Fluorescence in-situ hybridization (FISH) technique allows the detection of single RNA molecules in individual yeast cells. We use this method complemented with theoretical modeling to determine the rate of switching from OFF to ON (activation rate) and the average number of RNA molecules produced during each transcriptional burst (burst size).
Switching of the rDNA repeats between the inactive non-transcribing state and the active transcribing state occurs rapidly. On average, it takes an inactive repeat 8 2 min to transit to the active state.
As cell density increases, mean rRNA transcriptional activity decreases. This is due to the decreases in burst size. The activation rate remains constant.
In the rpd3 strain, activation rate doubles and the burst size is half that of wild-type cells in log-phase. This effect of Rpd3 on activation rate is independent of cell density. As the cell density increases, the burst size of the rpd3 strain becomes similar to that of wild-type cells.
Keywords: rDNA, single-molecule FISH, Rpd3
Abstract
Most eukaryotes contain many tandem repeats of ribosomal RNA genes of which only a subset is transcribed at any given time. Current biochemical methods allow for the determination of the fraction of transcribing repeats (ON) versus non-transcribing repeats (OFF) but do not provide any dynamical information and obscure any transcription activity at the single-cell level. Here, we use a fluorescence in situ hybridization (FISH) technique that allows the detection of single-RNA molecules in individual yeast cells. We use this method complemented with theoretical modeling to determine the rate of switching from OFF to ON (activation rate) and the average number of RNA molecules produced during each transcriptional burst (burst size). We explore how these two variables change in mutants and different growth conditions, and show that this method resolves changes in these two variables even when the average rDNA expression is unaltered. These phenotypic changes could not have been detected by traditional biochemical assays.
Introduction
Regulation of rDNA synthesis is important for cell growth control (Warner, 1999; Grummt, 2003). Yeast cells carry about 150 copies of tandemly repeated rRNA genes. The regulation of rDNA transcription occurs by controlling both the fraction of actively transcribing repeats and the transcription rate of the active genes (Sandmeier et al, 2002; French et al, 2003). During rapid growth, only about half of these genes are active (ON) and being transcribed (Conconi et al, 1989; Dammann et al, 1995). The remaining repeats are inactive (OFF) and are not transcribed. Two distinct chromatin states correlating with the difference in transcriptional activity has been observed (Dammann et al, 1993). It has been shown that OFF repeats are arranged into regular nucleosomal arrays that prevent access of the Pol I transcription machinery, whereas ON repeats have an ‘open conformation'. More recently, it has been shown that active repeats exist in a dynamic chromatin structure of unphased nucleosomes, associated with chromatin-remodeling enzymes that may act to facilitate efficient rRNA synthesis (Jones et al, 2007). Current biochemical techniques like in vivo psoralen (Dammann et al, 1993) and electron microscopy (EM) Miller spread (Miller and Beatty, 1969) assays are frequently used in the study of rDNA regulation. Psoralen selectively cross-links to ON repeats and slows their migration during electrophoresis, allowing the determination of the fraction of ON repeats. rDNA regulation studied using EM by the Miller spreading method allows visualization of active and inactive rDNA genes. These methods have greatly enhanced our understanding of rDNA regulation but they are tedious, require lysis of cells and do not give any information on the kinetics of regulation. Furthermore, these studies have primarily focused on population statistics. Here, we use a sensitive fluorescence in situ hybridization (FISH) method that allows visualization of single-RNA molecules in individual cells. We obtain distributions of rDNA transcriptional activity in single cells and use these to gain insight into the kinetics of regulation and to investigate how the kinetics changes as the cell adapts to different conditions.
Results and discussion
To monitor rDNA transcription in single-yeast cells, a synthetic construct containing 32 tandem copies of a 43–base-pair probe-binding sequence was inserted downstream of the 35S promoter into one of the repeats in the rDNA array (Figure 1A). These cells were then fixed and hybridized with a single-stranded oligodeoxyribonucleotide probe, labeled with five tetramethylrhodamine (TMR) fluorescent molecules, which was complementary to the tandem repeated sequences. The binding of many fluorophores to each RNA molecule allows its detection as a diffraction-limited spot in a conventional wide-field fluorescence microscope (Femino et al, 1998; Raj et al, 2006). We observed cells with no or few RNA molecules and also cells with many RNAs and bright transcription sites (Figure 1B, top-right panel). These correspond to cells with silenced and active tagged repeats, respectively. The bright transcription sites observed are consistent with EM Miller spreads that show many polymerases transcribing from an active gene producing many RNAs.
Figure 1.
Genetic manipulation, methods and modeling. (A) A synthetic construct is inserted behind the 35S promoter in one of the repeats in the array to report the transcriptional activity of that repeat. Probes labeled with TMR fluorphores are designed to bind to the RNA transcribed from this repetitive synthetic construct. (B) (Top-left) Bright-field image of the fixed yeast cells. (Bottom-left) A cell segmentation program is used to identify individual cells. (Top-right) Fluorescence images of cells that had been fixed and hybridized with fluorescent probes. Some cells have bright transcription sites and the individual RNA cannot be resolved as single dots. Other cells have fewer RNA that can be resolved. (Refer to Supplementary Figure S8 for more information.) (Bottom-right) Image obtained after processing the TMR image on top-right. First, a program is used to identify cells with bright transcription sites whose RNAs cannot be resolved. These cells are labeled red. The remaining cells are outlined in green and an image processing program is used to count the number of RNAs in them. The individual RNAs are shown in green. (C) Gene expression is modeled by having an ON state of high transcription and an OFF state of no transcription. Gene activation is modeled by the parameter α, which is the transition rate from the OFF state to the ON state. Gene deactivation is modeled by the parameter γ, which is the transition rate from the ON state to the OFF state. μ is the rate of transcription when the gene is in the ON state. RNA is degraded at a rate δ. (D) Simulation of the steady-state distribution obtained for different switching rates.
For cells with few RNAs, the RNAs are spatially resolved as individual spots. However, for cells with RNA counts above 40 or cells with bright transcription sites, the RNA molecules are too close to each other to be resolved. Using the cells with low RNA counts, we obtained a linear relationship between the total fluorescence and number of RNAs (Supplementary Figure S1) and estimated the number of RNA molecules in bright cells by linear extrapolation from their total fluorescence. This process is automated by a custom-designed MATLAB image processing code. The RNA distribution obtained for wild-type cells is shown in Figure 2A. To determine whether genome position within the rDNA locus has an important function in regulating transcription activity, five different colonies obtained after plasmid integration were examined. As the construct integrated randomly along the array, these colonies have the construct integrated at different positions. The distributions of the five different integrants were similar (Supplementary Figure S2), indicating that position effect does not affect transcription significantly. Colony 1 was selected for further studies.
Figure 2.
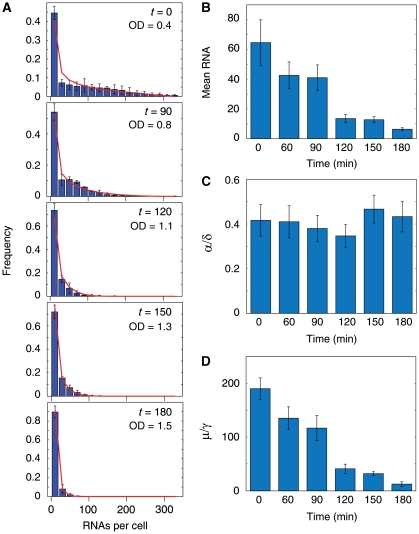
Transcriptional changes during diauxic shift. During diauxic shift, mean RNA production decreases. This decrease is due to a decrease in burst size. The gene activation rate remains constant. (A) RNA expression profiles of a single-rDNA repeat at t=0, 90, 120, 180 min after the culture reaches OD600=0.4 were determined using single-molecule FISH. Red lines are best fit from the burst model. Binsize is 20. (B–D) Mean RNA, normalized activation rates and burst sizes of the rDNA repeat at the different times. Error bars are standard errors from three independent experiments.
The experimentally determined distributions enable us to test dynamical models of transcription regulation and to determine their quantitative parameters. We assume that the burst model (Raj et al, 2006) holds for rDNA transcription. Each repeat can be either in the active state of high transcription or in the inactive rate of no transcription. This model is supported by Miller spread (French et al, 2003) data that shows that each rDNA gene is either silent or transcribing many RNA. This model is characterized by four parameters (Figure 1C). The activation rate α is the rate of switching from the inactive state to the active state. The inactivation rate γ is the rate of switching from the active state back to the inactive state and transcription rate μ refers to the rate of RNA synthesis in the active state. δ is the first-order rate constant of RNA decay. The three normalized rates, α/δ, γ/δ and μ/δ, characterize the steady-state distributions. Figure 1D displays calculated distributions obtained at different switching rates.
The model depicted in Figure 1C can be solved analytically (Raj et al, 2006) and was fit to the experimental steady-state distribution. Because of the long tail of the distributions, the normalized deactivation rate γ/δ and the normalized transcription rate μ/δ cannot be obtained independently. However, the normalized activation rate α/δ and the burst size μ/γ, which is the ratio of transcription rate over gene inactivation rate can be obtained accurately. The burst size μ/γ corresponds to the average number of RNA molecules that are transcribed in each gene activation event. We obtained an average of α/δ=(0.42±0.07) and μ/γ=(190±20). We determined the decay rate δ of the RNA directly in an independent experiment by using thiolutin, an inhibitor of RNA synthesis, and measuring the amount of RNA left at various times after transcriptional inhibition (Supplementary Figure S4). From these measurements, we obtained δ=(0.28±0.03) per min. This yields α=(0.12±0.02) per min, corresponding to an average time of (8±2) min for an inactive repeat to transit to the active state. Earlier, psoralens cross-linking results have shown that at log-phase in minimal medium, about 40% of the repeats are active (Dammann et al, 1995). Hence, an active repeat transcribes RNA for about 5 min before transiting back to the inactive state.
Next, we explored how rDNA synthesis responds to perturbations in external conditions. It is known that rRNA synthesis decreases in response to nutrient downshift (Sandmeier et al, 2002). To determine whether the decrease is due to a decrease in normalized activation rate α/δ or burst size μ/γ, a culture starting at OD600=0.4 was monitored for 180 min. We measured the RNA distribution of cells at t=0, 60, 90, 120, 150 and 180 min and fitted the distributions to the steady-state solution of the burst model. Because of fast decay rate of the RNA as compared with the rate of change of the other parameters, the distribution of RNA closely reflects that of the distribution at steady state. Hence, fitting the RNA distributions to the steady-state solution is a good approximation (see Materials and methods; Supplementary Figure S6). As expected, the mean RNA counts decreases as the cell density increases (Figure 2A and B). Using the model, we infer that the normalized activation rate α/δ remains fairly constant, whereas the burst size, μ/γ decreases (Figure 2C and D; Supplementary Table I). We measured the RNA half-life to be (3.4±0.8) min for the cells at t=180 min (Supplementary Figure S4). This decay rate is similar to the half-life (3.6±0.5) min measured at early log-phase, suggesting that activation rate α is constant throughout the 180 min of the experiment. Hence, the decrease in rRNA synthesis is due to the decrease in burst size. Earlier work has shown that the fraction of open repeats decreases during diauxic shift and that Rpd3, a histone deacetylase, is responsible for this decrease (Sandmeier et al, 2002). As our measurements show that the activation rate α remains constant, we deduced that Rpd3 decreases the fraction of open repeats by affecting the deactivation rate γ.
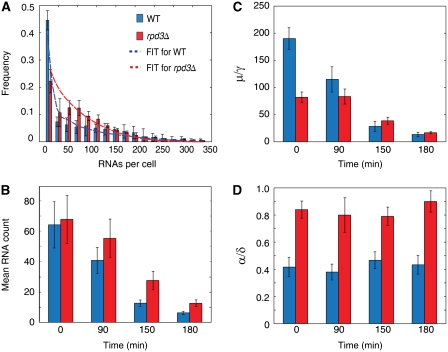
To further explore the role of Rpd3 in controlling rDNA transcription, we examined the effect of deleting Rpd3. Despite Rpd3's role in diauxic shift, little is known about its effect during log-phase. Indeed, it seems like Rpd3 does not affect rDNA transcription in the log-phase as both the fraction of open repeats and average rRNA concentration in wild-type and rpd3Δ strains are similar (Sandmeier et al, 2002). To determine its effect during the log-phase, rpd3Δ strains were grown to OD600=0.4 and fixed for hybridization. Consistent with earlier data, we observed that the mean of the rpd3Δ strain is similar to wild type (Figure 3B). However, the RNA distribution is different (Figure 3A). After analyzing the experimental distribution with the burst model, we find that the activation rate α in the rpd3Δ cells is twice that of wild type, and its burst size μ/γ is reduced by half (Figure 3C and D). The change in the activation rate α and burst size μ/γ in the rpd3Δ cells could not have been revealed by traditional biochemical methods, which measure population averages. We further measured the RNA distributions for the rpd3Δ strain at t=90, 150 and 180 min after it reached OD600=0.4. Unlike the increase in activation rate α, which is constant in the rpd3Δ strain at the different time points, the burst size μ/γ becomes more similar to that of wild type as cell density increases (Supplementary Figure S9A; Supplementary Table II).
Figure 3.
Effect of Rpd3 deletion. Although the rpd3Δ strain has similar mean RNA expression as wild type, its activation rate is double and its burst size is half that of wild type. (A) RNA expression profiles of a single-rDNA repeat in the rpd3Δ strain (red) and wild type (blue) at OD=0.4. Best fits from the model are shown in dash lines. Binsize is 20. (B–D) Mean RNA, burst sizes and normalized activation rates of the wild-type and rpd3Δ strains. Error bars are standard errors from two independent experiments.
Here, we used transcript counting to study the kinetics of rDNA transcriptional regulation. Our measured switching rate of the rDNA repeat is faster than reported switching rates of markers inserted into other regions that show gene silencing like the telomeres (Gottschling et al, 1990) and the mating locus (Xu et al, 2006). Our results suggest that each repeat spends about 5 min in the active state producing about 190 RNA molecules before switching off. This yields a Pol I reinitation interval of 300/190=1.6 s, which is comparable to the 1.2–2.2 s estimate obtained using EM Miller spreading (French et al, 2003). It has been suggested that active chromatin structure cannot be directly inherited as replication of transcriptionally active rDNA gene results in two newly replicated inactive coding regions packaged into nucleosomes (Lucchini and Sogo, 1995). Here, we suggest that not only the subset of active chromatin is not inherited but the set of active repeats is changing within one cell cycle. We ran a simulation to obtain the probability density for the amount of time that each repeat is open per cell cycle using different switching rates (Supplementary Figure S7). With the inferred switching rates, almost all the repeats will be active in each cell cycle. It has been shown that repair of active genes occurs faster than silenced genes (Conconi et al, 2002). Hence, one advantage of fast switching may be to allow damaged repeats to be repaired before it is inherited.
Population measurements performed on rpd3Δ and wild-type strains showed that the two strains have similar fractions of active genes and mean transcriptional activity during log-phase. However, our single-cell measurement shows that the activation rate doubles and the burst size is reduced by half in the rpd3Δ strain. As the fraction of open repeats in the rpd3Δ strain is reported to be similar to that of wild type (Sandmeier et al, 2002), we suspect that the deactivation rate would have also doubled. This would explain the reduction in burst size if transcription rate is unaffected by Rpd3. This suggests that Rpd3 functions to decrease both the activation and deactivation rates in log-phase. There are evidences showing that Rpd3 can function both as a transcriptional activator and deactivator (Deckert and Struhl, 2002; Sertil et al, 2007). If we assume that the transcription rate is unaffected by Rpd3, we can normalize the burst size in the wild-type strain to that of the rpd3Δ to obtain the ratio of their deactivation rates γrpd3Δ/γWT at different time points. This ratio becomes closer to 1 at later time points. This suggests that unlike the effect of Rpd3 on the activation rate α, which is independent of cell density, the effect of Rpd3 on the deactivation rate γ decreases with cell density. One hypothesis for how Rpd3 can regulate activation and deactivation rates independently is that Rpd3 uses different mechanisms to control them. For example, Rpd3 may decrease the activation rate α by deacetylating the H3 or H4 subunits of UAF, a transcription factor for Polymerase I recruitment (Keys et al, 1996) and increase the deactivation rate γ by affecting the nucleosomes surrounding the rDNA promoter. This suggests that in wild-type cells, Rpd3 does not inactivate ON repeats as cell density increases. Rather, Rpd3 actively acts to repress the deactivation rate γ at low cell density and stops repressing at high cell density, thus enabling the deactivation rate γ to increase (Supplementary Figure S9B). Future work is needed to elucidate the detailed molecular mechanism through which Rpd3 affects these rates.
Materials and methods
In situ probe
The probe used for the in situ hybridization is a DNA oligonucleotide synthesized on an Applied Biosystems (Foster City, CA) 394 DNA synthesizer using mild phosphoramidites (Glen Research, Sterling, VA). The oligonucleotide sequence is
5′-CGGCRGGTAAGGGRTTCCATARAAACTCCTRAGGCCACGA-3′;
The ‘R's represent locations where an amino-dT was introduced in the place of a regular dT. The oligonucleotides were synthesized on a controlled pore glass column (Glen Research) that introduced an additional amine group at the 3 end of the oligonucleotide. The probes' amine groups were then coupled to the TMR (Molecular Probes, Eugene, OR). The probes were purified on an HPLC column to isolate oligonucleotides displaying the highest degree of coupling of the fluorophore to the amine groups.
Strain construction
A plasmid with the promoter of 35S driving the synthetic repeat was linearized with HindIII, which cuts the plasmid within the promoter region. This plasmid was then integrated into haploid W303a by plasmid integration. Transformations were performed with different concentration of the plasmid using URA3 as the selectable marker. Colonies on the plate with the lowest plasmid concentration yielding successful transformants were selected.
To ensure that the RNA counts we obtained do not include partial degraded RNAs, a Northern blot was performed using the FISH probe sequence. As observed in Supplementary Figure S3A, the Northern blot yields a single band, showing that the transcripts detected by FISH are full length. We noticed that the length of the transcripts is about 3 kb, which is longer than the 2 kb reporter sequence in the plasmid. This is not surprising as Polymerase I transcriptional read-through of a polymerase II termination site has been reported earlier (Lo et al, 1998). The sequence downstream of the reporter sequence is part of the URA3 sequence in the plasmid downstream of the termination site.
To check that the reporter is integrated within the rDNA repeats, we performed a PCR using a primer binding to the AMP sequence on the plasmid and the other primer binding to a region of the 35S sequence. This PCR reaction detects a band of 2 kb, showing that the reporter is integrated within the rDNA repeats (Supplementary Figure S3B).
For confirming that a single plasmid has been integrated, quantitative PCR was performed using primers against URA3 and ACT1. W303a has a single copy of URA3 gene. Integration of the plasmid leads to additional copies of URA3 being integrated into the genome of W303a. This will lead to a reduction in the number of cycles needed to generate similar amount of the URA3 PCR product. We compared this cycle difference in the strains before and after plasmid integration. A change in cycle number of −0.9±0.3 is obtained after plasmid integration, showing that one copy of the plasmid has been inserted (Supplementary Figure S3C). ACT1 was used as a loading control.
To generate the rpd3Δ strain, primers with 50 bp homology to the beginning and end of the coding sequence of Rpd3 were used to PCR out a sequence containing the LEU2-coding sequence. This PCR product was then transformed into the wild-type strain and plated on plates lacking leucine. Colonies selected are those with the LEU2 replacing the RPD3 gene and were confirmed by PCR.
Fluorescence in situ hybridization
Yeast cells (haploid W303a) were grown in CSM-URA minimal media at 30°C to an OD600 of 0.4 (for log-phase experiments). Cells were fixed by adding formaldehyde to a final concentration of 4% (v/v) for 45 min at room temperature (23±2°C). The cell wall was digested with lyticase at 30°C for 20 min. After that, cells were stored in ethanol at 4°C. Before hybridization, cells were rehydrated in 30% (v/v) formamide, 2 × SSC for 10 min. Cells were hybridized in 30 μl of hybridization solution containing labeled DNA probes in 25% (v/v) formamide, 2 × SSC, 1 mg/ml BSA, 10 mM VRC, 0.5 mg/ml Escherichia coli tRNA and 0.1 g/ml dextran sulfate overnight at room temperature. Before imaging, cells were washed twice in 30% (v/v) formamide, 2 × SSC for 30 min. DAPI was added to the first washing step. The cells were then added to multichambered coverglass (Lab-Tek, Nalge Nunc, Rochester, NY) coated with concanavalain A (0.1 mg/ml, Sigma) for 10 min. Cells that did not stick to the coverglass were washed off.
Image acquisition
Images were taken with a Nikon TE2000 inverted fluorescence microscope equipped with a 100 × oil-immersion objective and a Princeton Instruments camera using MetaMorph software (Molecular Devices, Downington, PA). The TMR filter was obtained from Chroma (Rockingham, VT). Exposure time for TMR was 1 s. Stacks of images were taken automatically with 0.2 microns between the z-slices.
Image analysis
To segment the yeast cells, a watershed algorithm was used. Cell boundaries were obtained by running an edge detection algorithm on the bright-field image of the cells. After that, individual cell nuclei were identified in the DAPI images by (1) selecting pixels above a certain threshold, (2) connecting these pixels that are close to each other and (3) rejecting connected regions that are larger or smaller than certain thresholds. These nuclei serve as markers for running a marker-controlled watershed algorithm over the bright-field image. Next, a program was run to count the number of RNA molecules in each cell. The general procedure was to run a median filter followed by a Laplacian filter on each optical slice taken. This enhanced particulate signals. A threshold was then selected to pick-up individual spots in each plane. Next, the program counted the total number of isolated signals (i.e. connected components) in three dimensions. The particle count was robust for a range of threshold chosen. This particle count was accurate for cells with low number of RNA. For cells with RNA counts larger than 40 molecules, the RNAs were too close and could not be resolved as individual dots. To identify these cells, a program was run to determine cells with large connected regions of high fluorescence intensity. These regions consisted of the many individual RNAs that could not be resolved as individual dots. Next, the total cell fluorescence was obtained by summing up the fluorescence in each plane. By fitting the total cell fluorescence and the number of RNAs in cells with RNA counts smaller than 40 molecules, a linear relationship was obtained between cell fluorescence and RNA count. This relation was extrapolated to determine the number of RNAs in cells with high total cell fluorescence.
Fitting of parameters to experimental distribution
To determine the parameters of our model, we fit our experimental data to the steady-state solution to the master equation of the two-state model using a maximum likelihood method. We found that the likelihood value is similar for similar ratios of μ/β. This indicates that there was insufficient experimental data to resolve the two parameters. The error bars refer to 95% confidence interval. For more information on the details of the two-state models, please refer to Raj et al (2006).
Determination of RNA decay rate
A measure of 8 μg/ml of Thiolutin, an inhibitor of RNA synthesis, was added to the culture at time t=0 min. Thiolutin is a metabolite produced by Streptomyces luteroeticuli that inhibits all yeast RNA polymerases, mainly at the level of transcriptional initation. Cells were collected at t=0, 1, 2, 3, 4, 5, 10, 15, 20 and 40 min after thiolutin addition. The mean RNA count at each time point was measured using FISH and fitted to the equation m=m0e−dt+b, where m(t) is the mean RNA at different times after thiolutin addition. m0 + b is the initial amount of RNA before thiolutin addition. d is the rate constant for RNA decay and b is the amount of RNA transcription after thiolutin addition. The best fit was determined by the least square method with m0, d and b as free parameters. To determine the error in the fit, we randomized the RNA count at each time points by drawing random number from normal distributions with the measured mRNA and standard derivation for that time point. This set of RNA obtained from the randomization was fitted to the decay equation using the least square fit. This process was repeated 100 times and the standard derivation obtained from these 100 sets of parameters was used as the estimate of the error in the parameters.
We compared the decay rate measured using thiolutin inhibition to that measured using temperature inhibition in a cbf5-1 temperature sensitive strain (Cadwell et al, 1997). In the cbf5-1 strain, rDNA synthesis is inhibited at the non-permissive temperature of 38°C. To measure decay rate by thermal inactivation, the cbf5-1 strain was first grown to OD600=0.6 at 27°C, spun down and resuspended in an equal amount of medium preheated to 38°C. The cells were then removed and fixed at t=1, 2, 3, 4, 6, 10 and 20 min after transfer.
The half-lives measured using thermal inactivation and thiolutin inhibition are (1.5±0.2) min and (1.9±0.3) min, respectively (Supplementary Figure S5). The two half-lives are similar. The slight difference in the decay rate measured may be due to the fact that enzymes for decaying the RNA may be more reactive at 38°C as compared with 27°C. It is also interesting to note that the decay rate of the reporter construct is faster in this strain as compared with our experimental strain. This discrepancy may be due to the differences in strain backgrounds and the fact that our strain is a haploid whereas the cbf5-1 strain is a diploid.
Fitting diauxic shift experiments to steady-state distributions of the two-state model
For the diauxic shift experiments, cells were taken out of a culture at t=0, 60, 90, 120, 150 and 180 min after the culture reached OD600=0.4. The cells were then fixed and hybridized for imaging.
During diauxic shift, the mean mRNA decreases rapidly. To check whether the steady-state assumption is valid, we ran a simulation modeling the decrease in RNA expression and fitting the RNA distributions obtained to the steady-state equation.
To simulate the RNA decrease, we estimated the activation rates and burst sizes from the steady-state fits obtained for the experimental RNA distributions at t=0, 60, 90, 120, 150 and 180 min during the diauxic shift. As the activation rate remains relatively constant, we used a constant activation rate for the simulation. For the burst size, we assumed that it is piece-wise linear. Using these estimates for the activation rate and burst size, we were able to construct the time-dependent master equation matrix and run a discrete time Markov process to model the RNA distribution for the 180 min duration of the experiment. To fit the simulated RNA distribution at t=60, 90, 120, 150 and 180 min, 1000 cells were randomly drawn with RNA numbers reflecting the RNA distribution at that time point. The maximum likelihood method was then used to obtain the best fits for the distribution. This was repeated three times to obtain the average activation rate and burst size.
As seen in Supplementary Figure S6, the activation rates and burst sizes obtained from the steady-state fits are similar to the actual activation rates and burst sizes at each of the time points. This shows that the steady-state approximation can be used to fit the distributions.
Supplementary Material
Acknowledgments
We thank J Gore, G Neuert, B Pando, LF Cheow, M O'Kelly and the van Oudenaarden laboratory for discussions. We are grateful to A Raj for providing the TMR probes. RZ Tan was supported by A*STAR, Singapore. This work was supported by National Science Foundation grant NSF-0548484 and US National Institutes of Health grant R01-GM068957.
Footnotes
The authors declare that they have no conflict of interest.
References
- Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J (1997) The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol 17: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A, Bespalov VA, Smerdon MJ (2002) Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc Natl Acad Sci USA 99: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A, Widmer RM, Koller T, Sogo JM (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761 [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM (1995) Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol 15: 5294–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM (1993) Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res 21: 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Struhl K (2002) Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol Cell Biol 22: 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280: 585–590 [DOI] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL (2003) In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ (2007) RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol 14: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev 10: 887–903 [DOI] [PubMed] [Google Scholar]
- Lo HJ, Huang HK, Donahue TF (1998) RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol Cell Biol 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R, Sogo JM (1995) Replication of transcriptionally active chromatin. Nature 374: 276–280 [DOI] [PubMed] [Google Scholar]
- Miller OL Jr, Beatty BR (1969) Visualization of nucleolar genes. Science 164: 955–957 [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS (2002) Rpd3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 21: 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertil O, Vemula A, Salmon SL, Morse RH, Lowry CV (2007) Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol Cell Biol 27: 2037–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR (2006) Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell 23: 219–229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.