Abstract
Analysis of metal-protein interaction distances, coordination numbers, B-factors (displacement parameters), and occupancies of metal binding sites in protein structures determined by X-ray crystallography and deposited in the PDB shows many unusual values and unexpected correlations. By measuring the frequency of each amino acid in metal ion binding sites, the positive or negative preferences of each residue for each type of cation were identified. Our approach may be used for fast identification of metal-binding structural motifs that cannot be identified on the basis of sequence similarity alone. The analysis compares data derived separately from high and medium resolution structures from the PDB with those from very high resolution small-molecule structures in the Cambridge Structural Database (CSD). For high resolution protein structures, the distribution of metal-protein or metal-water interaction distances agrees quite well with data from CSD, but the distribution is unrealistically wide for medium (2.0 – 2.5 Å) resolution data. Our analysis of cation B-factors versus average B-factors of atoms in the cation environment reveals substantial numbers of structures contain either an incorrect metal ion assignment or an unusual coordination pattern. Correlation between data resolution and completeness of the metal coordination spheres is also found.
Keywords: Metalloprotein, protein structure, metal binding
1. Introduction
Metal ions are frequently observed in protein structures, and are often crucial for protein function, stability, or both. Moreover, in many cases metal ions are critical for crystal formation as the ions mediate crystal contacts between proteins. In the release dated February 20, 2007 of the Protein Data Bank (PDB) [1], approximately 30% of structures contained metal ions. Among 23,537 structures of proteins complexed with one or more small molecular ligands; 20% contained one or more metal ions close to the ligand binding site that are likely to interact either directly or indirectly with the ligand. 10% of the structures have a direct cation-ligand contact and the other 10% have a cation-ligand interaction bridged by an amino acid or ordered water molecules. This detailed analysis of the metal coordination architecture within proteins represents an important addition to the understanding of the biochemical functions of metalloproteins.
The ratio of the number of observed data to the number of parameters used in structure refinement depends on the data resolution and the number of atoms in a crystallographic asymmetric unit. For macromolecular structures, this ratio is usually low, due to the limited resolution of the data used to determine such structures. Therefore, the use of model restraints is a nearly universally applied technique in model building and structure refinement processes [2]. In addition to the stereochemical restraints for the macromolecule itself [3, 4], it is essential to apply restraints to the metal ion-binding site (and subsequently interpret the electron density) taking into account the coordination properties of the cation. In all the most popular programs used for macromolecular structure refinement, the restraints for metal-ligand interactions must be manually defined by the user. While the stereochemistry of proteins and nucleic acids is well understood, there is no universal approach to describe the geometry of metal ion binding sites. Alkaline earth cations such as calcium and magnesium are relatively easy to identify in electron density as the geometrical parameters (e.g. bond lengths and coordination number) of their binding sites are very well characterized [5–8]. Alkali metal ions such as sodium and potassium, however, are more difficult to identify because their coordination spheres are not as regular as those of alkaline earth metal ions [9]. Transition metals have even more complex binding patterns as not only can their coordination numbers vary but they can have different oxidation states. The bond lengths for transition metals depend on their oxidation state and even within the same oxidation state, different bond lengths are observed due to known geometrical distortions of the coordination spheres, for example due to the Jahn-Teller effect [10] or different spin state.
Studies describing the geometry of metal ion-binding sites within proteins and in small molecule structures were recently extensively discussed in a series of papers by Harding [5–9, 11]. Here, in contrast, our objective is to analyze the properties of metal ion binding sites in protein structures as a function of structure resolution and crystallographic methodology. In particular, we report a relational database approach to statistically analyze metal ion sites in protein structures present in the PDB [1], and compare them to high resolution small molecule structures obtained from the Cambridge Structural Database (CSD) [12]. We not only examined the distributions of bond lengths and coordination numbers but also the B-factors (displacement parameter sometimes referred as ‘temperature factor’) and relative occupancies of metal ions versus their coordinating atoms were analyzed. The distributions were cross-correlated with the computer programs used for structure refinement. Our results show some abnormally high or low values of bond lengths and B-factors in metal binding sites reported in the PDB. Despite many theoretical papers describing proper geometrical restraints for metal ion environments, our examination of recent structures indicates that those restraints are often not properly used in structure refinement.
2. Materials and methods
2.1. Data set under investigation
This work is based on the PDB database release of February 20, 2007 (41,814 structures). All structures in PDB which contain one or more Ca, Mg, Na, K, Mn, Co, Fe, Zn, Ni, Cu cations are included in the statistical analysis unless otherwise specified. In the analyses of structure resolution, B-factor or occupancy, only metal ion binding sites in protein structures solved by X-ray crystallography were included. For purposes of comparative analysis, the set was subdivided; structures with resolutions better than 1.5 Å were considered high resolution (8% of the X-ray structures in PDB) while structures with a resolution between 2–2.5 Å were considered medium resolution data (40% of X-ray structures in PDB). A third subset containing low resolution data (structures with resolution worse than 2.5 Å) was used only in the analysis of the coordination number. Structures with a resolution of 1.5–2.0 Å are likely mostly correct. They are, however, not such a good reference point as the high-resolution structures and therefore were not included in the present analysis. All calculations were performed without removing redundant structures except for analysis of atom and amino acid frequency profiles. These analyses, in addition, were performed on a non-redundant data set at 90% sequence identity cutoff. The clustering was done using a CD-hit program [13]. The highest resolution structure which contains a specific metal was chosen as the representative of each cluster for atom and amino acid profile analysis.
2.2. Calculation of frequency and p-value of the atom and amino acid profile
The normalized frequency of coordination by a given type of atom to a given cation is calculated using the formula . The relative probability that an atom of type X is bound to cations of type Y, Patom of type X bound to metal Y, is given by and simply represents the fraction of coordination. Thus, Patom of type X is the overall relative probability of atoms of type X observed in the data set (whether bound to metal or not) and is given by and represents the fraction of atoms. If the probability that a given type of atom is bound to a given metal is the same as the overall probability for the given type of atom, Patom of type X bound to metal Y = Patom of type X, then the normalized frequency Fatom X = 1. If a particular type of atom is seen relatively more frequently in the vicinity of a given metal atom than it is seen overall, Patom of type X bound to metal Y > Patom of type X and Fatom X will be greater than unity. Conversely, if a particular atom is seen relatively less frequently in the vicinity of a given metal atom than it is seen overall, Patom of type X bound to metal Y < Fatom of type X and Patom X will be smaller than unity.
For example, Fatom for main chain oxygen atoms is calculated using the formula . The numerator of this formula represents the relative frequency that those main chain oxygen atoms coordinate a given metal and the denominator is the ratio between the number of all main chain oxygen atoms (Natom MC_O) versus the number of all atoms (Nall atoms) in the whole PDB. The values for Natom MC_O or its equivalent for other types of atoms are also listed in the last column of Table 1a, 1b, and Table 2. The normalized frequency for residues are calculated in a similar manner by the formula:
Table 1.
Metal ion binding sites: elemental and chemical group composition
| (a) Normalized frequency Fatom for particular protein elements in metal ion environments Interactions with Fatom values between 1.5 and 3 are highlighted in light green. Values greater than 3 are highlighted in dark green (see the online version of this article for the colors). | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Atom | Alkali class | Imidazole class | Sulfur class | #atom (PDB) | |||||||
| Ca | K | Na | Mg | Mn | Co | Fe | Ni | Zn | Cu | ||
| Oxygen | 1.9 | 1.9 | 1.9 | 1.8 | 1.5 | 1.2 | 1.0 | 0.5 | 0.4 | 0.1 | 31662938 |
| Nitrogen | 0.0 | 0.0 | 0.1 | 0.2 | 0.5 | 0.8 | 0.9 | 1.2 | 0.8 | 1.5 | 28922420 |
| Sulfur | 0.0 | 0.0 | 0.1 | 0.1 | 0.6 | 4.0 | 5.7 | 14.2 | 32.5 | 18.5 | 740521 |
| # Interactions | 38396 | 6736 | 11994 | 22270 | 11126 | 2049 | 6681 | 1918 | 31588 | 6459 | |
| (b) Normalized frequency Fatom for particular chemical groups in metal ion environments. The interactions are colored as in (a). | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| atom | Ca | K | Na | Mg | Mn | Co | Fe | Ni | Zn | Cu | #atom (PDB) |
| MC_O | 1.1 | 1.8 | 2.0 | 0.6 | 0.1 | 0.1 | 0.0 | 0.1 | 0.0 | 0.1 | 21546919 |
| SC_O_amide | 3.4 | 2.5 | 1.9 | 2.5 | 1.1 | 0.6 | 0.2 | 0.2 | 0.1 | 0.2 | 1680883 |
| SC_O_carbo | 6.0 | 1.9 | 1.8 | 5.7 | 8.1 | 6.1 | 4.9 | 2.4 | 2.4 | 0.3 | 5178340 |
| SC_O_hydro | 0.4 | 2.2 | 1.3 | 2.6 | 0.2 | 0.5 | 1.7 | 0.2 | 0.1 | 0.1 | 3256796 |
| MC_N | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | 21559181 |
| SC_N_arg | 0.0 | 0.0 | 0.1 | 0.2 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3177032 |
| SC_N_lys | 0.0 | 0.0 | 0.1 | 0.3 | 0.1 | 0.0 | 0.0 | 0.2 | 0.1 | 0.0 | 1212067 |
| SC_N_amide | 0.2 | 0.0 | 0.3 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 1675900 |
| SC_N_ring | 0.1 | 0.6 | 0.5 | 2.2 | 11.0 | 16.4 | 19.8 | 21.6 | 17.9 | 33.5 | 1298240 |
| SC_S_cys | 0.0 | 0.0 | 0.1 | 0.1 | 1.3 | 7.6 | 13.9 | 31.8 | 79.0 | 31.4 | 304737 |
| SC_S_met | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 1.5 | 0.0 | 1.9 | 0.0 | 9.4 | 435784 |
Table 2.
Metal binding site amino acid residues environment. The normalized frequencies Fres for particular residues in metal ion environments are shown individually. The values are highlighted using the same color scheme as Table 1a (see the online version of this article for the colors).
| residue | Ca | K | Na | Mg | Mn | Co | Fe | Ni | Zn | Cu | #res (PDB) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLY | 0.9 | 0.8 | 0.8 | 0.3 | 0.0 | 0.1 | 0.0 | 0.2 | 0.0 | 0.2 | 1631940 |
| ALA | 0.2 | 0.9 | 0.4 | 0.2 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 1754173 |
| VAL | 0.4 | 0.8 | 1.1 | 0.3 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 1551140 |
| LEU | 0.1 | 0.5 | 0.7 | 0.2 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1927858 |
| ILE | 0.3 | 0.9 | 0.7 | 0.2 | 0.1 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 1220860 |
| SER | 0.4 | 1.5 | 1.6 | 1.2 | 0.1 | 0.3 | 0.0 | 0.1 | 0.1 | 0.1 | 1283333 |
| THR | 0.6 | 1.6 | 1.3 | 1.6 | 0.1 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 1218157 |
| CYS | 0.2 | 0.8 | 0.9 | 0.3 | 0.5 | 3.5 | 4.9 | 14.7 | 27.7 | 11.0 | 306466 |
| MET | 0.2 | 0.8 | 0.4 | 0.2 | 0.1 | 0.7 | 0.0 | 0.7 | 0.1 | 3.0 | 473167 |
| PRO | 0.1 | 0.3 | 0.5 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 995009 |
| ASP | 6.5 | 1.9 | 2.2 | 5.9 | 7.0 | 4.4 | 1.9 | 2.1 | 2.1 | 0.2 | 1229779 |
| ASN | 2.7 | 1.6 | 1.5 | 1.5 | 0.5 | 0.1 | 0.1 | 0.2 | 0.1 | 0.0 | 918078 |
| GLU | 2.7 | 2.2 | 1.1 | 2.5 | 4.3 | 4.2 | 4.6 | 1.3 | 1.2 | 0.3 | 1417858 |
| GLN | 0.9 | 1.4 | 1.3 | 1.2 | 0.3 | 0.4 | 0.1 | 0.0 | 0.1 | 0.1 | 792264 |
| LYS | 0.2 | 0.3 | 0.7 | 0.3 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 1267500 |
| ARG | 0.2 | 0.3 | 0.7 | 0.5 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1088414 |
| HIS | 0.5 | 1.5 | 1.0 | 2.3 | 10.1 | 14.9 | 18.0 | 21.8 | 16.2 | 30.3 | 502593 |
| PHE | 0.4 | 0.3 | 1.1 | 0.2 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 857598 |
| TYR | 0.8 | 0.8 | 1.1 | 0.3 | 0.1 | 0.1 | 2.4 | 0.0 | 0.0 | 0.2 | 759583 |
| TRP | 0.2 | 0.6 | 0.9 | 0.1 | 0.0 | 0.0 | 0.0 | 0.5 | 0.1 | 0.1 | 309945 |
A χ2 test is performed for both atom types and residues. For example, for each metal-to-atom-type interaction, a χ2 test is carried out with two degree of freedom in a 2×2 matrix of 4 values: the Fatom for atom type X bound to metal Y, the Fatom for all other atom types bound to metal Y, the Fatom for atom type X bound to all other metals, and the Fatom for all other atom types bound to all other metals. The significance of the χ2 test is then given in terms of a p-value which is an indication of the likelihood of obtaining a certain normalized frequency Fatom for atom type X bound to metal Y. The same analysis is performed for the normalized frequency Fres. The p-values for each type of metal-to-atom-type or metal-to-residue interaction are listed individually in Table 1c and Table 2b (supplemental material).
2.3. Analysis of metal ion binding sites
In the cases of Ca2+, Mg2+, Na+, and K+, only the interactions between the metal ions and oxygen atoms were considered. For the studied transition metals Mn, Co, Fe, Zn, Ni, and Cu, interactions with nitrogen or sulfur atoms were analyzed in addition to interactions with oxygen atoms. For the purpose of fast analysis, we created a relational database named NEIGHBORHOOD containing structural information about all residues, atoms, distances between residues, and distances between atoms. Distances were stored in the database as a property of an interaction between atoms while atom-related information such as B-factor and occupancy was stored as properties of an atom. Other entry-specific information such as resolution, R factor, deposition date, protomer chain, and sequence cluster information were stored as properties of the PDB entry to be cross-linked to the interaction properties or atom properties on-the-fly by SQL queries to this relational database. Intermolecular contacts between symmetry related molecules were calculated by the program CONTACT in CCP4 suite [14]. Additional derived data, such as the metal coordination number, is not stored in the database but calculated on-the-fly based on the pattern of interactions.
2.4. Comparison with small molecule structures
In most of the analyses, results from protein structures were compared to very high resolution data from structures in the Cambridge Structural Database (CSD version 5.29) release of November 2007 (423,752 structures) [12]. Structures that met certain templates were retrieved using the CSD interface program ConQuest [15]. Only data from structures with R factors less than 5% were retrieved for analysis. In the statistics for metal-ligand distance, the same distance cutoff (3 Å) was used as in the statistics for macromolecules. Data were first examined using the CSD analysis software VISTA [16], and then exported as text files for further analysis.
2.5. Correlation between metal ion B-factors and ligand B-factors
Displacement parameters (B-factors) and occupancy values for all atoms were taken from PDB files and pre-processed before being stored in the NEIGHBORHOOD database. B-factors lower than 2.0 or occupancies falling outside the range of 0.1–1 were considered erroneous and such data were not included in analysis. For example, there are 10 entries with calcium ion occupancies above 100% (1TRP, 1A0S, 1A8B, 1C8H, 1CLX, 1UEA, 1PEX, 1SAT, 1A8A, 1C8G). The B-factor for a metal ion environment was calculated as the mean B-factor for all atoms located within 4 Å of the metal ion of interest.
3. Results
3.1. Atom type and amino acid profiles of metal ion binding sites
A distribution of normalized frequencies Fatom of atoms located within 3 Å from the metal ion is shown in Table 1. The same table generated with a cut-off of 4 Å gives similar, but somewhat noisier, results. The non-redundant subset of structures, containing around 30% data of the complete data set, gives very similar results to the complete data set shown in Table 1. The number of interactions listed in the last row of both Tables 1a and 1b represents the number of pairs (in this case, a metal ion and an atom from amino acid) that are close enough to be considered a contact. Only types of metal ions with more than 1000 observed contacts were included in further statistical analyses. Interactions of each metal ion with each element in protein structures (oxygen, nitrogen, sulfur, and carbon) are shown in Table 1a. The interactions of each metal ion with a given protein element are further subdivided into classes reflecting different chemical moieties. For example, oxygen atoms are further differentiated into four subgroups: main chain oxygen, oxygen in amides (Asn/Gln), oxygen in carboxylates (Asp/Glu), and oxygen in hydroxyls (Ser/Thr/Tyr). Nitrogen atoms were subdivided into five subgroups: main chain, from Arg, from Lys, from amides (Asn/Gln), and from His. Sulfur atoms from Cys and Met residues were treated separately.
The values shown in Tables 1a and 1b are the normalized frequencies Fatom for each atom or atom type which is the likelihood that a particular atom interacts with a specific metal ion relative to its overall frequency in a given protein. A Fatom value around unity indicates that there is no preference for the atom in the chemical group being analyzed to be localized near the metal ion. If the Fatom is substantially lower than 1, it is unlikely that this type of atom will be near the metal ion when in a particular chemical group. For values higher than 1, Fatom shows the probability of finding a given type of atom near the metal ion is higher than the probability expected for a random distribution of atoms.
In order to show the significance level of the preference for particular interactions, the respective p-values for the Fatom values in Table 1b are listed in Table 1c (supplemental material). Due to the large sample size, most of the p-values are very significant even when the Fatom ratio is as low as 1.5, which is the value we used as the cutoff of a preferred interaction. In most cases, the p-value agrees very well with the normalized frequency Fatom so that the Fatom value alone accurately represents the degree of preference for a specific interaction. However, in a few cases, the Fatom value is less significant due to the variation of sample size for different interactions under analysis. For example, the Fatom value for magnesium – side chain amide oxygen interaction is 2.5 and for nickel – side chain methionine sulfur interaction is 1.9. While both these fall in the range of 1.5 to 3, their p-values (2×10−175 and 0.005, respectively) are very different. In such cases, the p-value has to be used in conjunction with the Fatom value to determine whether or not the degree of preference level is significant. In the previous example, magnesium – sidechain amide oxygen interaction is statistically significant but the nickel – sidechain methionine sulfur interaction is not.
In Table 1, the metal ions are ordered by decreasing normalized frequency of finding an oxygen atom coordinated to them. We classified metal ions into three classes based on coordination profile (as well as convenience for further analysis). The ‘alkali class’ (Ca, K, Na, Mg) consists of metals that interact almost exclusively with oxygen atoms. The ‘imidazole class’ (Mn, Co, Fe) and the ‘sulfur class’ (Ni, Zn, Cu) consist of metals that readily interact with oxygen, nitrogen, and sulfur. Members of both classes have a high degree of preference for imidazole rings but metals in the sulfur class have in addition a high degree of preference for thiol or thiolate moieties.
The normalized frequency Fres for particular amino acids at metal ion-binding sites is shown in Table 2 and the corresponding p-values are shown in Table 2b (supplemental material). The Fres value and p-value are analogous to the values used in the analysis of the atom profile described above. Again, it can be seen that the normalized frequency value Fres agrees with the p-value in most cases but the p-value is still a useful discriminator of the significance level of the interaction when the Fres value falls between 1 and 3. The distribution of preferences for particular amino acids to bind particular metal ions agrees with the trend observed in the distribution of atoms. For both the imidazole and sulfur classes of metals (Mn, Co, Fe, Ni, Zn, Cu), histidine is a very strongly preferred residue. The imidazole class of metals (Mn, Co, Fe) also shows a strong degree of preference for aspartic and glutamic acids. For the sulfur class of metals (Ni, Zn, Cu), cysteine is a very strongly preferred residue (as expected).
3.2. Metal ion-ligand distances
The average distances observed for metal ion-protein (or ordered water) interactions are listed in Table 3. Each element is listed separately and the distances are subdivided by the interacting atom. All atoms within 3 Å of a metal ion were considered to be interacting atoms. The mean values and standard deviations for metal ion coordination distances are listed together with the number of observations. For each metal ion coordination interaction that was investigated, the distances are listed separately for data from CSD, from PDB high-resolution structures, and from PDB medium-resolution structures. Whenever there were too few data to obtain reliable statistics, the values were replaced by the symbol “−”. In cases when two maxima were observed in the distance distributions derived from the CSD, the distances between the metal ion and coordinating atoms are marked as “doublets”. For these bimodal cases, the data were subdivided into “short” and “long” groups and the means and standard deviations were calculated separately as shown in Table 4.
Table 3.
Mean metal-ligand distances in Å (with standard deviations in parentheses), and number of observations for each metal, subdivided by coordinating atom and by data set (CSD, PDB-HR, or PDB-MR). CSD are data from the Cambridge Structural Database, and PDB-HR and PDB-MR are data from the high- and medium-resolution subsets of the PDB, respectively.
| (a) Metal-oxygen distances for the alkali class of metals | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ca | K | Na | Mg | |||||
| Distance (Å) |
Sample size |
distance | num | distance | num | distance | num | |
| M-OC (CSD) | 2.44(11) | 1901 | 2.81(8) | 10502 | 2.46(14) | 8398 | 2.10(8) | 1806 |
| M-OC (PDB-HR) | 2.37(12) | 2246 | 2.76(14) | 199 | 2.43(20) | 567 | 2.21(25) | 639 |
| M-OC (PDB-MR) | 2.43(19) | 13800 | 2.74(17) | 2109 | 2.50(21) | 2517 | 2.27(24) | 4759 |
| M-OH2 (CSD) | 2.42(6) | 688 | 2.80(10) | 636 | 2.41(9) | 1917 | 2.07(4) | 527 |
| M-OH2 (PDB-HR) | 2.42(15) | 1572 | 2.64(37) | 149 | 2.46(24) | 652 | 2.14(19) | 1677 |
| M-OH2 (PDB-MR) | 2.49(23) | 5112 | 2.72(22) | 701 | 2.58(24) | 2335 | 2.29(26) | 9184 |
| (b) Metal-oxygen and metal-nitrogen distances for the imidazole class of metals | ||||||||
|---|---|---|---|---|---|---|---|---|
| Mn | Co | Fe2+ | Fe3+ | |||||
| distance | num | distance | num | distance | num | distance | num | |
| M-N (CSD) | doublet | 6856 | doublet | 16049 | doublet | 1903 | doublet | 5848 |
| M-N (PDB-HR) | 2.20(13) | 65 | 2.07(13) | 40 | 2.18(7) | 75 | 2.16(13) | 93 |
| M-N (PDB-MR) | 2.32(21) | 852 | 2.21(16) | 216 | 2.20(14) | 344 | 2.25(15) | 888 |
| M-S (CSD) | doublet | 945 | 2.26(11) | 2314 | 2.27(9) | 76 | 2.28(8) | 4503 |
| M-S (PDB-HR) | 2.29(15) | 7 | 2.33(3) | 34 | 2.29(4) | 89 | ||
| M-S (PDB-MR) | 2.45(7) | 17 | 2.30(0) | 1 | 2.33(13) | 79 | ||
| M-OC (CSD) | doublet | 6775 | doublet | 6221 | 2.18(9) | 234 | 2.04(9) | 4652 |
| M-OC (PDB-HR) | 2.15(15) | 186 | 2.09(12) | 72 | 2.11(14) | 47 | 2.14(19) | 99 |
| M-OC (PDB-MR) | 2.26(21) | 2786 | 2.17(21) | 337 | 2.14(19) | 374 | 2.13(22) | 1184 |
| M-OH2 (CSD) | 2.19(6) | 1321 | 2.10(5) | 1685 | 2.10(4) | 125 | 2.10(6) | 271 |
| M-OH2 (PDB-HR) | 2.18(18) | 183 | 2.31(36) | 63 | 2.20(11) | 46 | 2.19(22) | 68 |
| M-OH2 (PDB-MR) | 2.29(24) | 1552 | 2.29(25) | 215 | 2.32(33) | 124 | 2.30(29) | 403 |
| (c) Metal-oxygen, metal-nitrogen and metal-sulfur distances for the sulfur class of metals | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ni | Zn | Cu2+ | Cu+ | |||||
| distance | num | distance | num | distance | num | distance | num | |
| M-N (CSD) | doublet | 14233 | 2.10(9) | 9621 | 2.02(9) | 19664 | 2.03(8) | 7525 |
| M-N (PDB-HR) | 1.99(14) | 68 | 2.07(11) | 661 | 2.04(9) | 216 | 2.04(15) | 47 |
| M-N (PDB-MR) | 2.18(17) | 424 | 2.14(16) | 4109 | 2.10(14) | 1251 | 2.11(16) | 114 |
| M-S (CSD) | doublet | 4495 | 2.38(13) | 1744 | 2.34(16) | 3833 | 2.33(12) | 612 |
| M-S (PDB-HR) | 2.24(15) | 20 | 2.32(6) | 221 | 2.34(23) | 53 | 2.36(27) | 51 |
| M-S (PDB-MR) | 2.30(15) | 99 | 2.33(13) | 3451 | 2.35(24) | 336 | 2.26(20) | 32 |
| M-OC (CSD) | doublet | 5728 | 2.15(26) | 7345 | 2.10(28) | 14489 | 2.12(28) | 1862 |
| M-OC (PDB-HR) | 2.17(22) | 30 | 2.08(20) | 434 | 2.24(34) | 31 | ||
| M-OC (PDB-MR) | 2.23(23) | 210 | 2.15(22) | 2553 | 2.38(37) | 131 | ||
| M-OH2 (CSD) | 2.08(6) | 1681 | 2.09(8) | 1109 | doublet | 1512 | doublet | 480 |
| M-OH2 (PDB-HR) | 2.19(21) | 79 | 2.19(28) | 429 | 2.23(30) | 75 | ||
| M-OH2 (PDB-MR) | 2.28(30) | 257 | 2.30(30) | 1499 | 2.44(34) | 188 | ||
Table 4.
Mean metal-ligand distances derived from the CSD for metals that produce two maxima on metal-ligand distance distribution.
| short distance | long distance | |||
|---|---|---|---|---|
| distance (Å) | # of obs. | distance (Å) | # of obs. | |
| Mn-N | 1.99(10) | 1626 | 2.29(16) | 5230 |
| Mn-S | 2.36(7) | 745 | 2.64(9) | 200 |
| Mn-OC | 1.91(4) | 1597 | 2.19(9) | 5178 |
| Co-N | 1.95(5) | 11552 | 2.14(6) | 4497 |
| Co-OC | 1.90(2) | 2018 | 2.10(9) | 4203 |
| Fe2+-N | 1.97(4) | 943 | 2.18(5) | 960 |
| Fe3+-N | 1.67(2) | 301 | 2.08(12) | 5547 |
| Ni-N | 1.89(4) | 3686 | 2.09(7) | 10547 |
| Ni-S | 2.18(3) | 3555 | 2.46(10) | 940 |
| Ni-OC | 1.86(4) | 941 | 2.07(7) | 4787 |
| Cu2+-H2O | 1.97(3) | 578 | 2.37(17) | 934 |
| Cu+-H2O | 1.98(3) | 152 | 2.33(13) | 328 |
3.3. Metal ion coordination numbers
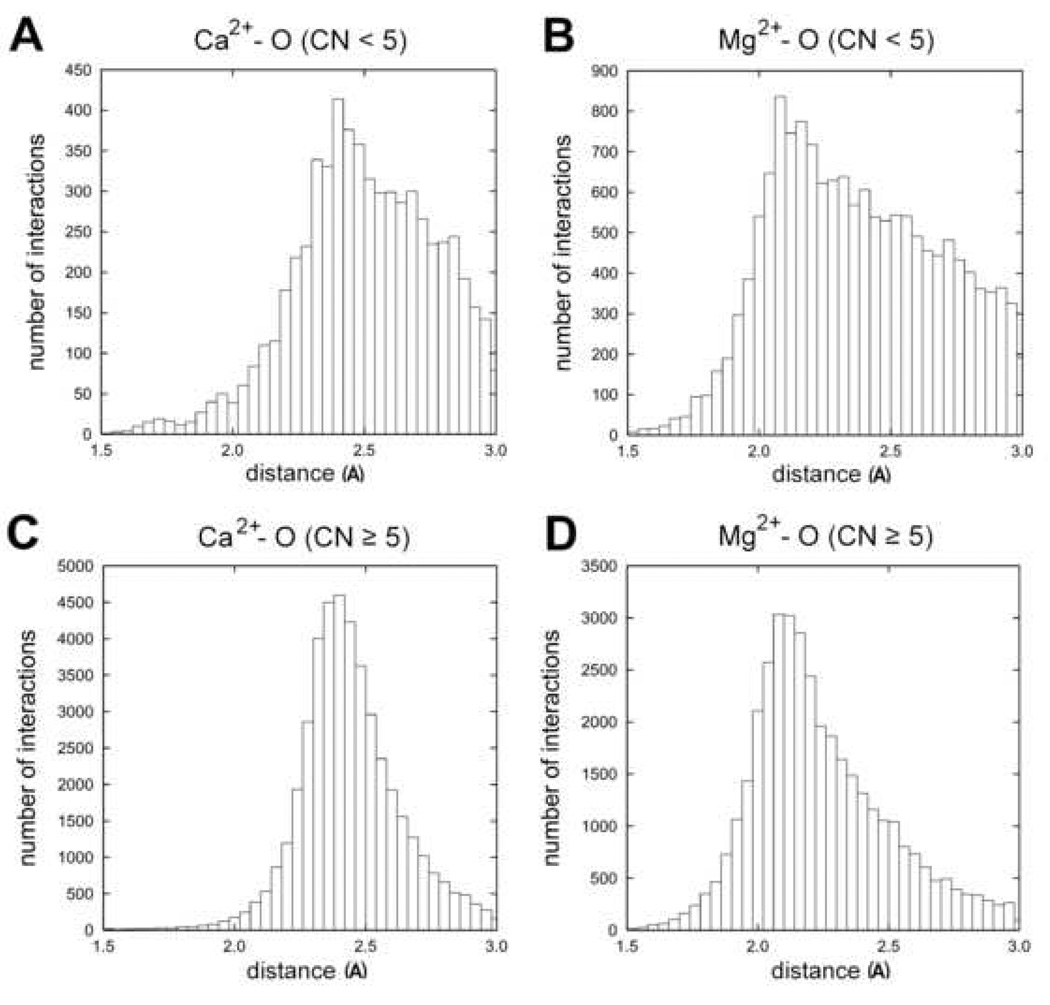
The distributions for incomplete and complete coordination spheres of calcium and magnesium ions, obtained with the assumption that only oxygen atoms form the first coordination sphere, are shown in Fig. 1. Given that Mg2+ and Ca2+ typically form octahedral coordination geometry, coordination spheres were considered (more or less) complete if the coordination number (CN) was 5 or more and incomplete if the CN was 4 or less. The data set used for CN calculation was processed a little differently as the distance cutoff of 3 Å was used explicitly to define neighboring residues that form the coordination sphere and the bidentate coordination of carboxyl groups was taken into account by considering such coordination as two contacts. The mean calcium ion-oxygen distance is 2.44(19) Å over 44,017 observations for metal ions with a complete coordination sphere and is 2.50(27) Å over 6125 observations for metal ions with an incomplete coordination sphere. The mean magnesium-oxygen distance is 2.24(26) Å over 37,371 observations for metal with a complete coordination sphere and is 2.37(32) Å over 15,180 observations for metal with an incomplete coordination sphere. The shapes of the distributions for incomplete coordination spheres are distorted. Especially for the distribution of magnesium-oxygen distances, there are more observations of distances larger than the peak of the distribution than smaller, skewing the mean towards longer distances.
Figure 1.
Calcium-oxygen and magnesium-oxygen distance distributions for complete and incomplete coordination spheres. (A) Calcium-oxygen distance distribution for incomplete coordination spheres (CN<5) and (C) calcium-oxygen distance distribution for complete coordination spheres (CN≥5). Magnesium-oxygen distance distributions for incomplete and complete coordination spheres are shown in B and D respectively. The vertical axes give the number of interactions and the horizontal axes the distance between metal and oxygen.
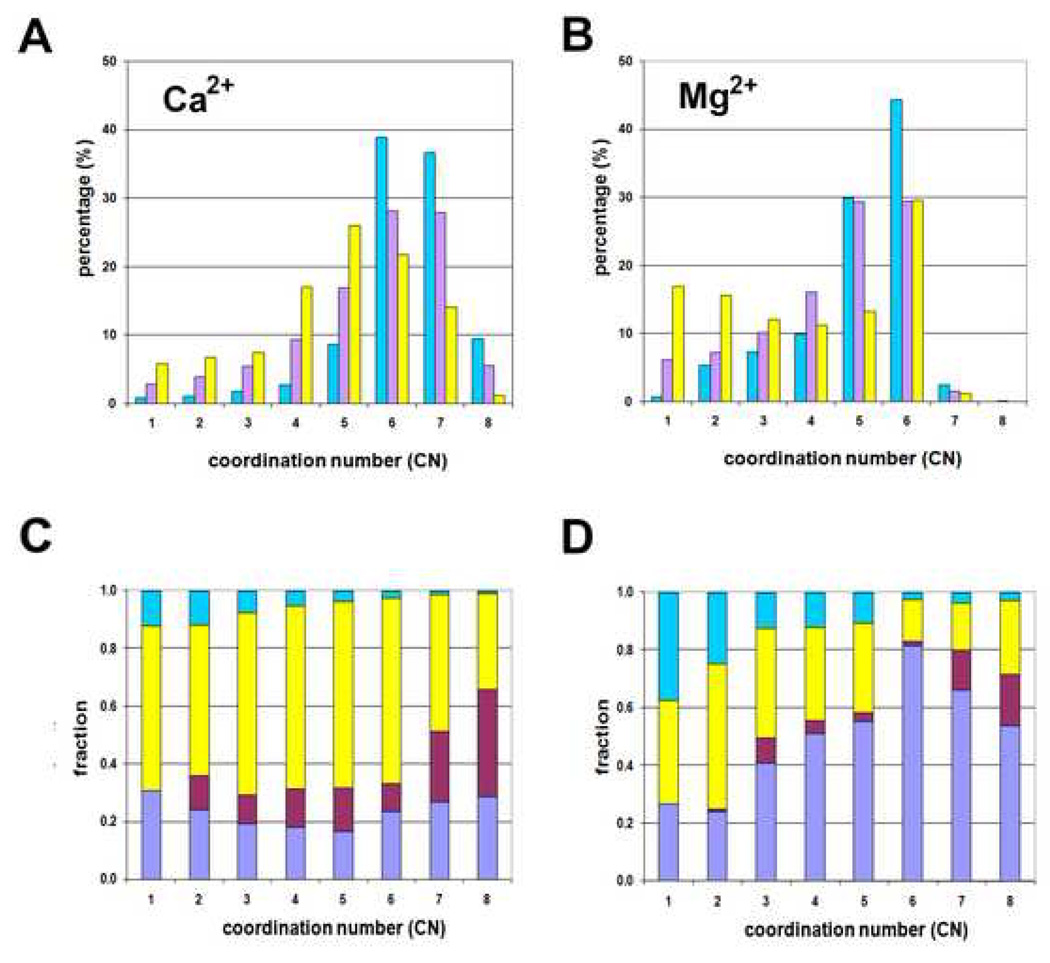
Completeness of the coordination sphere in structural models deposited to PDB for both calcium and magnesium ions is correlated with data resolution (Fig. 2). For calcium ions, the mean CN is 6.3(1.2) over 702 metal ion binding sites for high resolution data, 5.7(1.6) over 3803 sites for medium resolution data, and 4.8(1.7) over 1836 sites for low resolution data. While few high resolution structures had only 5 oxygen atoms coordinating calcium, we found many structures with only 5 oxygen atoms coordinating magnesium, even at a very high resolution. Thus, the mean CN for magnesium is 5.1(1.2) over 533 sites for high resolution data, 4.5(1.5) over 3886 sites for medium resolution data, and only 3.8(1.9) over 6364 sites for low resolution data (Figs. 2A, 2B).
Figure 2.
Metal-oxygen coordination sphere for various resolutions and coordination sphere components (calcium A,C and magnesium B,D). A,B: The horizontal axes give the coordination number and the vertical axes the percentage of structures for each data set. The cyan bars (left) correspond to structures with a resolution of 1.5Å or better, the violet bars (middle) correspond to structures with resolution between 2.0Å and 2.5Å, and the yellow bars (right) correspond to structures with a resolution worse than 2.5Å. C,D: the relative fractions of coordination sphere components. Cyan fractions (top) correspond to interaction with water, magenta (3rd from top, for 2–8) to bidentate coordination from a carboxyl group from Asp/Glu, yellow (2nd from top) to non-bidentate interaction with amino acid oxygen, and violet (bottom) to interaction with oxygen from a non-proteinaceous ligand (see the online version of this article for the colors).
3.4. Correlation between the environment of metal ions and their B-factors
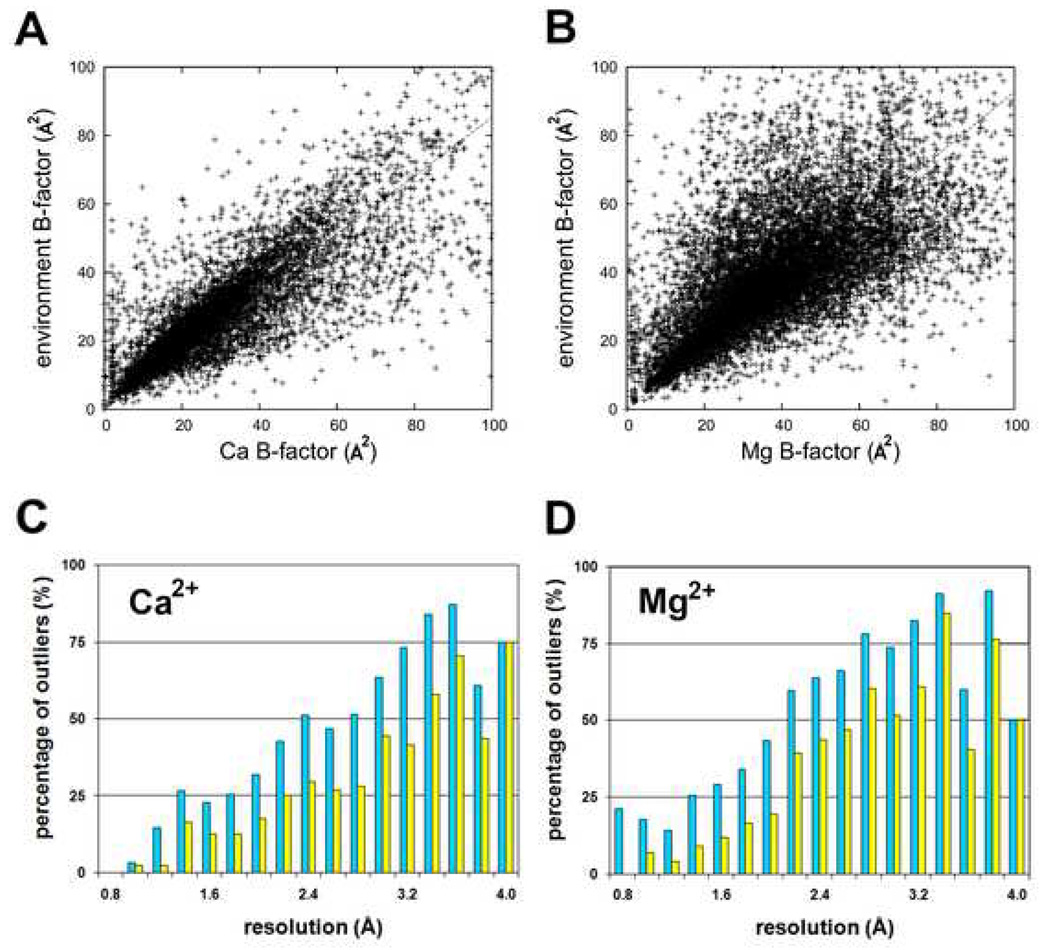
The B-factor of a properly determined and refined metal ion should be close to the B-factors of its coordinating atoms. However, as the B-factor and occupancy of an atom are strongly correlated, errors in occupancy affect the values of B-factors. We plotted the B-factor of metal ions versus the mean value of the B-factors of the atoms present in its coordination sphere (Figs. 3A, 3B). For the overwhelming majority of observations, the points are located near the line with a unity slope confirming the expected correlation of B-factors. However, there are points that deviate far from this line. In the calcium B-factor plot, a vertical collection of points at the left of the plot represents a number of metal binding sites where the calcium ion B-factors are around 2 Å2 while the average B-factors for the environments range between 2 and 55. A similar vertical line of points is also observed in the magnesium B-factor plot. There is also a cluster of sites for which the calcium ion environments are well ordered (with B-factors around 10–20 Å2) but the calcium ions have unreasonably high B-factors around 100 Å2.
Figure 3.
Scatter plots of mean B-factor of coordinating oxygens versus B-factor for (A) calcium and (B) magnesium ions. The histograms show the percentage of B-factor difference outliers for (C) calcium and (D) magnesium as a function of resolution. The cyan bars (left) show the percentage of points where the difference between the metal B-factor and the mean B-factor of its coordinating atoms is bigger than 5 Å2. The yellow bars (right) show the percentage of points where the difference is bigger than 10 Å2 (see the online version of this article for the colors).
The outliers for the differences in B-factors for metal ions minus the mean B-factors for the coordinating environments are plotted versus structure resolution of the structures in Fig. 3 (3C, 3D). At two different outlier difference cutoffs (±5 Å2 and ±10 Å2), the percentage of outliers increases as resolution decreases. For high resolution data (better than 1.5 Å), the B-factors for both metal ions and their coordination environments indicate most of the metal-binding sites are well-ordered. For data with resolution worse than 2 Å, the number of outliers begins to increase to an extent that half of the observations lay outside of both ±5 Å2 and ±10 Å2 difference cutoffs. Such an effect becomes saturated around a resolution of 3 Å, where the majority of the B-factor differences are outliers. This dependence on resolution must be an artifact of the refinement and/or data quality as the chemistry of the metal coordination is resolution independent.
4. Discussion
4.1. Atoms and amino acids participating in metal ion-binding
All analyzed metal ions except Cu show a preference for interaction with a side chain carboxylate group (Table 1). Alkaline earth metal ions (Ca2+, Mg2+) exhibit the highest preference for coordination by side chain carboxylate groups followed by a weaker preference for interaction with oxygen atoms from side chain amide groups. Alkali metal ions (Na+, K+) are preferred approximately equally by all types of oxygen atoms. Metal ions from both the imidazole class (Mn, Co, Fe) and the sulfur class (Ni, Zn, Cu) show a very strong preference to interact with imidazole nitrogens of histidines. While the metal ions in the imidazole class show some preference for interaction with thiol groups, metal ions from the sulfur class show a very strong preference for interaction with the thiol/thiolate moiety of cysteines (Table 1). The data in Table 1 also show that the sulfur atoms of methionines are relatively frequently in close contact with copper ions. This is not surprising as methionine residues are part of a well defined structural motif [17] responsible for Cu ion-binding in type 1 blue copper proteins.
For calcium and magnesium ions, the aspartate and glutamate are most strongly preferred ligands, as expected. Potassium ions show a weak preference for all amino acids containing oxygen atoms in their side-chains (Ser, Thr, Asp, Asn, Glu, Gln, Tyr). This is consistent with the data presented in Table 1b, in which alkali metal ions (Na+, K+) show a similar preference to interact with all oxygen atoms from protein, regardless if they are main chain or side chain oxygen atoms. However, in the case of sodium, the trend in the residue preferences becomes almost undetectable as sodium does not show a preference to interact with any particular amino acid. It is also found that there are different preferred interactions between calcium and magnesium ions. While both ions show strong preferences for carboxylate and amide oxygen atoms, magnesium ions relatively rarely reside close to main chain oxygen (Fatom=0.6), while calcium ion does not show such “rejection” (Fatom=1.1, p-value=3×10−11). This may be due to the fact that the typical magnesium-oxygen bond is almost 0.3 Å shorter than the calcium-oxygen bond and simultaneous formation of bonds to both main chain and side chain oxygen atoms may involve some geometrical hindrance. In addition, calcium and magnesium ions have completely different preferences to interact with sidechain hydroxyl oxygens (from Ser, Thr, or Tyr). While magnesium ions show a very significant preference for hydroxyl oxygen atoms (Fatom=2.6), calcium ions show a completely opposite effect (Fatom=0.4). It may be speculated that the smaller Mg2+ induces hydroxyl deprotonation, producing a more strongly binding O-group, while the larger Ca2+ does not. This hypothesis should be validated by neutron diffraction data. Magnesium ions also show some preference for interaction with nitrogen atoms from lysine and histidine while larger calcium, potassium, and sodium ions do not show any preference for interaction with nitrogen atoms.
Based on the results presented in Table 2, the twenty common amino acids may be divided into three groups according to their relative preference for interaction with metal ions. The first group consists of Asp, Cys, Glu, and His; these amino acids are frequently found to coordinate metal ions. The second group includes Asn, Met, Ser, Thr, Trp, and Tyr; these amino acids show some preferences for interaction with some metal ions, albeit less frequently. The third group includes Ala, Arg, Gln, Gly, Ile, Leu, Lys, Pro, and Phe; the relative frequency of finding these amino acids in the vicinity of metal ions is very low. This is not surprising in the case of Ala, Gly, Ile, Leu, Pro, and Phe because only their main chain moieties are capable of coordinating metal ions. The presence of Arg and Lys in this group is readily explained since the side chains of these amino acids are frequently positively charged and are thus unfavorable candidates to coordinate cations. The fact that Gln belongs to this group is more surprising while Asn is found in the favorable group of residues. A similar tendency, for calcium to interact preferentially with Asp over Glu, was observed for calcium ion binding motifs and was explained by the fact that Asp is often present in the Asx turn motif [18]. As most metals show a higher preference for Asp over Glu (Table 2), the shorter side chain is favored for metal ion binding. One explanation is that the restriction of conformational freedom upon cation binding, with an associated unfavorable entropy change, is less for the shorter side chain of Asx. Iron is the only metal which shows a higher preference for Glu than for Asp. This may be due to the fact that the data set used for calculating Fatom was a redundant one (in terms of metal-binding sites, not sequence similarity), and frequently observed motifs can slightly bias the analysis, such as for example di-iron sites [19].
Classification of amino acids based on their normalized frequency of interaction with metal ions may be used together with geometrical data to assist in the assignment of unknown metals in crystal structures or to verify the identity of “known” cations. Secondly, it could be used to predict potential metal ion binding sites in protein structures or even to engineer the addition or removal of metal ion binding sites from protein molecules.
4.2. Metal coordination in macromolecule and small molecule structures
In most cases, the metal-to-coordinating-atom distance distributions from the high-resolution data set agree quite well (in both mean and standard deviation) with the data from small-molecule structures (Table 3). However, some of the distance distributions for small molecule structures display two peaks which are not observed in the PDB data. In small-molecule structural data from CSD, bimodal distributions are observed for almost all metals in the imidazole and sulfur classes including interactions between Mn/Co/Fe/Ni and N/O, and for the Ni-S interaction (Table 4). The bimodal distributions observed in small-molecule structures reflect well-understood effects of ligands on the electronic and spin states of the metal ion. Depending on the location of coordinating groups in the spectrochemical series [20], they may favor either the low or high spin state of a metal ion. Groups coordinating metal ions in protein structures (except CO, CN−, and hemes) usually produce a weak ligand field; thus mostly only the high-spin state of the metal ions is present. In contrast, in small molecule structures, metal-coordinating ions and molecules producing stronger ligand fields are found more frequently, thus both high- and low-spin metal complexes are studied. In most cases, the longer distance mean from the CSD (the high-spin state) should be used as the reference value for metal-to-coordinating-atom distance when examining metal binding in proteins.
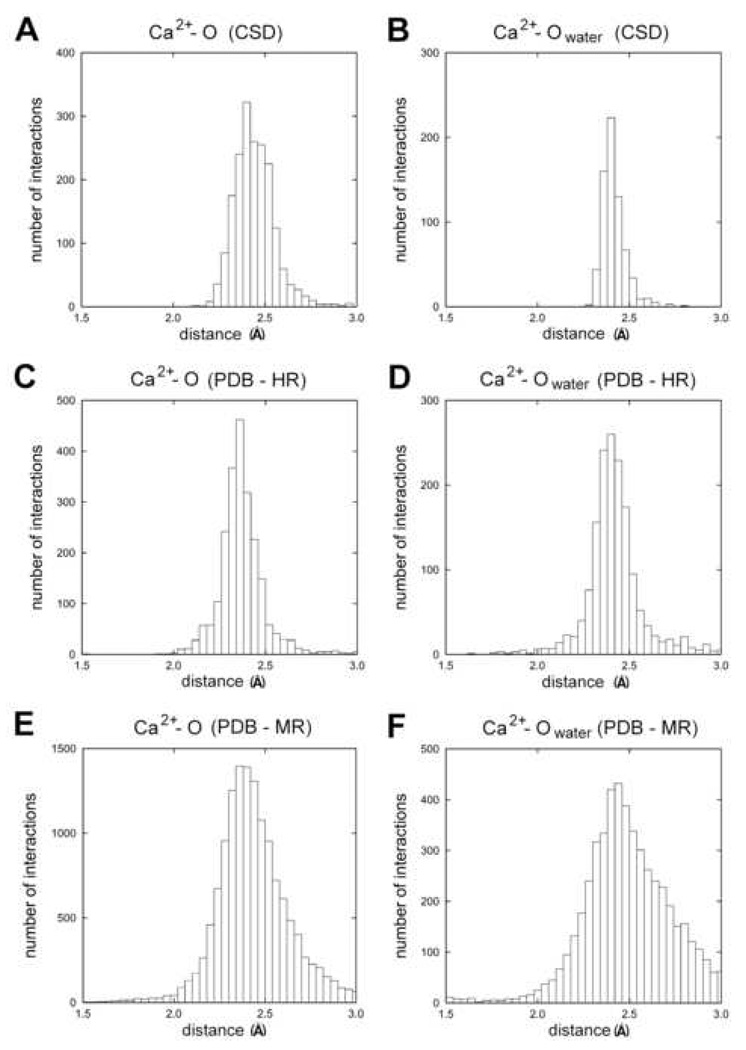
A comparison of calcium ion – oxygen distances is shown in Fig. 4 separately for water molecules and other atoms. It is apparent that for water molecules the distribution of distances broadens with lower resolution while the mean remains unchanged. The distribution of distances for high resolution protein structures is actually narrower than the distribution for structures from CSD. This most likely reflects a greater chemical diversity of ligands in CSD than in proteins.
Figure 4.
Distributions of calcium-to-protein-oxygen and calcium-to-water distances for different resolution ranges. The vertical axes give the number of interactions in each distance bin and the horizontal axes give the distance between calcium and oxygen. Distributions are made for both oxygen from protein (A, C, E) and oxygen from water (B, D, F). Data from the CSD (A, B), PDB high resolution data (C, D), and PDB moderate resolution data (E, F) are plotted individually.
4.3. Difference between high resolution and medium resolution
For high resolution data, the distributions of metal ion-ligand distances agree quite well with data from CSD as expected (Table 3). However, the distributions for medium resolution data are wider than those for high-resolution data indicating that for some of the structures, the geometric restraints around the metal ion used in the refinement were probably not properly set. For example, the mean value of calcium ion-oxygen distance is 2.37(12) Å for high-resolution data but the value for medium-resolution data is 2.43(19) Å (Fig. 4). The standard deviation for medium resolution data is 60% higher than the deviation for high-resolution data. Such a difference should not be observed if the restrained refinement of the metal binding site is properly carried out. It appears that most structures with unusual distances between calcium ions and oxygen atoms were refined without restraints as the use of restraints is a complex issue [4]. However, in some cases, the possibility that the unusual geometry is caused by the presence of two different metal ions with partial occupancy cannot be excluded. If we assume that the distance distributions derived from the CSD (where most structures are refined without geometric restraints) are error free, the distributions derived from macromolecule structures should have similar variations. However, the distribution of calcium to protein oxygen distances for medium-resolution macromolecular structures is much wider (Fig. 4E). Surprisingly, the high-resolution Ca-O distance distribution (Fig 4C) is narrower than the small molecule distribution (Fig. 4A) which can be explained by the use of too-tight protein geometry restraints combined with no restraints on the metal itself [4]. This does not apply to distances between Ca and water oxygen atoms as both the high-resolution and medium-resolution distributions (Figs. 4D, 4F) are broader than the small molecule distribution (Fig. 4B).
4.4. Calcium and magnesium ions coordination sphere
There is also an artificial correlation between data resolution and the mean coordination number for calcium or magnesium ions as low resolution diffraction data lead to models with an incomplete coordination sphere (Fig. 2). It is also apparent that in calcium or magnesium ion sites with complete coordination spheres, the calcium or magnesium ion-oxygen distances are much closer to values from CSD than in those sites with an incomplete coordination sphere. It is surprising that more than 5% of high-resolution structures report highly incomplete metal coordination spheres. Clearly, data resolution and R-factors alone cannot be used as the only criteria of the structure quality.
While the metal-binding sites in high resolution structures have mostly complete coordination spheres, a significant number of metal-binding sites in medium and low resolution structures contain highly incomplete coordination spheres (Figs. 2A, 2B). Very high coordination numbers (CN>6) are quite rare for magnesium ions but frequent in calcium ion binding sites, often due to bidentate coordination from a single carboxylate group. Calcium ions with coordination numbers higher than 6 can be explained by the fact that the frequency of bidentate coordination increases significantly for high coordination number calcium sites (7 or 8 oxygen atoms) (Figs. 2C, 2D). In both calcium and magnesium coordination sphere component plots, bidentate coordination exists roughly twice more frequently for CN=7 than for CN≤6 and roughly three times more frequently for CN=8. Occasionally, calcium ions coordinate only water molecules, forming hydrated calcium ions with a positive charge distributed over the complex. Such hydrated calcium ions are quite often observed in DNA structures. The highly negatively-charged surface of the major groove of double stranded DNA provides a suitable binding site for hydrated calcium ions with surface DNA atoms serving as the second coordination sphere. However, there are also hydrated calcium ions that do not fall into the DNA binding category; they are usually surrounded by negatively-charged residues (typically Asp or Glu) in the second coordination sphere. Hexaaquamagnesium ions are even more frequently observed.
Due to the irregular coordination of alkali metal ions in proteins, it is very difficult to identify a standard coordination model that describes most environments binding sodium or potassium ions in proteins. It is easier to generalize coordination properties of the imidazole and sulfur classes of metal ions in metalloproteins (Mn, Co, Fe, Ni, Zn, Cu) especially if the oxidation and spin states of the metal ion are taken into account. The coordination geometry preferences for some of these metal ions have been described previously [6, 21].
4.5. Unusual values related to metal ion binding sites present in PDB files
There are many unusual distance values between metal ions and protein atoms in structures reported in PDB (as compared to the CSD). As previously discussed, often metal-to-coordinating-atom distances are not properly restrained. The suspicious structures reported here have metal ion-ligand distances that were likely not restrained at all during refinement, resulting in physically impossible geometry. There are hundreds of PDB structures that include unusually small metal ion-ligand distances. For example, a number of calcium ion-oxygen distances are much smaller than 2.1 Å (the structure of cytochrome C oxidase assembly protein with PDB code 1XZO report a calcium-oxygen distance as short as 1.60 Å). Unusually short distances are obviously erroneous but it is also likely that many of the unusually long distances reported are also a result of improper application of refinement restraints.
While the majority of structures contain only a few metal ion binding sites, there are also structures with a large number of metal ions that do not interact directly with protein. This might indicate a problem in assignment of metal ions in the protein structure [22].
There are many unusual or suspicious values for occupancy and B-factor, particularly in entries deposited before the year 2000. These unusual values likely result from unintentional errors in the interpretation of refinement results or insufficient validation during deposition of the structures to the PDB, or both. As shown in Fig. 3, several dozen structures have unreasonably low metal ion B-factors, around 2 Å2. This is probably due to incorrect handling of metal ions where the B-factors were artificially set to the minimal value allowed by the refinement program. For example, 2 Å2 is the minimal allowed value for a B-factor in REFMAC. There are also many structures (2A3X, 1J0M, 1JIW, 1EAK, 1HEI, 1CLQ, 1SUS, 1N3C) that report unusually high B-factors, for some calcium ions (over 100 Å2), while the atoms in their environment have B-factors of less than 40 Å2. Such differences suggest either incorrect identification or partial occupancy of the metal cation.
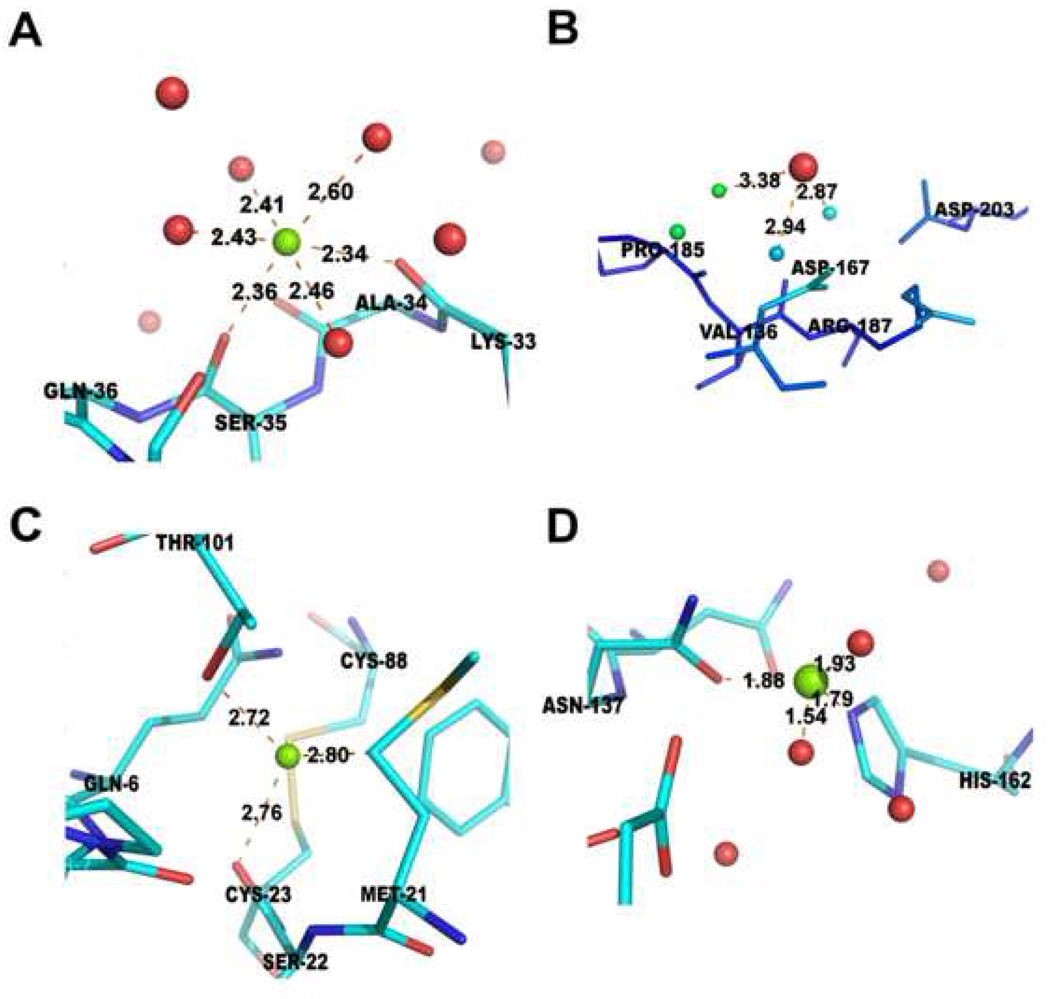
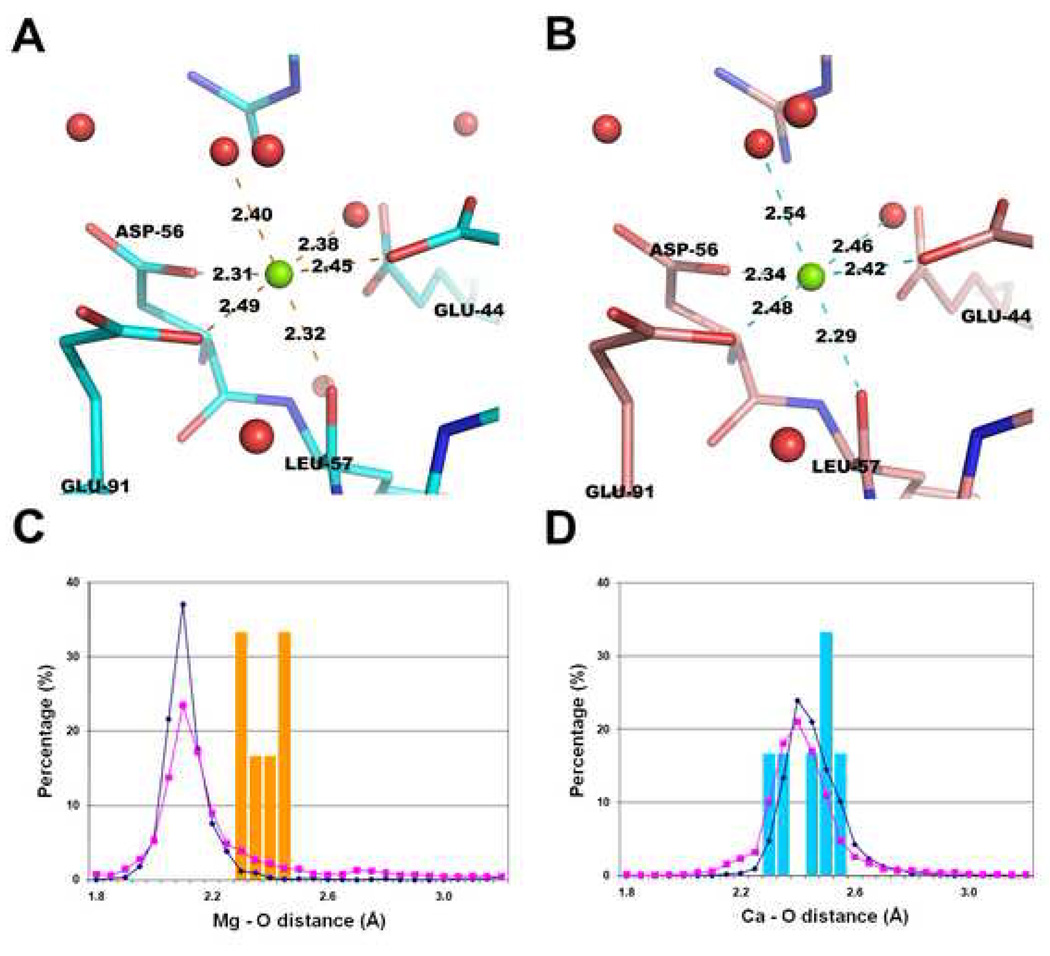
To illustrate applications of the statistics described above, we present analyses of some structures with potential errors. In some cases, it is apparent that the type of metal ion is misidentified. For example, there are cases when the magnesium ion-oxygen distance is unrealistically long while the coordination sphere is well-defined (Fig. 5A, Fig. 6A). For both magnesium ions shown (PDB code 2AS8, Mg 1001, and PDB code 1JUB, Mg A850), all magnesium ion-oxygen distances are about 0.3 Å longer than the reference distance (2.16 Å) based on CSD data, yielding very unfavorable coordination geometries for magnesium ions (Fig. 6A). In both cases, the B-factors of the magnesium ions are lower than the B-factors of coordinating atoms. To verify the correctness of cation assignment, we replaced the magnesium ions with calcium ions in one of the structures (2AS8) and re-refined the structure with or without distance constraints. We also re-refined the structure with magnesium ions using Mg-O distance constraints (Table 5). When the metal ion is identified as calcium, much better agreement with both the electron density map and geometry was obtained (Table 5) though the presence of Ca2+ over Mg2+ cannot be excluded conclusively without additional experiments. In the electron density for this structure, the magnesium ion is isoelectronic with water which makes its identification in the protein structure almost entirely dependent on binding site geometry. There are cases where the types of coordinating groups, coordination distances, and coordination numbers for magnesium ions are very unusual. One such case is a magnesium ion binding site in 1Q9Q (Fig. 5C) where there is a contact with a carbon atom (Mg-C distance of 2.80Å). The coordination number for this same site is also too small with two oxygen atoms (of distance to Mg of 2.72Å and 2.76 Å respectively). Putting a magnesium atom in such an environment is highly problematic not only in terms of binding geometry but also in terms of very unusual chemistry of the “metal ion binding site”. Another “magnesium site” in the same structure (Fig. 5D) has very small magnesium-oxygen and magnesium-nitrogen distances with all distances around 0.3 Å shorter than the reference distances derived from the CSD (2.16Å).
Figure 5.
Unusual metal atom model parameters. (A) An atom identified as magnesium with unusually long Mg-O distances (PDB code: 1JUB; Mg A850). (B) (C) Two atoms identified as magnesium in a structure with multiple geometry problems (PDB code: 1Q9Q) (see the online version of this article for the colors).
Figure 6.
Re-interpretation of a magnesium binding site as calcium (PDB code: 2AS8; Mg 1001). (A) The binding site of an atom identified as magnesium with unusually long Mg-O distances. (B) Re-refinement of the same structure, after identifying the metal atom as calcium. The histograms below show the distance distributions of the metal binding site before (C) and after (D) re-interpretation. The vertical axes give the percentage of structures and the horizontal axes the metal-oxygen distance in Å. The blue lines (diamond) represent the Mg-O (C) or Ca-O (D) distance distributions for CSD data. The magenta lines (square) represent the distance distributions for high resolution PDB data. The orange bars are the magnesium-oxygen distances in 2AS8 structure (C), while the cyan bars are the calcium-oxygen distances after re-interpretation of the structure (D) (see the online version of this article for the colors).
Table 5.
Comparison of restrained and unrestrained refinement of the structure 2AS8 with the metal identified either as calcium and magnesium. The rightmost column represents the original refinement of 2AS8. Two copies of the metal-binding site are found in the structure and the B-factors for each chain are reported.
| Metal identified as | Ca2+ | Ca2+ | Mg2+ | Mg2+(2AS8) |
|---|---|---|---|---|
| R [%] | 17.4 | 17.4 | 17.4 | 18.8 |
| Rfree [%] | 23.1 | 23.1 | 23.2 | 25.2 |
| Refinement | restrained | Unrestrained | restrained | unrestrained |
| Restraint distance | 2.4 Å | - | 2.1 Å | - |
| B factors [Å2] | ||||
| Metal | 24.1, 22.1 | 25.4, 23.4 | 14.2, 12.1 | 15.6, 17.2 |
| Water #1 | 20.1, 29.3 | 18.6, 36.6 | 18.9, 26.7 | 45.8, 21.9 |
| Water #2 | 24.6, 16.8 | 15.5, 25.7 | 15.7, 23.4 | 29.3, 17.2 |
| Glu #1 | 19.7, 24.8 | 18.1, 25.3 | 19.9, 26.4 | 20.3, 29.0 |
| Glu #2 | 19.9, 25.1 | 19.1, 27.1 | 20.7, 27.3 | 20.0, 27.1 |
| Asp | 20.2, 19.4 | 18.1, 17.7 | 20.5, 18.4 | 23.1, 21.7 |
| Leu | 18.6, 19.5 | 16.6, 18.1 | 19.5, 19.8 | 19.8, 19.1 |
| Mean (O atoms) | 22.3, 20.8 | 22.4, 20.4 | 21.8, 21.1 | 27.0, 22.1 |
| M-O distances | 2.36 – 2.42 Å | 2.29 – 2.54 Å | 2.13 – 2.28 Å | 2.26 – 2.49 Å |
5. Conclusion
Analysis of PDB structures that contain metal ions reveals that despite the several publications providing an excellent description of the geometry of metal ion environments, there are still many structures (even some solved very recently) that have quite unusual geometry. Often, the geometries of metal ion binding sites were not properly restrained, most probably due to the lack of mechanisms to automatically generate such restraints in all of the commonly used refinement programs. We suggest it is necessary to validate not only the macromolecular parts of the structure but also all non-proteinaceous moieties and their interactions with the macromolecule. We also present an analysis of the normalized frequencies of amino acids and chemical moieties involved in metal-protein (or metal-water) interactions. The analysis shows positive and negative preferences of some metals towards particular amino acids. Our approach may be used for fast identification of structural motifs that cannot be identified on the bases of sequence similarity alone.
Supplementary Material
Acknowledgment
We would like to thank Zbigniew Dauter, Andrzej Joachimiak, and Matthew Zimmerman for critically reading the manuscript and making valuable comments. The work was supported by NIH grants GM74942 and GM53163.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans PR. Acta Cryst. D. 2007;63:58–61. doi: 10.1107/S090744490604604X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engh R, Huber R. Acta Cryst. A. 1991;47:392–400. [Google Scholar]
- 4.Jaskolski M, Gilski M, Dauter Z, Wlodawer A. Acta Cryst. D. 2007;63:611–620. doi: 10.1107/S090744490700978X. [DOI] [PubMed] [Google Scholar]
- 5.Harding M. Acta Cryst. D. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 6.Harding M. Acta Cryst. D. 1999;55:1432–1443. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 7.Harding M. Acta Cryst. D. 2000;56:857–867. doi: 10.1107/s0907444900005849. [DOI] [PubMed] [Google Scholar]
- 8.Harding M. Acta Cryst. D. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 9.Harding M. Acta Cryst. D. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 10.Jahn H, Teller E. Proceedings of the Royal Society of London. Series A, Mathematical and Physical Sciences. 1937;161:220–235. [Google Scholar]
- 11.Harding M. Acta Cryst. D. 2004;60:849–859. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- 12.Allen FH. Acta Cryst. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Godzik A. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 14.Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 15.Bruno IJ, Cole JC, Edgington PR, Kessler M, Macrae CF, McCabe P, Pearson J, Taylor R. Acta Cryst. B. 2002;58:389–397. doi: 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]
- 16.CCDC. Vista - A Program for the Analysis and Display of Data Retrieved from the CSD. 12 Union Road, Cambridge, England: Cambridge Crystallographic Data Centre; 1994. [Google Scholar]
- 17.Kaufman Katz A, Shimoni-Livny L, Navon O, Navon N, Bock CW, Glusker JP. Helvetica Chimica Acta. 2003;86:1320–1338. [Google Scholar]
- 18.Pidcock E, Moore GR. J. Biol. Inorg. Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- 19.Kurtz DM. J. Biol. Inorg. Chem. 1997;2:159–167. [Google Scholar]
- 20.Zumdahl SS. In: Chemical Principles Fifth Edition. 5 ed. Zumdahl SS, editor. Boston: Houghton Mifflin Company; 2005. pp. 550–551. 957–964. [Google Scholar]
- 21.Rulisek L, Vondrasek J. J. Inorg. Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 22.Wlodawer A, Minor W, Dauter Z, Jaskolski M. FEBS J. 2007;275:1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.