–51. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnello V, De Rosa FG. Extrahepatic disease manifestations of HCV infection: some current issues. J Hepatol. 2004 Feb;40(2):341–52. doi: 10.1016/j.jhep.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Theoretical Biology and Biophysics. Los Alamos, NM: [Last accessed 30 January 2009]. 2003. Old and new HCV genotypes in the database. Available at: http://hcv.lanl.gov/content/sequence/HCV/classification/genotable.html. [Google Scholar]
- 5.Hepatitis C. World Health Organization; Switzerland: [Last accessed January 30, 2009]. 2000. Available at: http://www.who.int/mediacentre/factsheets/fs164/en/ [Google Scholar]
- 6.Theoretical Biology and Biophysics. HCV sequence database; Los Alamos, NM: [Last accessed 30 January 2009]. 2003. Distribution of HCV sequences: World. Available at: http://hcv.lanl.gov/components/sequence/HCV/new_geography/geography.comp?region=world&f orm=show&organism=HCV. [Google Scholar]
- 7.The Organ Procurement and Transplantation Network. Richmond, VA: [Last accessed January 30, 2009]. 2009. Waiting list candidates. Available at: http://www.optn.org/ [Google Scholar]
- 8.Package Insert PEG-Intron™ (Peginterferon alfa-2b) Powder for Injection. Schering Corporation; Kenilworth, NJ: 2001. [Google Scholar]
- 9.Package Insert PEGASYS® (peginterferon alfa-2a) Hoffmann-La Roche Inc.; Nutley, NJ: 2002. [Google Scholar]
- 10.Roferon-A (Interferon alfa-2a, recombinant) Hoffmann-La Roche Inc.; Nutley, NJ: 2008. [Google Scholar]
- 11.INTRON® A Interferon alfa-2b, recombinant For Injection. Schering Corporation; Kenilworth, NJ: 2008. [Google Scholar]
- 12.Reddy KR. Development and pharmacokinetics and pharmacodynamics of pegylated interferon alfa-2a (40 kD) Semin Liver Dis. 2004;24(Suppl 2):33–8. doi: 10.1055/s-2004-832926. [DOI] [PubMed] [Google Scholar]
- 13.Harris JM, Martin NE, Modi M. Pegylation: a novel process for modifying pharmacokinetics. Clin Pharmacokinet. 2001;40(7):539–51. doi: 10.2165/00003088-200140070-00005. [DOI] [PubMed] [Google Scholar]
- 14.Foster GR. A Guide to PEGylation, Pharmacokinetics and Pegylated Interferons. Roche; Basel, Switzerland: 2003. Better by Design. [Google Scholar]
- 15.Glue P, Fang JWS, Rouzier-Panis R, et al. Pegylated interferon-[alpha]2b: Pharmacokinetics, pharmacodynamics, safety, and preliminary efficacy data. Clinical Pharmacology and Therapeutics. 2000;68:556–67. doi: 10.1067/mcp.2000.110973. [DOI] [PubMed] [Google Scholar]
- 16.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002 Sep 26;347(13):975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 17.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001 Sep 22;358(9286):958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 18.Strader DB, Wright T, Thomas DL, et al. Diagnosis, management, and treatment of hepatitis C. Hepatology. 2004 Apr;39(4):1147–71. doi: 10.1002/hep.20119. [DOI] [PubMed] [Google Scholar]
- 19.Zeuzem S, Yoshida EM, Benhamou Y, et al. Albinterferon alfa-2b dosed every two or four weeks in interferon-naive patients with genotype 1 chronic hepatitis C. Hepatology. 2008 Aug;48(2):407–17. doi: 10.1002/hep.22403. [DOI] [PubMed] [Google Scholar]
- 20.Human Genome Sciences. Rockville, MD: [Last accessed January 30, 2009]. 2008. Albuferon® (albinterferon alfa-2b) Available at: http://www.hgsi.com/albuferona.html. [Google Scholar]
- 21.Forestier N, Reesink HW, Weegink CJ, et al. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology. 2007 Sep;46(3):640–8. doi: 10.1002/hep.21774. [DOI] [PubMed] [Google Scholar]
- 22.Kieffer TL, Sarrazin C, Miller JS, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007 Sep;46(3):631–9. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 23.Roberts SK, Cooksley G, Dore GJ, et al. Robust antiviral activity of R1626, a novel nucleoside analog: a randomized, placebo-controlled study in patients with chronic hepatitis C. Hepatology. 2008 Aug;48(2):398–406. doi: 10.1002/hep.22321. [DOI] [PubMed] [Google Scholar]
- 24.Sarrazin C, Kieffer TL, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007 May;132(5):1767–77. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 25.Sarrazin C, Rouzier R, Wagner F, et al. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology. 2007 Apr;132(4):1270–8. doi: 10.1053/j.gastro.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 26.Zeuzem S, Nelson DR, Marcellin P. Dynamic evolution of therapy for chronic hepatitis C: how will novel agents be incorporated into the standard of care? Antivir Ther. 2008;13(6):747–60. [PubMed] [Google Scholar]
- 27.Perelson AS, Neumann AU, Markowitz M, et al. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996 Mar 15;271(5255):1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 28.Ho DD, Neumann AU, Perelson AS, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995 Jan 12;373(6510):123–6. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 29.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002 Jan;2(1):28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 30.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998 Oct 2;282(5386):103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 31.Dixit NM, Perelson AS. The metabolism, pharmacokinetics and mechanisms of antiviral activity of ribavirin against hepatitis C virus. Cell Mol Life Sci. 2006 Apr;63(7-8):832–42. doi: 10.1007/s00018-005-5455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layden-Almer JE, Ribeiro RM, Wiley T, et al. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003 Jun;37(6):1343–50. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 33.Berg T, von Wagner M, Nasser S, et al. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology. 2006 Apr;130(4):1086–97. doi: 10.1053/j.gastro.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Davis GL, Wong JB, McHutchison JG, et al. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology. 2003 Sep;38(3):645–52. doi: 10.1053/jhep.2003.50364. [DOI] [PubMed] [Google Scholar]
- 35.Ferenci P, Laferl H, Scherzer TM, et al. Peginterferon alfa-2a and ribavirin for 24 weeks in hepatitis C type 1 and 4 patients with rapid virological response. Gastroenterology. 2008 Aug;135(2):451–8. doi: 10.1053/j.gastro.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 36.Jensen DM, Morgan TR, Marcellin P, et al. Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha-2a (40 kd)/ribavirin therapy. Hepatology. 2006 May;43(5):954–60. doi: 10.1002/hep.21159. [DOI] [PubMed] [Google Scholar]
- 37.Kau A, Vermehren J, Sarrazin C. Treatment predictors of a sustained virologic response in hepatitis B and C. J Hepatol. 2008 Oct;49(4):634–51. doi: 10.1016/j.jhep.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Zeuzem S, Buti M, Ferenci P, et al. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J Hepatol. 2006 Jan;44(1):97–103. doi: 10.1016/j.jhep.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Karino Y, Toyota J, Sugawara M, et al. Hepatitis C virus genotypes and hepatic fibrosis regulate 24-h decline of serum hepatitis C virus RNA during interferon therapy in patients with chronic hepatitis C. J Gastroenterol Hepatol. 2003 Apr;18(4):404–10. doi: 10.1046/j.1440-1746.2003.03009.x. [DOI] [PubMed] [Google Scholar]
- 40.Layden JE, Layden TJ, Reddy KR, et al. First phase viral kinetic parameters as predictors of treatment response and their influence on the second phase viral decline. J Viral Hepat. 2002 Sep;9(5):340–5. doi: 10.1046/j.1365-2893.2002.00377.x. [DOI] [PubMed] [Google Scholar]
- 41.Shudo E, Ribeiro RM, Talal AH, et al. A hepatitis C viral kinetic model that allows for time-varying drug effectiveness. Antivir Ther. 2008;13(7):919–26. [PubMed] [Google Scholar]
- 42.Talal AH, Ribeiro RM, Powers KA, et al. Pharmacodynamics of PEG-IFN α differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43(5):943–53. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 43.Sherman KE, Shire NJ, Rouster SD, et al. Viral kinetics in hepatitis C or hepatitis C/human immunodeficiency virus-infected patients. Gastroenterology. 2005 Feb;128(2):313–27. doi: 10.1053/j.gastro.2004.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torriani FJ, Ribeiro RM, Gilbert TL, et al. Hepatitis C virus (HCV) and human immunodeficiency virus (HIV) dynamics during HCV treatment in HCV/HIV coinfection. J Infect Dis. 2003 Nov 15;188(10):1498–507. doi: 10.1086/379255. [DOI] [PubMed] [Google Scholar]
- 45.Zeuzem S, Herrmann E, Lee JH, et al. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha2a. Gastroenterology. 2001 May;120(6):1438–47. doi: 10.1053/gast.2001.24006. [DOI] [PubMed] [Google Scholar]
- 46.Shudo E, Ribeiro RM, Perelson AS. Modelling hepatitis C virus kinetics during treatment with pegylated interferon alpha-2b: errors in the estimation of viral kinetic parameters. J Viral Hepat. 2008 May;15(5):357–62. doi: 10.1111/j.1365-2893.2007.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shudo E, Ribeiro RM, Perelson AS. Modelling the kinetics of hepatitis C virus RNA decline over 4 weeks of treatment with pegylated interferon alpha-2b. J Viral Hepat. 2008 May;15(5):379–82. doi: 10.1111/j.1365-2893.2008.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers KA, Dixit NM, Ribeiro RM, et al. Modeling viral and drug kinetics: hepatitis C virus treatment with pegylated interferon alfa-2b. Semin Liver Dis. 2003;23(Suppl 1):13–8. doi: 10.1055/s-2003-41630. [DOI] [PubMed] [Google Scholar]
- 49.Buti M, Sanchez-Avila F, Lurie Y, et al. Viral kinetics in genotype 1 chronic hepatitis C patients during therapy with 2 different doses of peginterferon alfa-2b plus ribavirin. Hepatology. 2002 Apr;35(4):930–6. doi: 10.1053/jhep.2002.32150. [DOI] [PubMed] [Google Scholar]
- 50.Formann E, Jessner W, Bennett L, et al. Twice-weekly administration of peginterferon-alpha-2b improves viral kinetics in patients with chronic hepatitis C genotype 1. J Viral Hepat. 2003 Jul;10(4):271–6. doi: 10.1046/j.1365-2893.2003.00446.x. [DOI] [PubMed] [Google Scholar]
- 51.Lake-Bakaar G, Ruffini L, Kuzmic P. Effect of ribavirin and amantadine on early hepatitis C virus RNA rebound and clearance in serum during daily high-dose interferon. Dig Dis Sci. 2003 Jan;48(1):126–39. doi: 10.1023/a:1021798701772. [DOI] [PubMed] [Google Scholar]
- 52.Gabrielsson J, Weiner D. Pharmacokinetic/pharmacodynamic Data Analysis: Concepts and Applications. 3rd ed Apotekarsocieteten; Stockholm, Sweden: 2000. [Google Scholar]
- 53.Welling PG. Pharmacokinetics : processes and mathematics. American Chemical Society; Washington, DC: 1986. [Google Scholar]
- 54.Wagner JG. Kinetics of pharmacologic response. I. Proposed relationships between response and drug concentration in the intact animal and man. J Theor Biol. 1968 Aug;20(2):173–201. doi: 10.1016/0022-5193(68)90188-4. [DOI] [PubMed] [Google Scholar]
- 55.Dixit NM, Perelson AS. Complex patterns of viral load decay under antiretroviral therapy: influence of pharmacokinetics and intracellular delay. J Theor Biol. 2004 Jan 7;226(1):95–109. doi: 10.1016/j.jtbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Bekkering FC, Neumann AU, Brouwer JT, et al. Changes in anti-viral effectiveness of interferon after dose reduction in chronic hepatitis C patients: a case control study. BMC Gastroenterol. 2001;1:14. doi: 10.1186/1471-230X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahari H, Lo A, Ribeiro RM, et al. Modeling hepatitis C virus dynamics: liver regeneration and critical drug efficacy. J Theor Biol. 2007 Jul 21;247(2):371–81. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007 Jul;46(1):16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 59.Herrmann E, Lee JH, Marinos G, et al. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003 Jun;37(6):1351–8. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 60.Dahari H, Feliu A, Garcia-Retortillo M, et al. Second hepatitis C replication compartment indicated by viral dynamics during liver transplantation. J Hepatol. 2005 Apr;42(4):491–8. doi: 10.1016/j.jhep.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Seber GAF, Wild CJ. Nonlinear regression. Wiley; New York: 1989. [Google Scholar]
- 62.Dahari H, Shudo E, Ribeiro RM, et al. Mathematical modeling of HCV infection and treatment. Methods Mol Biol. 2009:439–53. doi: 10.1007/978-1-59745-394-3_33. [DOI] [PubMed] [Google Scholar]
- 63.Dahari H, Shudo E, Ribeiro RM, et al. Modeling complex decay profiles of hepatitis B virus during antiviral therapy. Hepatology. 2009 Jan;49(1):32–8. doi: 10.1002/hep.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vertex Pharmaceuticals. Cambridge, MA: [Last accessed November 25, 2008]. 2008. Telaprevir (VX-950) Available at: http://www.vpharm.com/current-projects/drug-candidates/telaprevir-VX-950.html. [Google Scholar]
- 65.Investor update. Hoffmann-La Roche Ltd.; Nutley, NJ: [Last accessed November 25, 2008]. 2008. Available at: http://www.roche.com//investors/ir_update/inv-update-2008-04-28b.htm. [Google Scholar]
- 66.Liang Y, Ishida H, Lenz O, et al. Antiviral suppression vs restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology. 2008 Nov;135(5):1710–18. e2. doi: 10.1053/j.gastro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 67.Farley S. A double whammy for hep C. Nat Rev Drug Discov. 2003;2(6):419. [Google Scholar]
- 68.Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001 Sep;75(18):8516–23. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo JT, Sohn JA, Zhu Q, et al. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology. 2004 Jul 20;325(1):71–81. doi: 10.1016/j.virol.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 70.Guo JT, Zhu Q, Seeger C. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J Virol. 2003 Oct;77(20):10769–79. doi: 10.1128/JVI.77.20.10769-10779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feld JJ, Ko MS, Hra K, et al. Ribavirin improved second phase kinetics througth enhanced interferon signaling in genotype 1 HCV infection. Hepatology. 2008 Oct;48(4 Suppl):389A. [Google Scholar]
- 72.Neumann AU, Rozenberg L, Reesink HW, et al. Novel mode of viral decline during telaprevir (VX-950) and PEG-IFN combination treatment predicted by a new combined intracellular and cellular hepatitis C viral dynamics model. J Hepatol. 2007 Apr 01;46:S236. [Google Scholar]
- 73.DiStefano JJ., 3rd. Noncompartmental vs. compartmental analysis: some bases for choice. Am J Physiol. 1982 Jul;243(1):R1–6. doi: 10.1152/ajpregu.1982.243.1.R1. [DOI] [PubMed] [Google Scholar]