Abstract
What is the relationship between enzymes and metabolites, the two major constituents of metabolic networks? We propose three alternative relationships between enzyme capacity and metabolite concentration alterations based on a Michaelis–Menten kinetic; that is enzyme capacities, metabolite concentrations, or both could limit the metabolic reaction rates. These relationships imply different correlations between changes in enzyme capacity and metabolite concentration, which we tested by quantifying metabolite, transcript, and enzyme abundances upon local (single-enzyme modulation) and global (GCR2 transcription factor mutant) perturbations in Saccharomyces cerevisiae. Our results reveal an inverse relationship between fold-changes in substrate metabolites and their catalyzing enzymes. These data provide evidence for the hypothesis that reaction rates are jointly limited by enzyme capacity and metabolite concentration. Hence, alteration in one network constituent can be efficiently buffered by converse alterations in the other constituent, implying a passive mechanism to maintain metabolic homeostasis upon perturbations in enzyme capacity.
Keywords: design principle, metabolic network, metabolomics, proteomics, transcriptome
Introduction
Physiological behavior emerges from complex dynamic interactions between transcripts, enzymes, and metabolites, the constituents of metabolism and its regulatory network (Sauer, 2006). Our ability to monitor global abundance alterations of those network constituents has developed rapidly over the past decade, culminating in an impressive array of so-called omics methods (Domon and Aebersold, 2006; Ishii et al, 2007; Dunn, 2008; Bennett et al, 2009). As a relatively recent addition, metabolomics methods became available for broad coverage and quantitative analysis of intracellular metabolite concentrations (van der Werf et al, 2007; Bennett et al, 2008; Garcia et al, 2008; Buescher et al, 2009). With the generation of large data sets by these omics methods, data integration, in particular across different omics levels, becomes the key challenge (ter Kuile and Westerhoff, 2001; Stitt and Fernie, 2003; Sauer et al, 2007).
The problem of data integration is the unknown or complex relationships between the different data types. An example of a more straightforward relationship is the one between transcripts and proteins that are directly linked through translation, where lack of correlation between both quantities is taken as a measure for post-transcriptional regulation (Griffin et al, 2002; Stitt and Fernie, 2003). The general relationship between proteins and metabolites is much less obvious. Although studied extensively for decades, the focus was on single reactions or small sets of connected reactions to elucidate molecular mechanisms of enzymatic catalysis in great detail, thereby enabling their formalized description by kinetic laws (Fersht, 1974; Cornish-Bowden, 1976). On the foundation of separately characterized enzymatic reactions, kinetic models are often used for simultaneous analysis of connected reaction sets (Teusink et al, 2000; Bettenbrock et al, 2006). The determination of model parameters (Mendes and Kell, 1998) and the transfer of in vitro determined parameters to in vivo conditions (van den Brink et al, 2008) is essential but very difficult. To enable model description of larger metabolic networks, simplifications such as lumping of linear pathways and linearization of non-linear equations were introduced. This greatly reduced the number of parameters but largely preserved the predictive power, provided one assesses small deviations from the reference state (Visser and Heijnen, 2003; Fell, 2005; Kresnowati et al, 2005). Metabolic control analysis and its extensions, in particular regulation analysis, provide a coherent framework for sensitivity of the influence of enzyme activities and metabolite concentrations on the flux through linear pathways in steady state (Kacser and Burns, 1995; ter Kuile and Westerhoff, 2001). This framework has also been extended to incorporate non-infinitesimal deviations from steady state in linear and branched pathways (Small and Kacser, 1993a, 1993b; Hatzimanikatis, 1999) and to facilitate analysis of larger networks by modularization (Schuster et al, 1993). Less mechanistically, metabolome and expression data have been integrated by correlation analyses to infer enzyme-reactant relationships (Bradley et al, 2009) and metabolic flux rewiring (Moxley et al, 2009). Two other statistical approaches were partial least squares analysis to identify responses specific for environmental conditions and gene knockouts (Pir et al, 2006) and covariance analysis in the context of the known metabolic network to distinguish different types of enzyme activity regulation (Cakir et al, 2006). In all these examples, two or more inputs such as measured data sets or models were combined to infer biologically interesting outputs such as regulation events or enzymatic control over flux. However, the prediction of changes and their direction in the metabolome from expression data or vise versa remains unsolved.
Here, we attempt to identify a general relationship between fold-changes in metabolite concentrations and enzyme capacity in central carbon metabolism that would allow to predict changes in metabolite concentration based on changes in enzyme capacity and vise versa. On the basis of enzyme kinetics, as in many of the above studies (Fersht, 1974; Kacser and Burns, 1995; Teusink et al, 2000 ), we propose several mechanistic relationships between the constituents of metabolism and then use a metabolic model to translate them into observable predictions. A precondition for this analysis are highly quantitative and large-scale transcript, enzyme, and metabolite abundances that are rarely available (Ishii et al, 2007; Sauer et al, 2007). Hence, we performed genome-wide transcript as well as quantitative proteomics (Picotti et al, 2008, 2009) and metabolomics (Buescher et al, 2009; Ewald et al, 2009) analyses that are targeted to central metabolism of the yeast Saccharomyces cerevisiae. To test the various predictions, we perturb yeast metabolism by deletion of a key transcription factor and by modulating expression of single enzymes and subsequently monitor transcript, enzyme, and metabolite response in wild type and mutant. These data allow us to demonstrate the validity of the proposed relationship in vivo for large alterations in single or multiple enzymes in various pathways of central carbon metabolism.
Results
Hypothetical principles of enzyme–metabolite relationship
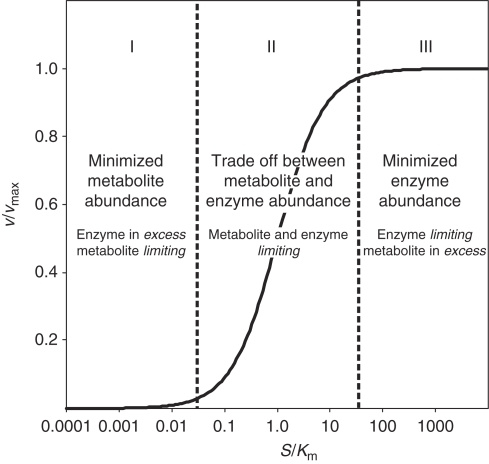
To elucidate whether general relationships exist between metabolite concentrations and enzyme capacities (i.e. the outcome of enzyme abundance combined with activity), we propose three hypothetical and alternative governing principles (Figure 1). The first hypothesis postulates a minimization of metabolite concentration at a given flux. In this case, in vivo substrate metabolite concentrations are much lower than Km of the enzyme that catalyze their reaction. Consequently, catalytic enzyme capacities are in excess and substrate metabolite concentrations limit the reaction rates. Thus, reaction rates are independent of small differences in enzyme capacity and small changes in reaction substrate metabolite concentrations are rapidly propagated to reaction product metabolites. As the metabolite concentrations would then be robust to small differences in enzyme abundances, alterations in metabolite concentrations are not expected to correlate with alterations in enzyme capacity.
Figure 1.
Alternative hypotheses on three different relationships between enzyme abundances and metabolite concentrations alterations based on a Michaelis–Menten enzyme kinetic.
The second hypothesis postulates a tradeoff between metabolite concentration and enzyme capacity. Hence, substrate metabolite concentrations are close to the Km values of the catalyzing enzymes, consequently both enzyme capacities and metabolite concentrations affect the reaction rate. In this case, a negative correlation between differences in concentrations of substrate metabolites and differences in enzyme capacity is expected. Three subclasses of such a tradeoff can be distinguished depending on whether substrates are metabolites that participate only in few reactions (e.g. glucose-6-P or succinate) or cofactors that participate in many reactions throughout the network (e.g. ATP or NADH). (a) Network-wide propagation of effects through cofactor reaction coupling would be minimal if cofactor concentrations are much higher than their Km values in the various enzymes. Thus, substrate metabolite concentrations, but not cofactor concentrations, correlate negatively with small differences in enzyme capacity. (b) Both, cofactor and metabolite concentrations are close to their Km values and influence the reaction rate. Thus, cofactors times metabolite concentrations correlates negatively with small differences in enzyme capacity. (c) Network-wide propagation of effects through cofactor reaction coupling would be maximal when substrate metabolite concentrations are much higher than the Km values. Thus, substrate cofactor concentrations, but not substrate metabolite concentrations correlate negatively with small differences in enzyme capacity.
The third hypothesis postulates a minimization of enzyme capacity at a given flux. Hence, substrate metabolite concentrations are much higher than the Km of their catalyzing enzymes, consequently metabolites are in excess and the enzyme capacities limit the reaction rates. Thus, the reaction rate is insensitive to small differences in metabolite concentrations and it is directly proportional to the enzyme capacity. Thus, one expects a positive correlation between differences in concentrations of reaction product metabolites and differences in enzyme capacity. The three subclasses (a–c) identified in hypothesis II can also be applied here.
As hypotheses I–III imply different relationships between enzyme capacities and metabolite concentrations, identification of the prevailing situation in microbial metabolism requires quantitative in vivo metabolite concentration and enzyme capacity data upon moderate changes in enzyme capacity. As a first test, we chose wild type S. cerevisiae and an otherwise isogenic mutant with a complete deletion of the transcription factor Gcr2p, an activator of glycolysis (Chambers et al, 1995). This mutant exhibits altered transcript abundances, enzyme activities, and metabolite concentrations within closely connected reactions in glycolysis and in the tricarboxylic acid cycle (Uemura and Fraenkel, 1990, 1999; Sasaki and Uemura, 2005). Although any other transcription factor that modulates expression of multiple genes could be used, Gcr2p has the advantage that its targets are primarily in central metabolism where metabolite and enzyme abundances are comparably high and therefore measurable at high accuracy and coverage. To quantify the relationship between metabolite concentrations and enzyme capacities, we determined transcript, enzyme, and metabolite abundances in wild type and GCR2 mutant in batch culture on glucose minimal medium. Transcript and enzyme abundances are used as surrogates for enzyme capacities. Certainly, this does not hold true if post-transcriptional or allosteric regulation takes place, and such cases are expected as outliers from the correlation.
Transcript abundances
As a first and global measure of the GCR2 deletion consequences, we determined 5649 mRNA abundances in wild type and mutant by microarray analysis (Supplementary Table 1). In the mutant, the expression of 257 and 165 genes was significantly increased and decreased, with a fold-change between 1.3 and 14.2 (P-value⩽0.05). As expression alterations beyond carbohydrate metabolism are either indirect effects of the reduced mutant growth rate (Supplementary Table 2) or results from so far unknown targets of Gcr2p, we focused our attention on central carbon metabolism.
Differential expression within central metabolism during growth in minimal medium was consistent with GCR2 mutant data in rich medium (Sasaki and Uemura, 2005). The abundance of the glycolytic gene GLK1 was 1.8 fold (P-value=0.003) increased and those of PGI1, GPM1, and ENO1 between 1.4 and 1.7 fold (P-value⩽0.0006) decreased in the mutant compared with the wild type (Supplementary Figure 1). ENO2 and CDC19 were only slightly decreased (fold-change between 1.2 and 1.3, P-value⩽0.0007). For 7 out of 9 tricarboxylic acid cycle reactions, we found expression of at least one encoding gene to be slightly increased (fold-change between 1.1 and 1.2, P-value ⩽0.04) in the mutant compared with the wild type. Given the highly coordinated response, even this subtle increase seems to be biologically relevant. On rich medium, only four genes of the tricarboxylic acid cycle were expressed at higher level (Sasaki and Uemura, 2005), which might be related to the additional influx of amino acids into the cycle.
Enzymes abundances
As a precondition for this work, the transcript data confirmed the primary regulatory targets of Gcr2p in central metabolism, and now enabled us to specifically target the relevant proteins and metabolites for further analysis. To obtain a more quantitative readout on altered in vivo enzyme capacities that are expected to result from this differential gene expression, we quantified the abundance of 50 central metabolic enzymes by targeted mass spectrometry-based proteomics (Supplementary Table 3). The abundance of 9 out of 15 measured glycolytic enzyme was lower in the GCR2 mutant than in the wild type (fold-change between 1.3 and 5.3, P-value⩽0.05), which is in agreement with reported glycolytic in vitro enzyme activities in rich medium (Uemura and Fraenkel, 1999). Likewise, enzyme abundances in the acetate and ethanol formation pathways were decreased in the mutant. The abundance of 3 out of 7 measured enzymes within the pentose phosphate pathway and 6 out of 19 within the tricarboxylic acid cycle were increased in the mutant (fold-change between 1.3 and 3.0, P-value⩽0.05). Generally, the observed differences in enzyme abundances were more pronounced than the respective differences in mRNA abundances (Supplementary Figure 1).
Metabolite concentrations
Intracellular concentrations of metabolites in the vicinity of differentially expressed enzymes were quantified by targeted metabolomics. Specifically, we determined absolute concentrations of 24 metabolites and relative concentration for an additional three metabolites, thereby covering 80% of central carbon metabolism (Supplementary Figure 2; Supplementary Table 4). All measurements were performed with liquid chromatography–mass spectrometry analysis except for the tricarboxylic acid metabolites that were determined by gas chromatography–mass spectrometry. Overall, metabolite concentrations in the GCR2 mutant were mostly higher in glycolysis and lower in the tricarboxylic acid cycle compared with the wild type. The greatest fold-change was observed for P-glycerate, as reported earlier (Uemura and Fraenkel, 1999).
We used two approaches to further confirm the validity of our metabolite data set. First, exponentially growing cells are expected to exhibit an adenylate energy charge of 0.80 or higher (Wiebe and Bancroft, 1975). As the calculated adenylate energy charge on glucose was 0.89±0.02 (0.75±0.02 on ethanol) and 0.78±0.02 (0.73±0.02 on ethanol) for wild type and mutant, respectively, we have evidence that the highly sensitive energy cofactor concentrations are a faithful representation of the in vivo situation. Second, we used network-embedded thermodynamic analysis (Kümmel et al, 2006; Zamboni et al, 2008) to demonstrate that all measured metabolite concentrations were thermodynamically consistent with the expected direction of flux.
Integration of metabolite concentrations with transcript and enzyme abundances
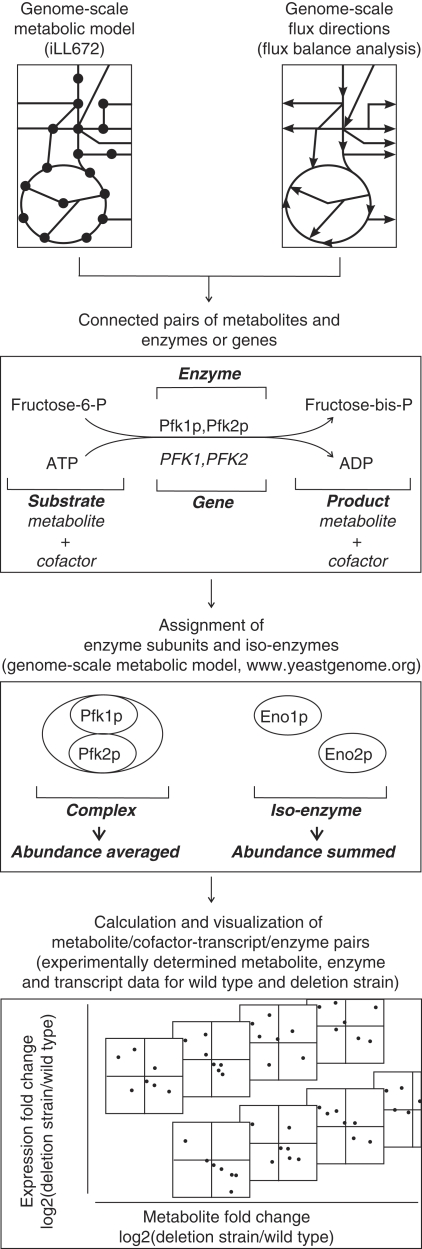
To determine whether one of the above-proposed hypotheses (Figure 1) between enzymes and metabolite prevails in vivo, we quantified the various metabolite and transcript/enzyme correlations. To maintain the functional metabolic context, we defined pairs of metabolites/cofactors with their connected genes/enzymes, using the genome-scale metabolic model iLL672 and flux balance-derived flux directions (Küpfer et al, 2005) (Figure 2). For most cases, we obtained clearly defined pairs of reaction substrate or reaction product metabolites/cofactors with their corresponding enzymes/genes. Where multiple genes were assigned to a single reaction, we used the yeast genome database (Cherry et al, 1998) and the genome-scale model to distinguish between iso-enzymes and subunits of enzymatic complexes. For iso-enzymes, the individually determined abundances were summed into a single reaction abundance value before calculating fold-change between mutant and wild type. For multisubunit complexes, we calculated the average complex abundance by averaging over all subunits, weighted with their stoichiometric participation in the complex.
Figure 2.
Workflow to test potential relationships between enzyme abundances and metabolite concentration alterations. Metabolite alterations were linked to alterations in the connected enzymes/transcripts using the metabolic network of S. cerevisiae (Küpfer et al, 2005).
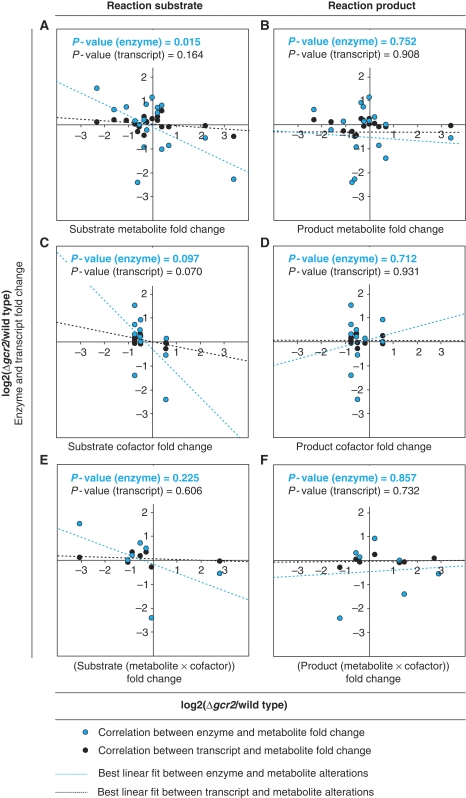
The implied fold-change correlations were then evaluated by the P-value for the null hypothesis that the data are not correlated (Figure 3). If metabolite abundance is minimized (hypothesis I), enzyme and metabolite abundances should be independent of each other and there would be no correlation in Figure 3. If a tradeoff between metabolite and enzyme abundance exists (hypothesis II): (a) substrate metabolite fold-changes could correlate with transcript/enzyme fold-changes (Figure 3A) and thus network-wide propagation of effects through cofactor reaction coupling is minimal; (b) substrate metabolite times substrate cofactor fold-changes could correlate with transcript/enzyme fold-changes (Figure 3E) and thus both are equally relevant; and (c) substrate cofactor fold-changes could correlate with transcript/enzyme fold-changes (Figure 3C) and thus network-wide propagation of effects through cofactor reaction coupling is maximal. Finally, if enzyme abundance is minimized (hypothesis III), product metabolite/cofactor correlate with transcript/enzyme fold-changes (Figure 3B and D) and/or product metabolite times product cofactor fold-changes correlate with transcript/enzyme fold-changes (Figure 3F).
Figure 3.
Correlation of transcript and enzyme fold-changes with fold-changes in substrate metabolites (A), product metabolites (B), substrate cofactors (C), product cofactors (D), substrate metabolites times substrate cofactors (E), and product metabolites times product cofactors (F) based on exponentially growing GCR2 mutant and wild type batch cultures on glucose minimal medium (data are found in Supplementary Tables 1, 3, and 4A).
Although functionally related, neither enzyme nor transcript fold-changes were significantly correlated with fold-changes in metabolites/cofactors when these were reaction products (Figure 3B, D, and F corresponding to hypothesis IIIa–c). Likewise, no correlation of enzyme or transcript fold-changes with substrate metabolite times substrate cofactor was found (Figure 3E corresponding to hypothesis IIb). Although reasonable P-values of 0.097 and 0.070 were found for the correlation between fold-changes of substrate cofactors and fold-changes of enzymes or transcripts, respectively (Figure 3C corresponding to hypothesis IIc), we did not consider them further because they were defined by only two point clouds that resulted from similar cofactor concentrations in wild type and GCR2 mutant.
The most significant correlation was observed for fold-changes in substrate metabolite concentrations with fold-changes in enzyme abundance (Figure 3A corresponding to hypothesis IIa). Not unexpectedly, enzyme abundances were a significantly better approximation for enzyme capacities than transcript abundances; as seen by the lower P-value for the enzyme-metabolite correlation than the transcript–metabolite correlation. A further improved correlation was achieved by considering all diverging enzymes that react upon a given substrate metabolite simultaneously rather than considering them as a separate reaction (Figure 4). This improvement by summing the abundances before calculating fold-changes supports the intuitive notion that divergent branches of enzymes simultaneously affect substrate metabolite concentrations. The high correlation between substrate metabolite and enzyme fold-changes suggests a tradeoff between enzyme capacity and metabolite concentrations in central metabolism. These results also indicated that reaction coupling by cofactors does not occur in vivo, since the best correlation was obtained by solely considering substrate metabolites without cofactors. In general, allosterically regulated enzymes might not fit into this correlation because their enzyme capacity can be regulated independently of their abundance. Although several allosteric enzymes occur in central carbon metabolism, only pyruvate kinase (Cdc19p) was identified as an outlier of the correlation (Figure 4).
Figure 4.
Correlation between substrate metabolite and enzyme fold-changes when considering all enzymes that share a given reaction substrate metabolite simultaneously (P-value=0.012) (data are found in Supplementary Table 3 and 4A).
Validity of negative correlation between substrate metabolite and enzyme abundances
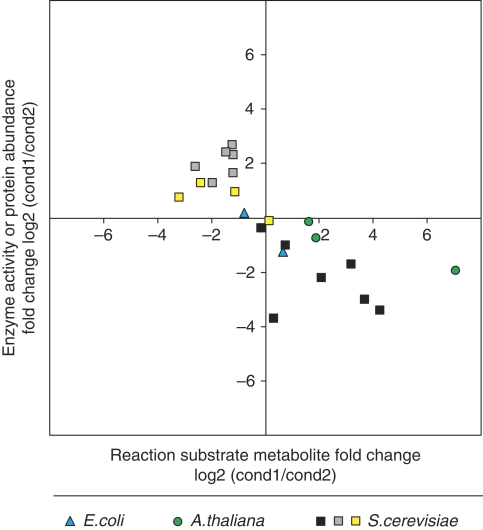
To test the general validity for central carbon metabolism of the above identified tradeoff between reaction substrate metabolite concentrations and enzyme abundances, we performed four independent validations: a statistical, a literature based, and two experimental ones. Statistically, we verified that the correlation between substrate metabolites and enzymes could not have been found by chance. For this purpose, we calculated P-values for 108 correlations of 17 metabolite–enzyme pairs that were randomly picked from all measured metabolite and enzyme abundances. The smallest observed P-value was 0.174 (Bonferroni corrected for 108 random samples). On this basis, we conclude that by chance occurrence of the proposed correlation is highly unlikely. On the basis of the literature data, we performed the above correlation analysis with data from the bacterium Escherichia coli (Rahman et al, 2006), the plant Arabidopsis thaliana (Rohde et al, 2004), and three data sets from the yeast S. cerevisiae (Uemura and Fraenkel, 1999; Castrillo et al, 2007; Tai et al, 2007) (Figure 5). All available data followed the proposed correlation, thus providing further evidence for the general validity of this relationship. Different theoretical analysis of optimal enzyme properties suggested substrate concentrations either one order of magnitude below Km (Fersht, 1974) or within one order of magnitude of Km (Cornish-Bowden, 1976), the latter was also found experimentally in E. coli (Bennett et al, 2009). This supports our hypothesis of a tradeoff between metabolite and enzyme efficiency. The finding that cofactor concentrations are predominantly above their respective Km values in E. coli (Bennett et al, 2009) strongly supports our finding that reaction coupling in the metabolic network is rather loose.
Figure 5.
Correlation between metabolite and enzyme activity/protein abundance fold-changes based on literature data of central carbon metabolism (Uemura and Fraenkel, 1999; Rohde et al, 2004; Rahman et al, 2006; Castrillo et al, 2007; Tai et al, 2007).
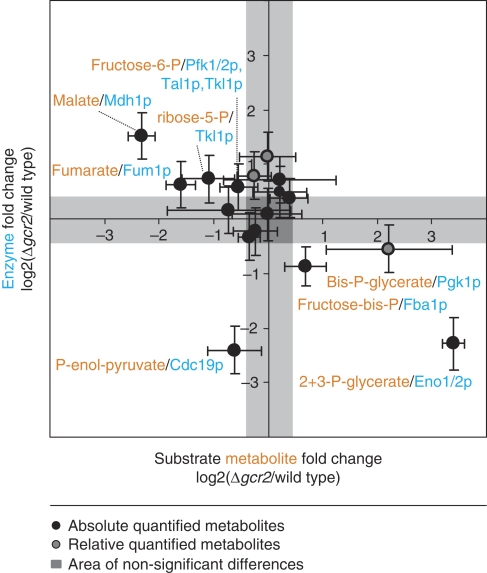
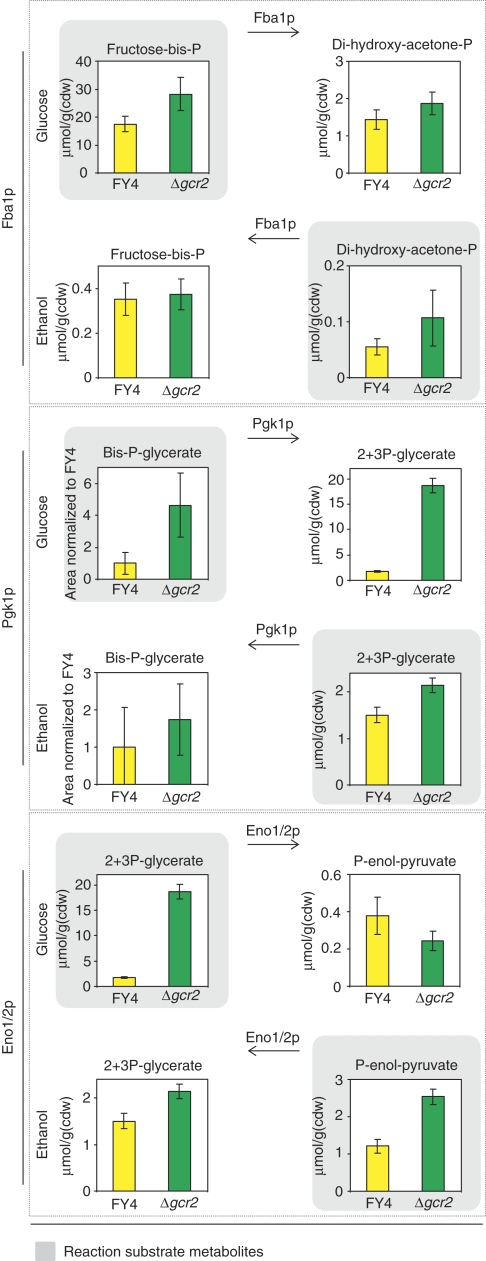
As a more serious challenge of the identified correlation, we designed an experiment where the absolute flux alterations are large and additionally the flux directions are altered. For this purpose, we quantified intracellular metabolite concentrations in both wild type and GCR2 mutant during growth on ethanol as sole carbon source. Under this condition, the flux through the glycolytic pathway is reversed relative to glucose growth and the fluxes are also much lower (Küpfer et al, 2005). Reported in vitro enzyme capacities demonstrated that fold-changes in capacity between the wild type and the GCR2 mutant are qualitatively identical and quantitatively similar on glycolytic and gluconeogenic substrates (Uemura and Fraenkel, 1999). Thus, we expected the new substrate metabolites (which were product metabolites on glucose) to occur at higher concentrations in the mutant than in the wild type. We can test this for the reactions catalyzed by Fba1p, Eno1/2p, and Pgk1p, because flux directions are reversed and enzyme abundances are significantly altered (Figure 4). The expectation was fulfilled by the experimental data in all cases, thereby further corroborating the negative correlation between enzyme capacity and metabolite concentrations (Figure 6).
Figure 6.
Substrate and product metabolite concentrations for reactions catalyzed by Eno1/2p, Fba1p, Pgk1p in the exponentially growing GCR2 mutant (green) and wild type (yellow) on glucose or on ethanol as sole carbon source. Eno1/2p, Fba1p, and Pgk1p exhibit lower enzyme activities in the mutant compared with the wild type (Uemura and Fraenkel, 1999) (data are found in Supplementary Table 4A).
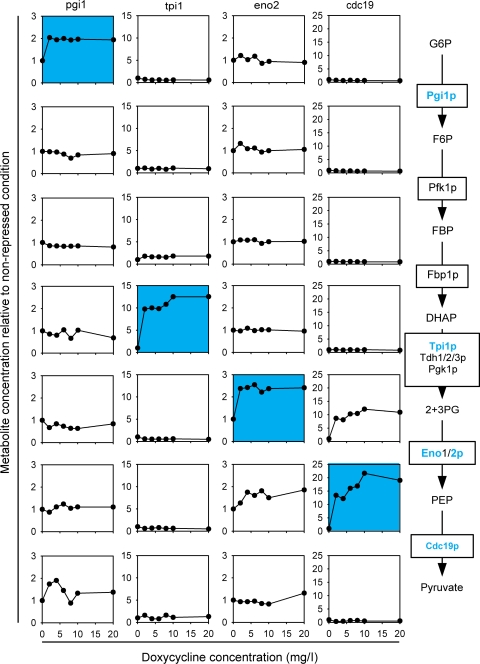
So far, our experimental evidence was based on perturbing multiple enzyme abundances through a transcription factor mutant. To ensure that our findings are also valid for single-reaction perturbations, we modulated individual abundances of the four glycolytic enzymes Pgi1p, Tpi1p, Eno2p, and Cdc19p using strains whose endogenous genomic promotor was replaced by a Tet-controlled promotor (Mnaimneh et al, 2004). The Tet-controlled promotor is repressed by addition of doxycycline (or tetracycline). Such strains are available for about 13% of all yeast genes, and we tested the four strains for which we could measure metabolites surrounding the perturbed reaction. The strains were grown on glucose minimal medium, supplemented with leucine, methionine, uracil, histidine, and lysine at different amounts of doxycycline to modulate expression. We then determined intracellular metabolite concentrations during exponential growth and normalized them to the concentrations without doxycycline (Figure 7). The identified relationship between substrate metabolite and enzyme abundance alterations predicts an exclusive increase for the substrate metabolite of downregulated enzymes, while all other metabolite concentrations should remain constant. This prediction was indeed verified with two minor exceptions. Reduced Eno2p abundance, led also to a minor increase of the product metabolite P-enol-pyruvate, which might be related to allosteric regulation of Cdc19p that further catalyzes P-enol-pyruvate. Furthermore, reduced Cdc19p abundance, not only increased the expected P-enol-pyruvate concentration but also 2+3-P-glycerate, albeit somewhat dampened, which indicates that the adjacent enolase reaction operates close to thermodynamic equilibrium (Kümmel et al, 2006). We thus conclude that metabolic homeostasis in central carbon metabolism is achieved by local metabolite responses, independent of whether the perturbations are at single enzymes or more coordinated at multiple enzymes.
Figure 7.
Metabolite concentrations in S. cerevisiae strains with independently downregulated protein abundance. Protein abundances were modulated using yeast strains whose endogenous promotor was replaced by a Tet-controlled one. Blue background highlights the substrate metabolite for the perturbed enzyme abundance. Enzyme depicted in blue were modulated. G6P=glucose-6-P, F6P=fructose-6-P, FBP=fructose-bis-phosphate, DHAP=di-hydroxy-acetone-P, 2+3PG=2+3 P-glycerate, PEP=P-enol-pyruvate (data are found in Supplementary Table 4B).
Discussion
We demonstrate here that global or local alterations in enzyme abundance correlate negatively with enzyme reaction substrate concentration at least in central carbon metabolism. This implies a tradeoff between enzyme and metabolite efficiency in metabolic networks. As the correlation was specific to substrate metabolites that are connected to few reactions, but did not extent to highly connected cofactor metabolites, there was no reaction coupling such that the metabolite response remained relatively local.
These findings can be interpreted as a passive network mechanism to maintain close-to-wild-type homeostasis of central carbon metabolism upon perturbations that alter the enzyme capacity (Cornish-Bowden, 1976). Such alterations in enzyme capacity are buffered by converse changes in substrate metabolite concentration, thereby minimizing the difference in metabolic flux that is caused by the alteration. It seems that cells locally sacrifice metabolite homeostasis to maintain fluxes and global metabolite homeostasis. This finding is in line with earlier findings, suggesting small influence of single enzymes on metabolic flux (Kacser and Burns, 1995), and further explains why metabolic fluxes are surprisingly robust to local (e.g. enzyme deletions) but also global (e.g. transcription factor deletions) genetic perturbations (Blank et al, 2005; Fischer and Sauer, 2005; Perrenoud and Sauer, 2005; Tang et al, 2009). Furthermore, this mechanism has two intuitive advantages for the cell. First, environmental stresses that drain reduction cofactor metabolites (e.g. oxidative stress) (Hampsey, 1997; Grant, 2008) are not propagated throughout the network, thus minimizing the impact on metabolic homeostasis. Second, global environmental perturbations that affect enzyme capacity are buffered by increased metabolite concentrations, thus minimizing growth effects. Only when alterations in central carbon metabolism exceed the passive metabolite–enzyme capacity buffering the reaction flux will change. If this passive buffering mechanism is also present in secondary metabolism, which has an inherent lower overall flux remains open. Although we demonstrated the relationship in yeast, it remains to be tested whether it generally holds for other organisms. Yet, in the small data sets of a plant (Rohde et al, 2004) and a bacterium (Rahman et al, 2006) that we used for our empirical validation, we found no evidence against it.
Finally, the identified relationship between metabolite concentrations and enzyme capacities provides a theoretical basis for the use of metabolomics as fast screening method in functional genomics. As mass spectrometry-based intracellular metabolomics is amenable for high-throughput analysis (Ewald et al, 2009), large data sets could be generated rapidly. Our results indicate that, at least to some extent, an altered metabolite concentration can be interpreted as a converse change in its catalyzing enzyme capacity.
Materials and methods
Strains, medium, and cultivation conditions
S. cerevisiae wild type FY4 Mata (Winston et al, 1995) (kindly provided by Fred Winston) was used as a reference. The GCR2 mutant was constructed as whole gene deletion by using a KanMX4 cassette in the prototroph background of FY4 Mata (Winston et al, 1995) (kindly provided by Charlie Boone). Tet-titratable promotor strains were obtained from openbiosystem (Mnaimneh et al, 2004).
Liquid pre-cultures were inoculated from freshly plated YPD plates. Pre-cultures were always grown in glucose minimal medium as described earlier with 10 g/l glucose (Blank and Sauer, 2004). Ethanol (5 g/l) as carbon source was only added to the final cultures. Cultivations of 25–50 ml were performed in 500 ml shake flasks at 30°C and 250 r.p.m. (metabolite and proteome measurement), or in 96-deep-well plates (Kuehner AG, Birsfeld, Switzerland) (Duetz et al, 2000) with a culture volume of 1.2 ml, at 30°C and 300 r.p.m. (transcriptome measurement). All shakers had a 50 mm amplitude. To improve mixing, a single 4 mm diameter glass bead (Sigma-Aldrich, Buchs, Switzerland) was added to each well. For the metabolome measurement, we reduced the medium potassium hydrogen phthalate concentration from 100 mmol/l to 10 mmol/l with no significant change in physiology. Experiments with Tet-controlled promotor strains were performed in reaction tubes with 3.5 ml filling volume. The cultivation medium was glucose minimal medium supplemented with leucine (0.24 g/l), methionine (0.02 g/l), uracil (0.02 g/l), histidine (0.02 g/l), and lysine (0.03 g/l). To titrate expression alterations, the doxycycline concentration (Sigma-Aldrich) was varied between 0 mg/l and 20 mg/l.
All samples were taken at an OD600 between 0.8 and 1.2 from cultures growing exponentially on minimal medium. To ensure comparability among the samples, only cultures that followed a standardized growth curve and thus exhibited reproducibly the determined physiology were used (Supplementary Table 2).
Physiological parameters
Specific growth rates were determined from at least three independent cultures and at least six OD600 points during the exponential growth. Uptake and secretion rates were determined as described elsewhere (Heer and Sauer, 2008).
Transcriptome analysis
Harvesting, extraction of mRNA, and on-column DNase digestion were performed by the mechanical disruption protocol of the RNeasy Mini Kit (50) (Qiagen, Rapperswil, Switzerland).
Total RNA samples were reverse transcribed with One-Cycle cDNA Synthesis Kit (Affymetrix Inc., P/N 900431, Santa Clara, CA, USA). The double-stranded cDNA was purified using the Sample Cleanup Module (Affymetrix Inc., P/N 900371). The purified double-stranded cDNAs were in vitro transcribed with biotin-labeled nucleotides using the IVT Labeling Kit (Affymetrix Inc., P/N 900449). The biotinylated cRNA was purified using the Sample Cleanup Module, and NanoDrop ND 1000 and Bioanalyzer 2100 were used to determine quality and quantity. Biotin-labeled cRNA samples were fragmented randomly to 35–200 bp at 94°C in fragmentation buffer (Affymetrix Inc., P/N 900371) and suspended in 100 μl of hybridization mix (Affymetrix Inc., P/N 900720), containing a hybridization control and control oligonucleotide B2 (Affymetrix Inc., P/N 900454). Samples were hybridized to GeneChip Yeast Genome 2.0 arrays for 16 h at 45°C. Arrays were then washed using an Affymetrix Fluidics Station 450 FS450 0003 protocol. An Affymetrix GeneChip Scanner 3000 (Affymetrix Inc.) was used to determine the fluorescent intensity emitted by the labeled target.
Affymetrix CEL files were processed using R (version 2.8.0) and the Bioconductor affy package (Gautier et al, 2004). Probe intensities were normalized for background by using the robust multiarray average method (RMA) (Irizarry et al, 2003), using only perfect match (PM) probes. Normalization was performed using the qsplines algorithm (Workman et al, 2002). Gene expression values were calculated from the PM probes using the expression index calculation method (Li and Wong, 2001). Raw data are stored in GEO (series number GSE19569, NCBI tracking system 15756402).
Proteome analysis
The targeted proteomics protocol based on single-reaction monitoring as described by Picotti et al (2008, 2009) was applied. The majority of the proteins is quantified based on two or more peptides. Exceptions with single peptides were Adh2p, Fba1p, Gnd2p, Gpd2p, Hxk2, Lat1p, Pfk2p, Rki1p, Sdh2p, Sdh4p, and Tdh2p. All the coordinates of the single-reaction monitoring assays used are listed in Supplementary Table 5. Raw tandem mass spectrometry data have been deposited in the publicly accessible repository of proteomic data PeptideAtlas (S. cerevisiae—MRMAtlas build, http://www.mrmatlas.org; Picotti et al (2008)) and can be browsed using the yeast genome database accession name of each protein/ORF measured.
Metabolome analysis
Sampling and cold methanol quenching were performed by de Koning and van Dam method (de Koning and van Dam, 1992) and its extensions (Buescher et al, 2009; Ewald et al, 2009). Liquid chromatography separation of compounds was achieved by an ion pairing-reverse phase method developed for ultra high performance systems, based on previously published high pressure methods (Luo et al, 2007; Buescher et al, 2009; Ewald et al, 2009) and implemented on a Waters Acquity UPLC (Waters Corporation, Milford, MA, USA) using a Waters Acquity T3 end-capped reverse phase column with dimensions 150 mm × 2.1 mm × 1.8 μm (Waters Corporation). Selective and sensitive detection of compounds was achieved by coupling liquid chromatography to a Thermo TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) using a heated electrospray ionization source (Thermo Fisher Scientific). The mass spectrometer was operated in negative mode with multiple reaction monitoring. Fragmentation parameters were optimized individually for all compounds (Supplementary Table 6). Both acquisition and peak integration were performed with the Xcalibur software version 2.07 SP1 (Thermo Fisher Scientific) and in-house integration software (Begemann and Zamboni, unpublished). Peak areas were normalized to fully 13C-labeled internal standards (Wu et al, 2005) and the amount of biomass.
Tricarboxylic acid cycle intermediates were determined with gas chromatography-time-of-flight mass spectrometry (Ewald et al, 2009). Raw data are available as Supplementary Table 4.
Statistical analysis
To validate correlations, we calculated P-value, with the Matlab function ‘corrcoef’ (The MathWorks Inc., Natick, MA, USA). First, the correlation coefficient matrix R is calculated
where x denotes the log2 fold-changes of metabolites and y denotes the log2 fold-changes of transcripts/enzymes. The P-value is then computed by transforming the correlation to create a t-statistic.
Supplementary Material
Log2 ratios of transcript fold changes between the GCR2 mutant and the wild-type. An average of three biological replicas is reported. (see text file)
Log2 ratios of enzyme fold changes between the GCR2 mutant and the wild-type. An average of two biological replicas is reported. (see text file)
Metabolite concentration. Determined in exponentially grown wild-type FY4 and GCR2 mutant batch cultures on A) glucose minimal medium and ethanol minimal medium. The cdw to OD correlation was determined for wild-type FY4 and applied also to the GCR2 mutant. B) Metabolite fold changes of four strains with Tet-controlled promotor (Tet-controlled enzymes: Pgi1p, Tpi1p, Eno2p, Cdc19p) determined in glucose minimal medium with amino acids and varying concentrations of doxycycline. Data are normalized to the metabolite concentration determined without the addition doxycycline. (see text files)
Metabolite concentration. Determined in exponentially grown wild-type FY4 and GCR2 mutant batch cultures on A) glucose minimal medium and ethanol minimal medium. The cdw to OD correlation was determined for wild-type FY4 and applied also to the GCR2 mutant. B) Metabolite fold changes of four strains with Tet-controlled promotor (Tet-controlled enzymes: Pgi1p, Tpi1p, Eno2p, Cdc19p) determined in glucose minimal medium with amino acids and varying concentrations of doxycycline. Data are normalized to the metabolite concentration determined without the addition doxycycline. (see text files)
SRM assays used in this study for measuring enzyme abundance alterations. Q1m/z, mass-to-charge ratio for the peptide ion used as Q1 value; Q3m/z mass-to-charge ratio for the fragmentation used as Q3 value; H/L, heavy/light version of the peptide; CE, collision energy. (see text file)
SRM assays used in this study for measuring metabolite concentrations with liquid chromatography mass spectrometry. Q1m/z, mass-to-charge ratio for the metabolite ion used as Q1 value; Q3m/z mass-to-charge ratio for the fragmentation used as Q3 value. (see text file)
Supplementary figures S1–2, Supplementary table legends S1–6, Supplementary table 2
Acknowledgments
We thank Owen Ryan from Charlie Boone's Lab (University of Toronto) for constructing and providing the GCR2 mutant. We are also thankful to Ana Paula Oliveira from Uwe Sauer's Lab (ETH Zurich) for the help in transcript data analysis. For financial support, SMF is grateful to the Competence Center for Systems Physiology and Metabolic Diseases, JMB to the EU project BaSysBio (LSHG-CT-2006-037469), and FR to the FWF Schrödinger Stipendium. PP is recipient of an intra-European Marie Curie Fellowship. Furthermore, the Swiss initiative of systems biology, SystemsX.ch is greatly acknowledged.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bennett BD, Kimball EH, Gao M, Osterhout R, van Dien SJ, Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Yuan J, Kimball EH, Rabinowitz JD (2008) Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenbrock K, Fischer S, Kremling A, Jahreis K, Sauter T, Gilles ED (2006) A quantitative approach to catabolite repression in Escherichia coli. J Biol Chem 281: 2578–2584 [DOI] [PubMed] [Google Scholar]
- Blank LM, Küpfer L, Sauer U (2005) Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol 6: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank LM, Sauer U (2004) TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiol 150: 1085–1093 [DOI] [PubMed] [Google Scholar]
- Bradley PH, Brauer MJ, Rabinowitz JD, Troyanskaya OG (2009) Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. PLoS Comput Biol 5: e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher JM, Czernik D, Ewald JC, Sauer U, Zamboni N (2009) Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal Chem 81: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Cakir T, Patil KR, Onsan Z, Ulgen KO, Kirdar B, Nielsen J (2006) Integration of metabolome data with metabolic networks reveal reporter reactions. Mol Syst Biol 2: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DCJ, Cornell MJ, Petty J, Hakes L, Wardleworth L, Rash B, Brown M, Dunn WB, Broadhurst D, O’Donoghue K, Hester SS, Dunkley TPJ, Haert SR, Swainston N, Li P et al. (2007) Growth control of the eukaryote cell: a systems biology study in yeast. J Biol 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A, Packham EA, Graham IR (1995) Control of glycolytic gene expression in budding yeast (Saccharomyces cerevisiae). Curr Genet 29: 1–9 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D (1998) SGD: Saccharomyces genome database. Nucleic Acids Res 26: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A (1976) The effect of natural selection on enzymic catalysis. J Mol Biol 101: 1–9 [DOI] [PubMed] [Google Scholar]
- de Koning W, van Dam K (1992) A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal Biochem 204: 118–123 [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R (2006) Mass spectrometry and protein analysis. Science 12: 212–217 [DOI] [PubMed] [Google Scholar]
- Duetz WA, Rüedi L, Hermann R, O’Conner K, Büchs J, Witholt B (2000) Methods for intense aeration, growth, storage and replication of bacterial strains in microtiter plates. Appl Environ Microbiol 66: 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WB (2008) Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys Biol 5: 11001. [DOI] [PubMed] [Google Scholar]
- Ewald JC, Heux S, Zamboni N (2009) High-throughput quantitative metabolomics: workflow for cultivation, quenching and analysis of yeast in a multi-well format. Anal Chem 81: 3623–3629 [DOI] [PubMed] [Google Scholar]
- Fell DA (2005) Enzymes, metabolites and fluxes. J Exp Bot 56: 267–272 [DOI] [PubMed] [Google Scholar]
- Fersht AR (1974) Catalysis, binding and enzyme-substrate complementary. Proc R Soc Lond B 187: 397–407 [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U (2005) Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet 37: 636–640 [DOI] [PubMed] [Google Scholar]
- Garcia DE, Baidoo EE, Benke PI, Pingitore F, Tang YJ, Villa S, Keasling JD (2008) Separation and mass spectrometry in microbial metabolomics. Curr Opin Microbiol 11: 233–239 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy-analysis of Affymetrix Gene Chip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Grant CM (2008) Metabolic reconfiguration is a regulated response to oxidative stress. J Biol 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 232–233 [DOI] [PubMed] [Google Scholar]
- Hampsey M (1997) A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133 [DOI] [PubMed] [Google Scholar]
- Hatzimanikatis V (1999) Nonlinear metabolic control analysis. Metab Eng 1: 75–87 [DOI] [PubMed] [Google Scholar]
- Heer D, Sauer U (2008) Identification of furfural as the key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microbial Biotechnol 1: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y et al. (2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316: 593–597 [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1995) The control of flux. Biochem Soc Trans 23: 341–366 [DOI] [PubMed] [Google Scholar]
- Kümmel A, Panke S, Heinemann M (2006) Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol Syst Biol 2: 2006.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresnowati MT, van Winden WA, Heijnen JJ (2005) Determination of elasticities, concentration and flux control coefficients from transient metabolite data using linlog kinetics. Metab Eng 7: 142–153 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of ologonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M (2007) Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A 1147: 153–164 [DOI] [PubMed] [Google Scholar]
- Mendes P, Kell D (1998) Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics 14: 869–883 [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Moxley JF, Jewett MC, Antoniewicz MR, Villas-Boas SG, Alper H, Wheeler RT, Tong L, Hinnebusch AG, Ideker T, Nielsen J, Stephanopoulos G (2009) Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proc Natl Acad Sci USA 106: 6477–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187: 3171–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 38: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold R (2008) A database of mass spectrometric assays for the yeast proteome. Nat Methods 5: 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pir P, Kirdar B, Hayes A, Onsan ZY, Ülgen K, Oliver SG (2006) Integrative investigation of metabolic and transcriptomic data. BMC Bioinformatics 7: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Hasan MR, Oba T, Shimizu K (2006) Effect of rpoS gene knockout on the metabolism of Escherichia coli during exponential growth phase and early stationary phase based on gene expressions, enzyme activities and intracellular metabolite concentrations. Biotechnol Bioeng 94: 585–595 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, de Rycke R, Kushnir S, van Doorsselaere J, Joseleau JP, Vuvlsteke M, van Driessche G, van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Uemura H (2005) Influence of low glycolytic activities in GCR1 and GCR2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 22: 111–127 [DOI] [PubMed] [Google Scholar]
- Sauer U (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Sys Biol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Heinemann M, Zamboni N (2007) Genetics. Getting closer to the whole picture. Science 316: 550–551 [DOI] [PubMed] [Google Scholar]
- Schuster S, Kahn D, Westerhoff HV (1993) Modular analysis of the control of complex metabolic pathways. Biophys Chem 48: 1–17 [DOI] [PubMed] [Google Scholar]
- Small JR, Kacser H (1993a) Responses of metabolic systems to large changes in enzyme activities and effectors. 1. The linear treatment of unbranched chains. Eur J Biochem 213: 613–624 [DOI] [PubMed] [Google Scholar]
- Small JR, Kacser H (1993b) Responses of metabolic systems to large changes in enzyme activities and effectors. 2. The linear treatment of branched pathways and metabolite concentrations. Assessment of the general non-linear case. Eur J Biochem 213: 625–640 [DOI] [PubMed] [Google Scholar]
- Stitt M, Fernie AR (2003) From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol 14: 136–144 [DOI] [PubMed] [Google Scholar]
- Tai SL, Daran-Lapujade P, Luttik MA, Walsh MC, Diderich JA, Krijger GC, van Gulik WM, Pronk JT, Daran JM (2007) Control of the glycolytic flux in Saccharomyces cerevisiae grown at low temperature: a multi-level analysis in anaerobic chemostat cultures. J Biol Chem 282: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Tang YJ, Martin HG, Deutschbauer A, Feng X, Huang R, Llora X, Arkin A, Keasling JD (2009) Invariability of central metabolic flux distribution in Shewanella oneidensis MR-1 under environmental or genetic perturbations. Biotech Progr 25: 1254–1259 [DOI] [PubMed] [Google Scholar]
- ter Kuile BH, Westerhoff HV (2001) Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett 500: 169–171 [DOI] [PubMed] [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329 [DOI] [PubMed] [Google Scholar]
- Uemura H, Fraenkel DG (1990) GCR2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol Cell Biol 10: 6389–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H, Fraenkel DG (1999) Glucose metabolism in GCR mutants of Saccharomyces cerevisiae. J Bacteriol 181: 4719–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink J, Canelas AB, van Gulik WM, Pronk JT, Heijnen JJ, de Winde JH, Daran-Lapujade P (2008) Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl Environ Microbiol 74: 5710–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf MJ, Overkamp KM, Muilwijk B, Coulier L, Hankemeier T (2007) Microbial metabolomics: toward a platform with full metabolome coverage. Anal Biochem 370: 17–25 [DOI] [PubMed] [Google Scholar]
- Visser D, Heijnen JJ (2003) Dynamic simulation and metabolic re-design of a branched pathway using linlog kinetics. Metab Eng 5: 164–176 [DOI] [PubMed] [Google Scholar]
- Wiebe WJ, Bancroft K (1975) Use of the adenylate energy charge ratio to measure growth state of natural microbial communities. Proc Natl Acad Sci USA 72: 2112–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3: 0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, van Winden WA, van Gulik WM, Heijnen J (2005) Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal Biochem 336: 164–171 [DOI] [PubMed] [Google Scholar]
- Zamboni N, Kümmel A, Heinemann M (2008) anNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data. BMC Bioinformatics 9: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Log2 ratios of transcript fold changes between the GCR2 mutant and the wild-type. An average of three biological replicas is reported. (see text file)
Log2 ratios of enzyme fold changes between the GCR2 mutant and the wild-type. An average of two biological replicas is reported. (see text file)
Metabolite concentration. Determined in exponentially grown wild-type FY4 and GCR2 mutant batch cultures on A) glucose minimal medium and ethanol minimal medium. The cdw to OD correlation was determined for wild-type FY4 and applied also to the GCR2 mutant. B) Metabolite fold changes of four strains with Tet-controlled promotor (Tet-controlled enzymes: Pgi1p, Tpi1p, Eno2p, Cdc19p) determined in glucose minimal medium with amino acids and varying concentrations of doxycycline. Data are normalized to the metabolite concentration determined without the addition doxycycline. (see text files)
Metabolite concentration. Determined in exponentially grown wild-type FY4 and GCR2 mutant batch cultures on A) glucose minimal medium and ethanol minimal medium. The cdw to OD correlation was determined for wild-type FY4 and applied also to the GCR2 mutant. B) Metabolite fold changes of four strains with Tet-controlled promotor (Tet-controlled enzymes: Pgi1p, Tpi1p, Eno2p, Cdc19p) determined in glucose minimal medium with amino acids and varying concentrations of doxycycline. Data are normalized to the metabolite concentration determined without the addition doxycycline. (see text files)
SRM assays used in this study for measuring enzyme abundance alterations. Q1m/z, mass-to-charge ratio for the peptide ion used as Q1 value; Q3m/z mass-to-charge ratio for the fragmentation used as Q3 value; H/L, heavy/light version of the peptide; CE, collision energy. (see text file)
SRM assays used in this study for measuring metabolite concentrations with liquid chromatography mass spectrometry. Q1m/z, mass-to-charge ratio for the metabolite ion used as Q1 value; Q3m/z mass-to-charge ratio for the fragmentation used as Q3 value. (see text file)
Supplementary figures S1–2, Supplementary table legends S1–6, Supplementary table 2