Abstract
Sustainable biofuel alternatives to fossil fuel energy are hampered by recalcitrance and toxicity of biomass substrates to microbial biocatalysts. To address this issue, we present a culture-independent functional metagenomic platform for mining Nature's vast enzymatic reservoir and show its relevance to biomass conversion. We performed functional selections on 4.7 Gb of metagenomic fosmid libraries and show that genetic elements conferring tolerance toward seven important biomass inhibitors can be identified. We select two metagenomic fosmids that improve the growth of Escherichia coli by 5.7- and 6.9-fold in the presence of inhibitory concentrations of syringaldehyde and 2-furoic acid, respectively, and identify the individual genes responsible for these tolerance phenotypes. Finally, we combine the individual genes to create a three-gene construct that confers tolerance to mixtures of these important biomass inhibitors. This platform presents a route for expanding the repertoire of genetic elements available to synthetic biology and provides a starting point for efforts to engineer robust strains for biofuel generation.
Keywords: biofuels, biomass inhibitor tolerance, functional metagenomics, strain engineering, synthetic biology
Introduction
Global environmental problems related to the combustion of fossil fuels and increasing concerns about their supply underscore the importance of developing renewable fuel alternatives with a reduced environmental footprint. The application of synthetic biology (Baker et al, 2006; Ro et al, 2006) to engineer biocatalysts that produce biofuels from diverse lignocellulosic materials including waste and low agricultural intensity biomass holds promise to deliver one such sustainable alternative (Farrell et al, 2006; Tilman et al, 2006; Fargione et al, 2008; Searchinger et al, 2008). However, bioconversion of lignocellulose to biofuels is currently limited by biomass recalcitrance (Himmel et al, 2007) and toxicity of non-fermentable compounds in the original substrate and formed as byproducts of biomass pretreatment (Klinke et al, 2004). Although the identity and inhibitory concentrations of these compounds have been characterized, their mechanisms of toxicity are poorly understood, and genes conferring tolerance to most of these compounds have not been identified. A synthetic biology approach to design efficient biocatalysts for biofuel generation requires a diverse inventory of functional genetic machinery allowing usage of or conferring tolerance toward these compounds.
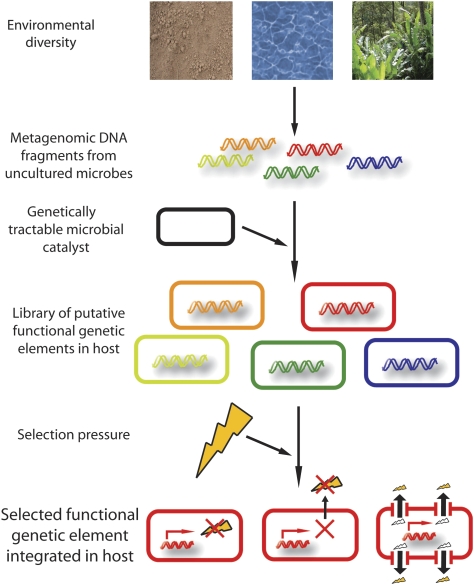
As plant biomass is constantly recycled in the environment (Kirk and Farrell, 1987), a reservoir of enzymatic machinery must exist in the soil microbiome that allows for the tolerance and complete processing of its constituent chemicals. However, the majority of this microbial processing machinery has remained inaccessible to synthetic biology and metabolic engineering, as a majority of microbes in the soil are recalcitrant to culturing (Torsvik et al, 1998). We show the utility of culture-independent metagenomic functional selections for discovery of novel functional genes from the soil microbiome, enabling expansion of the synthetic biology toolbox for lignocellulosic biomass conversion and tolerance. The key steps of this method involve extraction of metagenomic DNA from arbitrary environmental sources (Rondon et al, 2000), transformation of environmental metagenomic libraries into the microbial host of interest, and selection of functional genetic elements conferring the desired phenotype compatible with the chosen host (Figure 1). This method is well suited for biomass catalysis, as the functional genes that allow the host to use recalcitrant substrates or tolerate toxic chemicals can be directly selected from arbitrary metagenomic libraries.
Figure 1.
Functional metagenomic platform for discovery of novel functional genetic elements from diverse environmental microbiomes. Shown is a schematic detailing the key steps required for selecting functional genetic elements from diverse environments that confer a desired selective advantage to a microbial catalyst. Metagenomic DNA is directly extracted from arbitrary environmental samples without earlier culturing steps, purified, and transformed into a microbial host of interest. The entire library of putative functional genetic elements is subjected to a selection pressure (e.g. chemicals at inhibitory concentrations or recalcitrant substrates) that only allows survival of hosts containing functional genetic elements, which counteract the selection pressure (e.g. by allowing usage of the recalcitrant substrates or by conferring tolerance by intracellular or extracellular inactivation or efflux of the inhibitory compound). This scheme is ideally suited for discovery of novel functional genetic elements for biomass conversion to biofuels.
Results and discussion
Engineered Escherichia coli strains have been shown to harbor many advantages as biocatalysts for biofuel production, including the ability to ferment a majority of plant-derived monosaccharides, no requirements for complex growth factors, and earlier industrial use, but still suffer from lower tolerance to biomass inhibitors when compared with other candidate biofuel producing organisms (Dien et al, 2003). Therefore, as a proof of concept, we applied metagenomic functional selections to select a number of functional genetic elements from diverse soil microbiomes that confer resistance in E. coli to different classes of biomass inhibitors. We extracted metagenomic DNA from four different soil microbiomes (Table I), with an optimized protocol to carefully purify high molecular weight DNA (Materials and methods; Supplementary methods). We chose to create large-insert (40–50 kb) libraries to allow for the potential discovery of phenotypes requiring multiple genes. Four metagenomic libraries of sizes ranging from 0.2 to 2.5 Gb were created in a single-copy fosmid vector, and transferred into an E. coli host using phage transduction (Table I; Materials and methods). The concentrations of seven important biomass chemicals that inhibit the growth of the wild-type E. coli host were determined using growth assays on Luria Broth (LB) agar media with sparse concentration range screening (Supplementary Table I) based on previously published results (Zaldivar and Ingram, 1999; Zaldivar et al, 1999, 2000). These chemicals span the three major classes of biomass inhibitors (organic acids, alcohols, and aldehydes), which accumulate during the pretreatment of biomass and agricultural and municipal waste, resulting in the inhibition of microbial biofuel fermentation (Klinke et al, 2004). Selection of the four metagenomic libraries on solid media containing each of the seven biomass compounds at their determined inhibitory concentrations yielded metagenomic library clones that survived in 15 of the 28 combinations (Table I). Tolerant clones were identified against all seven inhibitors. Methylcatechol was the only compound for which tolerant clones were identified from all four soil libraries. Hydroquinone and furfural tolerant clones were identified in one library. Interestingly, the medium-sized (1 Gb) dairy farm soil library yielded clones tolerant to the most inhibitors (six out of seven), whereas the pH 5.5 bog soil library with the largest size (2.5 Gb) yielded clones tolerant to two of the seven inhibitors. These phenotypic results show the utility of metagenomic functional selections for transferring lignocellulosic inhibitor tolerance phenotypes to E. coli. As these phenotypes are encoded on large DNA inserts, it is important to identify which and how many specific genetic elements within these operon-sized stretches are responsible for these phenotypes. As a proof of concept, we chose to focus on a subset of clones for more in-depth analysis of growth kinetics in the presence of inhibitors, sequencing of the full-length inserts, and sub-cloning of functionally annotated genes.
Table 1. Soil metagenomic libraries from which inserts conferring tolerance in E. coli to seven lignocellulosic compounds were selected.
| Library # | Source | Library size (Mb) | Metagenomic inserts selected encoding resistance to lignocellulosic inhibitors | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hydroquinone | 4-methylcatechol | 4-hydroxy- benzaldehyde | Syringaldehyde | 2-furoic acid | Furfural | Ethanol | |||
| 1 | pH 4.5 bog soil (NH) | 200 | X | X | X | X | |||
| 2 | pH 5.5 bog soil (NH) | 2500 | X | X | |||||
| 3 | Urban park soil (MA) | 1000 | X | X | X | ||||
| 4 | Dairy farm soil (MA) | 1000 | X | X | X | X | X | X | |
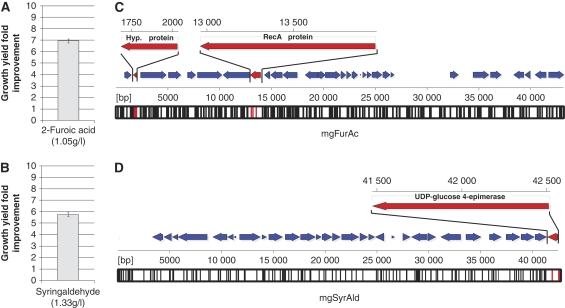
Inhibitory chemicals are derived from two major sources during biomass pretreatment—depolymerization of complex lignin polymers and degradation of biomass sugars (Klinke et al, 2004). Accordingly, we chose clones conferring tolerance to one lignin monomer, syringaldehyde, and one biomass sugar degradation product, 2-furoic acid, for further phenotypic and genetic analysis. For each inhibitor, 20 metagenomic clones with improved tolerance on solid selection media (Supplementary Table I) were retested in liquid culture at three concentrations spanning the range of previously reported inhibitory concentrations (Materials and methods), and for all clones improved phenotype was confirmed (Supplementary Figure 1). From this set, one clone for syringaldehyde (mgSyrAld) and one clone for 2-furoic acid (mgFurAc), with the greatest difference in cell growth when compared with control at 1.55 g/l syringaldehyde and 0.8 g/l 2-furoic acid, respectively, were selected for analysis of growth kinetics (Materials and methods). Metagenomic inserts from these clones were extracted and retransformed into wild-type E. coli to confirm that the improved phenotype was due to the presence of the metagenomic insert (Materials and methods). The phenotypic improvements were 5.7-fold for syringaldehyde and 6.9-fold for 2-furoic acid, expressed as fold improvements in cell growth at an inhibitor concentration that results in a 90% reduction of wild-type E. coli cell growth (Figure 2A and B).
Figure 2.
Sequence annotation and functional analysis of selected genetic elements improving biomass inhibitor tolerance in E. coli. (A, B) Improvements in inhibitor tolerance toward 2-furoic acid and syringaldehyde because of metagenomic inserts. Inhibitor concentrations resulting in 90% reductions in growth yield were determined for wild-type E. coli as 1.05 g/l for 2-furoic acid and 1.33 g/l for syringaldehyde. Improvements in E. coli growth yield at these concentrations because of metagenomic inserts were 6.9-fold for 2-furoic acid and 5.7-fold for syringaldehyde, showed here as the mean (and standard deviation) of triplicate readings after 24 h of growth. (C, D) The metagenomic inserts conferring tolerance to 2-furoic acid (mgFurAc) and syringaldehyde (mgSyrAld) were sequenced at 3 × coverage and annotated (Supplementary Tables II and III). Annotated genes for (C) mgFurAc and (D) mgSyrAld are shown as filled arrows, with the orientation denoting the relative direction of transcription based on an arbitrary sense strand. Transposon mutagenesis, followed by reselection of the tolerance phenotypes, was used to identify functional genetic elements in mgFurAc and mgSyrAld that contribute to the selected phenotypes (genes colored red and labeled) (Supplementary information). Vertical bars along the bottom of each sequence–position axis denote positions of transposon insertion in the loss-of-function study (black denotes no effect, red denotes loss-of-function).
The mgSyrAld and mgFurAc metagenomic inserts were sequenced at three-fold coverage, assembled, and annotated (Figure 2C and D) (Materials and methods; Supplementary Tables II and III). Regions of the metagenomic sequences with the highest detectable homology to the NCBI non-redundant nucleotide database using BLAST (Altschul et al, 1990) are 7% of mgFurAc with 79% identity to a region of the Pelobacter propionicus DSM 2379 genome and 1% of mgSyrAld with 73% identity to a region of the Burkholderia ambifaria AMMD chromosome 2, indicating that the selected metagenomic sequences are largely novel. Based solely on the sequence and computational annotation of the inserts, it is difficult to predict which genes are responsible for the improved tolerance especially as the mechanism of toxicity is poorly characterized for these compounds. Therefore, we performed a loss-of-function study with mgSyrAld and mgFurAc using transposon mutagenesis to identify the functional genetic elements contributing to the selected phenotypes (Figure 2C and D) (Materials and methods). The 192 transposon-inserted clones per inhibitor created for sequencing of the mgSyrAld and mgFurAc fosmids were individually subjected to kinetic growth survival assays in the presence of 1.4 g/l syringaldehyde and 0.8 g/l 2-furoic acid, respectively.
For mgSyrAld, 3 of the 192 unique transposon insertions resulted in a knockdown of the improved syringaldehyde tolerance, all mapping to either the promoter or the N-terminal coding region of a 348 amino acid gene product annotated to be a UDP-glucose-4-epimerase (Figure 2D) (Materials and methods). For mgFurAc, 7 of the 192 unique transposon insertions resulted in a knockdown of the improved 2-furoic acid tolerance, with three hits mapping to the coding region of a 342 amino acid gene product annotated to be a RecA protein, and four hits mapping to a 111 amino acid gene product with predicted membrane-spanning domains but of unknown function (Figure 2C) (Materials and methods; Supplementary information). Interestingly, these two genes are more than 10 kb apart in the mgFurAc metagenomic fragment, and none of the annotated genes between these locations appear to contribute to the selected phenotype based on the transposon mutagenesis results. Although the mechanism of inhibition in E. coli by syringaldehyde and 2-furoic acid is unknown, the gene hits identified in our selection may provide starting points for discovery of the underlying modes of inhibition and rescue (Supplementary information).
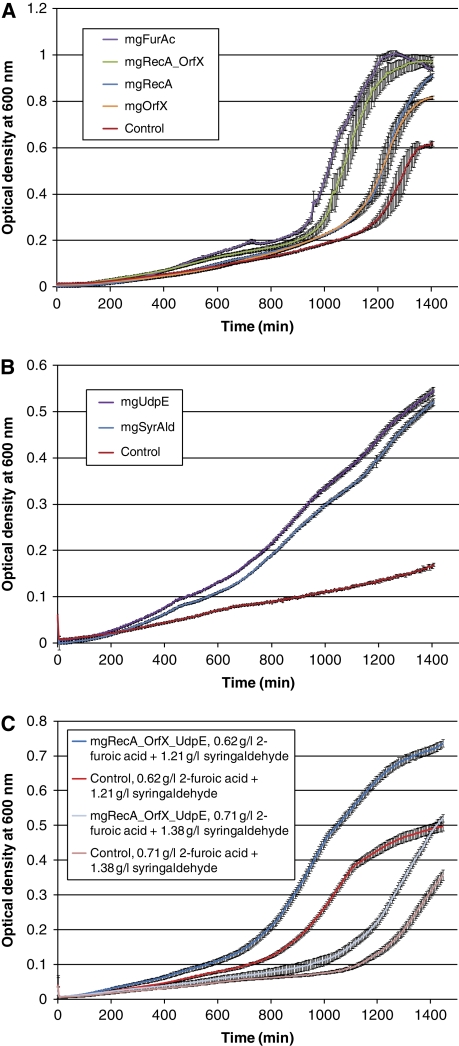
To verify that the three genes implicated in the loss-of-function studies were necessary and sufficient for the syringaldehyde and 2-furoic acid tolerance phenotypes, we PCR amplified each gene from the corresponding selected metagenomic fosmid (mgUdpE from mgSyrAld, and mgRecA and mgOrfX from mgFurAc), sub-cloned them into an expression vector, transformed these into wild-type E. coli, and repeated the tolerance assays (Materials and methods; Supplementary information). In all three cases, the individual genes exhibited improved tolerance to the inhibitors when compared with wild-type E. coli (Figure 3). The improved syringaldehyde tolerance because of the mgUdpE gene was very similar to mgSyrAld (Figure 3B). In contrast, both mgRecA and mgOrfX were individually unable to completely recapitulate the level of mgFurAc tolerance (Figure 3A), which might be expected if both genes are required for the full observed phenotype of mgFurAc. To test this hypothesis, we created a bicistronic construct (mgRecA_mgOrfX) by sub-cloning the mgOrfX gene and its upstream ribosome-binding site (RBS) between the mgRecA gene and the transcription terminator site. The bicistronic mgRecA_mgOrfX construct exhibited improved 2-furoic acid tolerance when compared with both individual genes, and more closely resembled the growth behavior of mgFurAc (Figure 3A). This shows the utility of metagenomic selections using large-insert metagenomic libraries to identify multiple genetic elements, which can together contribute to improved phenotypes despite not being immediately co-localized in gene sequence. For instance, a metagenomic library smaller than 10 kb would have been unable to capture the improved phenotype derived from the action of both genes.
Figure 3.
Inhibitor tolerance phenotypes encoded by sub-cloned metagenomic genes for (A) 0.8 g/l 2-furoic acid, (B) 1.4 g/l syringaldehyde, and (C) mixtures of 2-furoic acid and syringaldehyde; 24 h kinetic growth curves are shown for E.coli clones harboring different combinations of sub-cloned metagenomic tolerance genes. Error bars represent standard deviation from triplicate kinetic readings. (A) For 2-furoic acid, the constructs containing the individual genes mgOrfX and mgRecA recapitulate only part of the tolerance phenotype; however, a bicistronic construct (mgRecA_mgOrfX) fully recapitulates the phenotype of the full-length selected fosmid (mgFurAc), showing that the two genes are necessary and sufficient for conferring the 2-furoic acid tolerance phenotype. (B) For syringaldehyde, the sub-cloned metagenomic UDP glucose-4-epimerase gene (mgUdpE) fully recapitulates the phenotype of the full-length selected fosmid (mgSyrAld). (C) A tri-cistronic construct (mgRecA_mgOrfX_mgUdpE) enables improved tolerance to mixtures of 2-furoic acid and syringaldehyde showing that the identified tolerance genes can be combined to generate multifunctional constructs.
One of the goals of synthetic biology is to improve microbial phenotypes by integrating multiple functional genetic elements from arbitrary genetic or engineered sources. To test whether the selected genes that confer tolerance to E. coli when exposed to the biomass inhibitors syringaldehyde and 2-furoic acid individually could also confer tolerance to a mixture of these inhibitors, we created a tri-cistronic construct (mgRecA_mgOrfX_mgUdpE) by sub-cloning the mgUdpE gene and its upstream RBS just downstream of the bicistronic mgRecA_mgOrfX construct. The tri-cistronic construct exhibited improved growth phenotypes in the presence of mixtures of syringaldehyde and 2-furoic acid (Figure 3C; Supplementary Figure S4) showing that genes selected using this metagenomic selection platform against individual inhibitors can be combined to create constructs that confer tolerance toward inhibitor mixtures.
Strain optimization has previously been achieved through adaptive evolution (Yomano et al, 1998; Herring et al, 2006; Liu, 2006) where rare beneficial genomic mutations can be selected without earlier knowledge about the mode of inhibition. Adaptive evolution is ideally suited for optimization of the genomic inventory of functions in a given strain, but the timescale for evolving entirely new functions is generally prohibitive, as they typically require a substantial number of specific mutations. One of the strengths of metagenomic functional selections for strain improvement is that its success in discovering functional genetic elements is similarly independent of earlier knowledge regarding the mode of inhibition of selected chemicals, while sampling a large reservoir of novel genes from the metagenome.
The genetic machinery that we can evolve, select, and engineer into microbial biocatalysts for tolerating or degrading biomass inhibitory chemicals can be considered mechanistically analogous to genetic machinery encoding microbial antibiotic resistance. Both biomass-derived inhibitors and most antibiotic chemicals are produced naturally in the environment (Walsh, 2003), and microbial communities have likely evolved similar biochemistries to tolerate and process these xenobiotics. Mechanisms of antibiotic resistance include (a) adaptive genomic mutation that obscure the cellular target of the drug without compromising the native cellular function, (b) acquired mechanisms to degrade or expel the drug, often gained through horizontal transfer through plasmid exchange, and (c) adjusting expression in other pathways to increase target production or bypass it with redundant functions (Davies, 1994; Walsh, 2003). Metagenomic functional selections for biomass tolerance and conversion are likely to uncover genetic machinery that parallels the second and third mechanisms. Indeed, we hypothesize that the metagenomically selected proteins homologous to RecA and UDP-glucose-4-epimerase encoding resistance to 2-furoic acid and syringaldehyde, respectively, may function to complement or rescue the activity of their putatively compromised native cellular counterparts (Supplementary information). In future applications of this methodology, mechanisms to enzymatically metabolize biomass inhibitors may have enhanced utility because they would increase the nutrient yield and net carbon flux, though these mechanisms would be undesirable when the intended tolerance phenotype is against the biofuel product (e.g. ethanol).
The screening of metagenomic clone libraries from diverse environmental sources has previously yielded numerous biomolecules including novel proteases, amylases, cellulases, and antibiotics (Brady and Clardy, 2000; Rondon et al, 2000; Daniel, 2005; Warnecke et al, 2007), and the yields of these methods appear primarily limited by the number of clones that can feasibly be screened (Daniel, 2005). In comparison, a library-wide selection scheme allows for exhaustive interrogation of the enzymatic reservoir encoded within metagenomic libraries that can be made using current techniques (⩽5 × 1010 bp) (Riesenfeld et al, 2004; Daniel, 2005). Functional selections have been designed for hundreds of anabolic, catabolic, and resistance phenotypes in E. coli (Neidhardt et al, 1996; Sommer et al, 2009), and opportunities exist for the design of selections for other biotechnologically relevant phenotypes including controlled flocculation, surface adhesion, natural competency, and cell–cell communication (Williamson et al, 2005).
We have shown that metagenomic functional selections can successfully discover functional genetic elements encoding chemical tolerance relevant to biomass conversion. The same platform can be applied to select for microbial usage and production of specific biomass chemicals. The repertoire of biomass substrates that can be used by a microbial biocatalyst has been expanded by transfer of specific genetic machinery for substrate metabolism from other microbes with these properties (Jin et al, 2005). Similarly, substrate usage machinery may be selected from a metagenomic clone library by providing the substrate as the sole source of a required nutrient (e.g. carbon, nitrogen), only allowing clones expressing functional genes enabling substrate usage to grow selectively. Functional selections for chemical production can be achieved in a biocatalyst metagenomic clone library that contains a biochemical circuit that links the presence of the desired product to a selectable resistance or usage phenotype. For instance, a circuit can be designed where a transcription factor responsive to the stoichiometric presence or absence of the product controls the expression of an antibiotic resistance gene (Desai and Gallivan, 2004).
A distinguishing feature of synthetic biology is the emphasis on integrating arbitrary genetic elements to generate robust and predictable biocatalysts to solve multiple biological, chemical, and engineering problems including fuel generation, environmental remediation, and pharmaceutical production (Baker et al, 2006). Our work shows that metagenomic functional selections enable the direct discovery of novel genetic elements from Nature's enzymatic catalogue, providing a route for expanding the synthetic biology tool box.
Materials and methods
Environmental metagenomic DNA library construction
Soil samples (200–500 g) were collected from urban parks (MA), farm land (MA), and bogs (NH). Metagenomic DNA was extracted from 10 g of soil using the PowerMax Soil DNA Isolation kit (Mobio Laboratories Inc.), using a modified protocol (Supplementary information). High molecular weight (40–50 kb) DNA was size selected and purified using a pulse-field gel apparatus (Supplementary information). Size-selected gel-purified DNA was blunt-end repaired using the End-It DNA End-Repair kit (Epicentre Biotechnologies), purified by two serial phenol/chloroform extractions, and concentrated by ethanol precipitation (Supplementary information). Libraries of purified end-repaired 40–50 kb metagenomic DNA in E. coli were created using the CopyControl Fosmid Library Production kit (Epicentre Biotechnologies). For each library, ∼250 ng of DNA was ligated to 0.5 μg of the linearized fosmid pCC1FOS vector, packaged using replication-deficient phage extract, infected into E. coli strain EPI-300, and library size determined by dilution titering on LB agar plates containing 12.5 μg/ml chloramphenicol. E. coli-infected metagenomic DNA libraries were grown to mid-log phase in 10 ml LB–12.5 μg/ml chloramphenicol, and frozen down at −80°C in 1 ml aliquots in 15% glycerol. Each frozen stock was subsequently confirmed to have ∼1–5 × 108 colony forming units per ml.On the basis of the determined library sizes, each library aliquot saved contained over 100 cell copies per individual 40–50 kb metagenomic DNA fosmid library clone.
Selection of functional genetic elements from metagenomic libraries
Growth selections were performed on metagenomic libraries from four different soils covering ∼4.7 Gb of genetic material at the determined inhibitory concentrations of seven biomass inhibitors (Table I; Supplementary Table I; Supplementary information). On the basis of the determined library sizes and titers of the frozen library stocks, inocula were prepared to yield ∼100 cell copies of each metagenomic DNA library clone per selection (e.g. 2 × 106 cells were plated out from a library originally assayed to contain 2 × 104 clones). Cells were spread on LB agar plates containing 12.5 μg/ml chloramphenicol and the inhibitor at the selective concentration (Supplementary Table I). Growth of colonies was assayed after 48 h of growth at 37°C.
In all, 40 metagenomic DNA library clones, each conferring tolerance to syringaldehyde or 2-furoic acid, were chosen for further analysis of encoded functional parts from the selected inhibitor plates. For 2-furoic acid, six to seven tolerant clones each were chosen from selected libraries 1, 2, and 3 (Table I). For syringaldehyde, 10 tolerant clones each were chosen from selected libraries 3 and 4 (Table I). Each colony was grown to saturation (16–18 h) at 37°C with shaking in liquid LB medium containing 12.5 μg/ml chloramphenicol (hereon referred to as LB-chlor). Saturated cultures were diluted 1:40 in fresh LB-chlor and grown to mid-log phase (1–2 h) at 37°C with shaking. Log-phase cultures were inoculated (1:40) into LB-chlor containing one of three concentrations of the relevant inhibitor (2-furoic acid: 0.8, 7.9, and 15 g/l; syringaldehyde: 1.55, 1.775, and 2 g/l), with concentrations chosen to sparsely span the range of previously reported inhibitory concentrations of these compounds (Zaldivar and Ingram, 1999; Zaldivar et al, 1999). Cell growth after 24 h at 37°C with shaking was determined by end-point turbidity measurements at 600 nm using a Versamax microplate reader (Molecular Devices) (Supplementary Figure 1).
One metagenomic clone each, with the greatest difference in cell growth when compared with control for syringaldehyde (mgSyrAld) and 2-furoic acid (mgFurAc) (Supplementary Figure I) was chosen for further analysis. To confirm that the observed tolerance in the selected clones was a result of the selected metagenomic DNA, fosmids from the metagenomic clones were extracted using the FosmidMAX DNA Purification kit (Epicentre Biotechnologies). Purified fosmids were then retransformed into an electrocompetent version of the same control E. coli strain using a standard electroporation protocol. The retransformed clones recapitulated the improved inhibitor tolerance compared with the E. coli control. Growth kinetics for these clones were measured at 11 concentrations per inhibitor, evenly spanning the concentration ranges 0–1.5 g/l for 2-furoic acid and 0–3 g/l for syringaldehyde. Kinetic measurements were carried out in triplicate for each metagenomic clone and E. coli control by turbidity measurements at 600 nm every 5 min over 24 h at 37°C with shaking in a Versamax microplate reader. Inhibitor concentrations resulting in 90% reductions in cell growth after 24 h of growth at 37°C were determined for the control E. coli as 1.05 g/l for 2-furoic acid and 1.33 g/l for syringaldehyde (Supplementary Figure 2).
Sequencing of metagenomic inserts
The metagenomic inserts from mgFurAc and mgSyrAld were chosen for DNA sequencing and analysis. Sequencing clone libraries were created by in vitro insertion of a transposon carrying unique sequencing primer sites and a kanamycin resistance cassette into random positions in the purified metagenomic DNA fosmids, followed by transformation into the control E. coli strain, using the EZ-Tn5 <KAN-2> Insertion kit (Epicentre Biotechnologies). In all, 192 single transposon-inserted clones per fosmid were sequenced bi-directionally to yield approximately three-fold sequence coverage of the ∼40 kb inserts. Sequences were assembled into contigs using Phred/Phrap (University of Washington; http://www.phrap.org/phredphrapconsed.html). Each assembly yielded 2–5 contigs. Primers were designed to close gaps between contigs and sequences resulting from this additional round of primer walking yielded sufficient sequence information for complete assembly of single full-length contigs for both metagenomic DNA inserts. The Rapid Annotation using Subsystem Technology Server version 2.0 (Aziz et al, 2008) was used to annotate both full-length contigs, and annotation information for mgFurAc and mgSyrAld are tabulated in Supplementary Tables II and III, respectively.
Loss-of-function study by transposon mutagenesis
To identify the genes within the ∼40 kb metagenomic DNA inserts responsible for the improved tolerance, we performed a loss-of-function study on mgFurAc and mgSyrAld. The 192 transposon-inserted clones created for sequencing of the mgFurAc and mgSyrAld fosmids were individually subjected to growth survival assays in the presence of 0.8 g/l 2-furoic acid and 1.4 g/l syringaldehyde, respectively. Kinetic measurements were carried out for each transposon-inserted clone, along with triplicate measurements for the original metagenomic clone and E. coli control at these concentrations, by 600 nm turbidity measurements every 5 min over 24 h at 37°C with shaking in a Versamax microplate reader. Three separate transposition events in mgSyrAld and seven separate transposition events in mgFurAc resulted in knock down of the relevant tolerance phenotypes. The inhibitor tolerance growth kinetics of these transposon-inserted clones was retested in triplicate to confirm the knockdown phenotype. The exact sequence position for each transposition event was mapped by sequence comparison of the unique 19 bp Mosaic-End sequence from the EZ-Tn5 <KAN-2> Transposon found in each raw sequence read to the fully assembled and annotated mgFurAc and mgSyrAld contigs, using BLAST (Altschul et al, 1990).
Sub-cloning metagenomic genes implicated in tolerance phenotypes
The three genes (mgUdpE, mgRecA, mgOrfX) implicated in the syringaldehyde and 2-furoic acid phenotypes were PCR amplified from the mgSyrAld and mgFurAc fosmids using primers designed to be homologous to at least 18 bp to the 5′ or 3′ regions of the annotated genes, and adding KpnI and HindIII restriction sites to the 5′ or 3′ regions, respectively (Supplementary information). These amplicons were digested with KpnI and HindIII (New England Biolabs), ligated into the KpnI and HindIII sites of the multiple cloning site (MCS) of the pZE21-MCS1 vector (Lutz and Bujard, 1997), and transformed into electrocompetent EPI-300 E. coli using a standard electroporation protocol. The bi-cistronic mgRecA_mgOrfX construct was created by cloning the RBS of pZE21-MCS1 and the mgOrfX gene into the EcoRV and MluI sites of the pZE21-MCS1 construct containing mgRecA (Supplementary information). An additional region containing an XbaI site (which is unique to all three genes and the original pZE21-MCS1 vector) was added to the 3′ end of the mgOrfX gene in this construct. The tri-cistronic mgRecA_mgOrfX_mgUdpE construct was created by sub-cloning the RBS of pZE21-MCS1 and the mgUdpE gene into the new XbaI site and the MluI site of the pZE21-MCS1 construct containing the bicistronic mgRecA_mgOrfX (Supplementary information). Inhibitor tolerance growth kinetic assays were performed as before, with LB medium supplemented with 12.5 μg/ml chloramphenicol for constructs in the fosmid vector (large-insert metagenomic clones and control) or 50 μg/ml kanamycin for constructs in the pZE21 vector (sub-cloned genes and control). The individual 2-furoic acid and syringaldehyde assays were performed at a concentration of 0.8 and 1.4 g/l, respectively. The fosmid control (Supplementary Methods) used as a control to select the original metagenomic clones was again used as a control in these recapitulation assays. The mixed inhibitor assays were performed along an even nine-step gradient formulated between a maximum concentration mixture of 0.80 g/l 2-furoic acid and 1.55 g/l syringaldehyde and a minimum concentration mixture of media lacking inhibitors. E. coli EPI-300 harboring the pZE21-MCS1 vector without an insert was used as a control for the mixed inhibitor assays.
Supplementary Material
Supplementary information, Supplementary figures S1–4, Supplementary tables SI–III
Acknowledgments
We acknowledge the expert assistance of H Daum and F Hyde for library construction; D Libuda for pulse-field gel electrophoresis; J Aach for helpful discussions regarding the paper, and the US Department of Energy GtL Program, Harvard Biophysics Program, The Hartmann Foundation, and Det Kongelige Danske Videnskabernes Selskab for funding.
Footnotes
The authors declare that they have no conflict of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R (2006) Engineering life: building a fab for biology. Sci Am 294: 44–51 [DOI] [PubMed] [Google Scholar]
- Brady SF, Clardy J (2000) Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J Am Chem Soc 122: 12903–12904 [Google Scholar]
- Daniel R (2005) The metagenomics of soil. Nat Rev Microbiol 3: 470–478 [DOI] [PubMed] [Google Scholar]
- Davies J (1994) Inactivation of antibiotics and the dissemination of resistance genes. Science 264: 375–382 [DOI] [PubMed] [Google Scholar]
- Desai SK, Gallivan JP (2004) Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc 126: 13247–13254 [DOI] [PubMed] [Google Scholar]
- Dien BS, Cotta MA, Jeffries TW (2003) Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol 63: 258–266 [DOI] [PubMed] [Google Scholar]
- Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P (2008) Land clearing and the biofuel carbon debt. Science 319: 1235–1238 [DOI] [PubMed] [Google Scholar]
- Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM (2006) Ethanol can contribute to energy and environmental goals. Science 311: 506–508 [DOI] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO (2006) Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38: 1406–1412 [DOI] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- Jin YS, Alper H, Yang YT, Stephanopoulos G (2005) Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl Environ Microbiol 71: 8249–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk TK, Farrell RL (1987) Enzymatic ‘combustion': the microbial degradation of lignin. Annu Rev Microbiol 41: 465–505 [DOI] [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66: 10–26 [DOI] [PubMed] [Google Scholar]
- Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotechnol 73: 27–36 [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) (1996) Escherichia Coli and Salmonella: Cellular and Molecular Biology. American Society of Microbiology Press, Washington, USA [Google Scholar]
- Riesenfeld CS, Goodman RM, Handelsman J (2004) Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol 6: 981–989 [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943 [DOI] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66: 2541–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searchinger T, Heimlich R, Houghton RA, Dong F, Elobeid A, Fabiosa J, Tokgoz S, Hayes D, Yu TH (2008) Use of US croplands for biofuels increases greenhouse gases through emissions from land-use change. Science 319: 1238–1240 [DOI] [PubMed] [Google Scholar]
- Sommer MO, Dantas G, Church GM (2009) Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325: 1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Hill J, Lehman C (2006) Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 314: 1598–1600 [DOI] [PubMed] [Google Scholar]
- Torsvik V, Daae FL, Sandaa RA, Ovreas L (1998) Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol 64: 53–62 [DOI] [PubMed] [Google Scholar]
- Walsh C (2003) Antibiotics: Actions, Origins, Resistance. ASM Press: Washington, DC [Google Scholar]
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E et al. (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–565 [DOI] [PubMed] [Google Scholar]
- Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J (2005) Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol 71: 6335–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomano LP, York SW, Ingram LO (1998) Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J Ind Microbiol Biotechnol 20: 132–138 [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Ingram LO (1999) Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng 66: 203–210 [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Martinez A, Ingram LO (1999) Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65: 24–33 [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Martinez A, Ingram LO (2000) Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 68: 524–530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Supplementary figures S1–4, Supplementary tables SI–III