Abstract
Post-transcriptional regulation by microRNAs and siRNAs depends not only on characteristics of individual binding sites in target mRNA molecules, but also on system-level properties such as overall molecular concentrations. We hypothesize that an intracellular pool of microRNAs/siRNAs faced with a larger number of available predicted target transcripts will downregulate each individual target gene to a lesser extent. To test this hypothesis, we analyzed mRNA expression change from 178 microRNA and siRNA transfection experiments in two cell lines. We find that downregulation of particular genes mediated by microRNAs and siRNAs indeed varies with the total concentration of available target transcripts. We conclude that to interpret and design experiments involving gene regulation by small RNAs, global properties, such as target mRNA abundance, need to be considered in addition to local determinants. We propose that analysis of microRNA/siRNA targeting would benefit from a more quantitative definition, rather than simple categorization of genes as ‘target' or ‘not a target.' Our results are important for understanding microRNA regulation and may also have implications for siRNA design and small RNA therapeutics.
Keywords: microRNA, post-transcriptional regulation, siRNA
Introduction
Small RNAs, such as microRNAs and siRNAs, can downregulate hundreds of target genes. Targeting determinants include many site-specific factors, such as base-pair complementarity (Enright et al, 2003; Lewis et al, 2003, 2005; Stark et al, 2003; John et al, 2004), local context factors (Grimson et al, 2007; Hammell et al, 2008), and other destabilization signals (Sun et al, 2010). However, system-level factors also influence small RNA-mediated mRNA degradation. For instance, transfected microRNAs and siRNAs cause global de-repression of genes regulated by endogenous microRNAs, most likely through competition for RNA-induced silencing complex (RISC) (Khan et al, 2009). Other system-level factors include the cellular concentrations of the target transcripts and small RNAs loaded in RISC, determining the kinetics of the regulation. We hypothesize that microRNAs and siRNAs that have a higher number of available target transcripts will downregulate each individual target gene to a lesser extent than those with a lower number of targets (Figure 1A and B): we call this the dilution effect as those small RNAs with many targets have their effect diluted across many molecules. It follows that the competition between target genes for a limited number of active small RNAs may determine how much a small RNA can downregulate each of its target mRNAs.
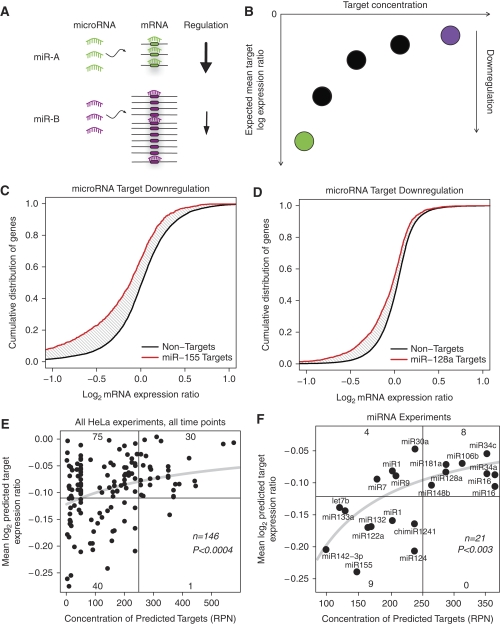
Figure 1.
Mean downregulation is correlated with target abundance. (A) Schematic of the hypothesis that target abundance determines mean downregulation of individual targets. Micro/siRNAs with many targets downregulate their targets to a lesser extent than micro/siRNAs with few targets. (B) Expected correlation between target abundance and log expression ratio. This can also be considered an anti-correlation between downregulation and target abundance. (C, D) Differential downregulation by miR-155 and miR-128, where miR-155 targets are more downregulated than the targets for miR-128. (E) Predicted target concentration and mean log expression ratio across 146 micro/siRNA transfection experiments in HeLa cells. (F) Predicted target concentration and mean downregulation across 21 independently measured, single time point microRNA transfection experiments. Curves were fit to log(1−a/(x−b)), where a and b were determined by least squares error.
Earlier work supports the hypothesis that target abundance can alter small RNA regulation dynamics. Serial dilution experiments in Drosophila embryo lysates show that the siRNA-loaded RISC enzyme can be sequestered by competing target molecules (Haley and Zamore, 2004). Similarly, sequestration can be artificially induced in living cells by expressing transfected microRNA ‘sponges' to soak-up endogenous microRNA molecules (Ebert et al, 2007). Furthermore, siRNAs with a larger number of off-targets show decreased toxicity, perhaps due to a lower effect on each individual gene (Anderson et al, 2008). Finally, there is evidence that target abundance has a significant functional role in an environmental response in plants; namely, phosphate starvation induces a non-coding RNA that sequesters a specific microRNA and de-represses its targets (Franco-Zorrilla et al, 2007).
Given this evidence, we reasoned that the effect of microRNAs and siRNAs on each individual gene may be modulated by target transcript abundance. Therefore, this effect should be seen in the large number of available small RNA perturbation experiments where genome-wide expression changes are measured.
Results and discussion
Target abundance affects average downregulation by small RNAs
The mean downregulation of target mRNAs for different transfected small RNAs is highly variable. For instance, when miR-155 and miR-128 are separately transfected into HeLa cells, the targets of miR-155 are more downregulated than the targets of miR-128 (Figure 1C and D). We hypothesized that this variability is due, in part, to variable abundance of target mRNAs. To examine this phenomenon more systematically, we analyzed the mRNA expression changes after transfection for a panel of 146 small RNA transfection experiments in HeLa cells and 32 small RNA transfection experiments in HCT116 cells (Lim et al, 2005; Jackson et al, 2006; Schwarz et al, 2006; Grimson et al, 2007; He et al, 2007; Kittler et al, 2007; Linsley et al, 2007; Anderson et al, 2008; Selbach et al, 2008) (Supplementary Table 1).
To analyze the proposed dilution effect, we quantified the abundance and mean downregulation of target transcripts for each small RNA transfection. Target mRNA abundance in HeLa cells is quantified using RNA-seq data in units of reads per nucleotide (RPN) (Morin et al, 2008; Mortazavi et al, 2008). Downregulation is computed from the ratio of expression levels after transfection to the levels before transfection, where the expression levels are measured by hybridization on microarrays. To determine the match between small RNAs and their targets, we used 7mer seed matches in the 3′ UTR of mRNAs. We explored several different ways of quantifying abundance and downregulation and of predicting targets, with similar results (Materials and methods; Supplementary Figures 1 and 2; Supplementary Tables 2 and 3).
We examined all 146 HeLa experiments and found a significant correlation (P<0.0004, Pearson's r=0.31, Spearman's ρ=0.30) between the concentration of predicted targets of the transfected small RNA and the average log expression ratio of the target mRNAs (Figure 1E). This can also be considered an anti-correlation between abundance of predicted targets and mean downregulation. This anti-correlation reflects the increased dilution of a limited pool of transfected small RNAs over an increased number of available target sites on targeted transcripts. For example, the predicted targets of miR-155, which are relatively few in number, are more downregulated than the targets of miR-128, which are more numerous.
An alternative statistical measure of the relationship between low target abundance and high downregulation can be obtained by dividing the experiments into quadrants by mean log expression ratio and target concentration (Figure 1E; cutoff for mean log expression ratio is −0.11 and cutoff for predicted target concentration is 250 RPN). Strikingly, when target abundance is low, the fraction of experiments with high downregulation is 10 times larger than when target abundance is high (40/115=34.8%, 1/31=3.2%; P<0.0003, Fisher's exact test).
We analyzed 32 transfection experiments in a second cell type, HCT116 (Dicer−/− or Dicer+/+) and found that high mean downregulation is associated with low predicted target abundance (P<0.054, Fisher's exact test; Supplementary Figure 3). Furthermore, the fraction of transfections that induce large downregulation is very similar in HCT-116 as it was in HeLa. Namely, 33% (n=4) of the transfection experiments whose small RNA had low target abundance (n=12) resulted in predicted targets being highly downregulated, whereas there was only one exception experiment whose small RNA had high downregulation when there was high abundance of predicted targets (n=20).
Downregulation by transfected microRNAs is a function of target abundance
Up to this point, the analysis has examined both siRNA and microRNA transfection experiments pooled together. We then wanted to examine whether the effect was in any way different for the subset of transfection experiments using microRNAs alone. We found that a representative set of microRNA experiments (Materials and methods) show a significant rank correlation between target abundance and mean log expression ratio (Figure 1F; P<0.003, ρ=0.62, n=21). Beyond correlation measures, we also quantify the dynamic range of the dilution effect. For instance, miR-142-3p, the microRNA with the lowest target abundance can downregulate its targets 83% more (on average in log2 mRNA expression ratio) than miR-16, the microRNA with the highest target abundance.
As microRNAs can inhibit translation while showing little or no change in mRNA expression (Filipowicz et al, 2008), we tested whether target mRNA abundance had an effect on the amount of protein downregulation. Using mass spectrometry measurements after microRNA transfection into HeLa cells (Selbach et al, 2008), we found mean downregulation of protein products is consistent with our hypothesis (ρ=0.60, n=5; Supplementary Figure 4). Although the correlation does not reach statistical significance due to low sample size, the trend is similar to the significant dilution effect in mRNA microarray experiments. This trend is relevant as there is no guarantee that changes in mRNA are reflected in the changes in functional protein.
Our results establish a significant anti-correlation between mean downregulation and target abundance; however, this could be the result of other variables that correlate with downregulation as well as target abundance. We explored three possible predictors of downregulation: A+U content, 3′-UTR length, and expression level of individual transcripts. Neither the A+U content of the seed nor of the entire microRNA are significantly correlated with downregulation (at a threshold of 0.05; Supplementary Figure 5). The 3′-UTR length is weakly correlated with target abundance and mean log expression ratio (Supplementary Figures 6 and 7) and individual genes with higher expression levels tend to be more downregulated on average when microRNAs are transfected (Supplementary Figure 8). We have shown that the A+U content is not correlated with mean downregulation, but the other two predictors might contribute to the dilution effect. However, further analysis can control for these predictors, as we describe next.
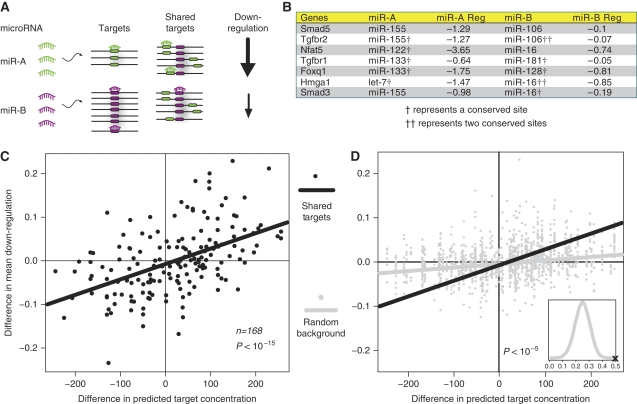
To test our hypothesis while controlling for the potential biases, increase statistical power, and examine differential regulation for individual genes, we performed a second type of analysis. We determined the expression changes of all genes that are targeted by any pair of microRNAs and compared the difference in average downregulation with respect to the difference in the total number of targets transcripts (Figure 2A). This shared target comparison controls for various sources of bias, including A+U content and 3′-UTR length distributions. Figure 2B shows a table of selected genes where each gene is targeted by both miR-A, which has fewer total targets in the cell, and miR-B, which has more. For example, Smad5 and Tgfbr2 are far less downregulated when miR-106 is transfected compared with miR-155; similarly Nfat5 is downregulated much less with miR-16 when compared with miR-122 transfections (Figure 2B). We also found a highly significant correlation between the difference in target abundance and difference in downregulation (Figure 2C; P<10−15, ρ=0.59). We determine an empirical P-value of P<10−5, which uses an empirical background distribution using a set of randomly selected non-targets the same size as the set of shared targets for each point (Figure 2D). Finally, to control for possible laboratory-specific artifacts, we specifically examined microRNA pairs from a single laboratory and found similar results (nominal P<10−6, ρ=0.74; Supplementary information). The full set of microRNA pairs considered in our calculations together with differential downregulation of shared targets are provided in Supplementary Table 4. In summary, this more refined analysis controlled for potentially confounding variables and supported the dilution effect for microRNAs.
Figure 2.
Analysis of shared targets of microRNAs. (A) Schematic of pairwise hypothesis: Given miR-A and miR-B, which both target the same gene, they will regulate differentially in proportion to their individual overall target abundance. (B) Examples of genes that may be differentially regulated (log2 expression ratio) as a result of total target abundance. (C) All pairs of microRNAs are used to search for shared targets. Each point is the difference in mean downregulation and difference in target abundance for a given pair of microRNAs. (D) Background distribution (gray) as a result of randomizing targets. Inset shows empirical P-value of P<10−5. Real correlation line is black.
Target abundance affects primary target and off-target average downregulation by siRNAs
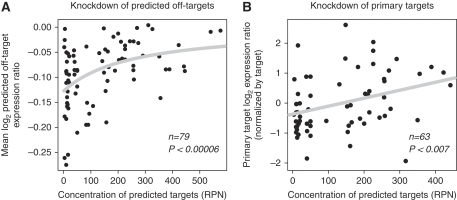
We also show that siRNA off-target downregulation is correlated with on- and off-target abundance. First, we found highly significant correlation between mean log expression ratio of off-targets and off-target abundance (Figure 3A; P<0.0001, ρ=0.37). However, siRNAs are designed to degrade a single primary target gene. Therefore, we analyzed correlation between each siRNA's downregulation of its primary target and abundance of off-targets. We normalized the downregulation by each siRNA with the same primary target by subtracting the mean and dividing by standard deviation, as different primary targets can be knocked down with highly different efficiencies (Supplementary Figure 9). We found a significant rank correlation between log expression ratio of primary target and abundance of off-targets (Figure 3B; P<0.007, ρ=0.34). This indicates that off-target abundance should be considered in the design of siRNA to maintain high knockdown efficiency.
Figure 3.
Target abundance determines siRNA primary target and predicted off-target downregulation. (A) Off-targets for a siRNA experiments show significant correlation. Curve was fit to log(1−a/(x−b)), where a and b were determined by least squares error. (B) The log expression ratio of the primary siRNA target is correlated with predicted off-target concentration. The downregulation for each primary target is normalized such that multiple siRNAs for different targets can be compared.
Recent work has noted that siRNAs with many off-targets may reduce RNAi-induced toxicity (Anderson et al, 2008). However, if this strategy were used, our results suggest one would face a trade-off between reduced siRNA toxicity (more off-targets may lead to decreased toxicity) and increased knockdown of direct siRNA target (fewer off-targets lead to increased knockdown).
Michaelis–Menten kinetics describes total transcripts degraded
As we found a significant anti-correlation between downregulation and total target abundance, we wanted to explore the kinetics that determines the total number of mRNA transcripts degraded. Note that the total transcripts degraded is related to downregulation, but it is an absolute number of molecules and not a ratio. Given the pre-transfection target transcript concentration x(0), we computed the post-transfection concentration x(T), at T=1 day, as follows. We used RNA-seq target transcript abundance for x(0) and determined post-transfection abundance using log expression ratio after transfection from microarray experiments: if we denote xi (0) as the pre-transfection abundance of target gene i and yi(T=1)/yi(0) as the change in expression, then we obtained an estimate for the post-transfection abundance for each target i as
We then estimated the initial velocity, that is the time rate of decrease of transcript concentration, as v=x(0)−x(T=1), to express the velocity as a function of the initial target concentration using Michaelis–Menten kinetics. Our assumption is that T=1 day is sufficiently early to approximate the initial velocity.
We computed v for each of the 146 transfection experiments in HeLa. Empirically, v is significantly dependent on target concentration and fits the Michaelis–Menten model better than linear or constant models (Supplementary Figure 10; Supplementary information). That is, we can fit values of Vmax and Km such that v=Vmax x(0)/(Km+x(0)) and express:
Therefore, this kinetic relationship can be used to predict the expected log expression change of target genes as a function of target abundance.
This relationship provides an explanation of the observed anti-correlation between mean downregulation and concentration of predicted targets. These results motivate a more quantitative framework for studying small RNA regulation and kinetics.
Conclusions
We find evidence that the activity of a small RNA can depend on the abundance of its targets. This in turn allows for potential crosstalk between targets through dilution of microRNAs/siRNAs. This may have implications for research and clinical applications of siRNA technology. Furthermore, it may help explain endogenous microRNA dynamics.
In the clinic, our observed dilution effect may have important implications for the design and efficacy of therapeutic siRNAs, suggesting a key design criterion for therapeutic siRNAs is minimizing the number of off-target sites on highly expressed genes. This may improve efficient direct target downregulation at moderate RNAi concentrations and thereby avoid unwanted saturation effects (Khan et al, 2009). In research applications, siRNAs, such as those used in functional genomic screens, will be more likely to function optimally in cells where off-transcript numbers are low. This may be particularly relevant when designing constructs for cell types that express very few genes at very high levels, such as hepatocytes (Ramsköld et al, 2009).
Endogenous microRNAs may have their effects diluted by highly abundant target transcripts in particular cell types or states. The activity of endogenous microRNAs on specific targets may be significantly altered in contexts where target concentrations change dramatically, such as shifts in environment during differentiation, development, and evolution. Specifically, particular pairs of microRNAs and target genes may be functionally constrained to co-evolve to maintain a constant strength of downregulation. This is different from (though not inconsistent with) the anti-target hypothesis (Farh et al, 2005; Stark et al, 2005) in which mRNAs that dominate a cell type are thought to specifically avoid microRNA targeting.
Finally, quantifying downregulation suggests a departure from considering microRNA targets based on RNA sequence alone. Imbalances in the relative concentrations of microRNAs and gene targets might exaggerate or compensate for sequence mismatches between the two species. For example, highly expressed microRNAs with very few target transcripts may well effectively downregulate mRNAs with ‘weak sites,' such as those containing G:U wobbles. Conversely, weakly expressed microRNAs with many potential mRNA target transcripts, may fail to downregulate species with excellent sequence matches. This will be important to consider in a new generation of microRNA target prediction methods.
Our work demonstrates that small RNA activity can be influenced by the concentrations of the target mRNA molecules, a phenomenon that may alter the effectiveness of therapeutic, investigational, and endogenous small RNAs.
Materials and methods
Quantification of transcript abundance
To quantify transcript abundance, we used RNA-seq measurements from HeLa S3 cells (Morin et al, 2008). We aligned 31 bp reads, allowing two mismatches, to the reference genome (UCSC genome browser hg18), requiring matches to be unique. This resulted in a total of 17.8 million aligned reads. We examined genes by their AceView 3′-UTR annotations (Thierry-Mieg and Thierry-Mieg, 2006) and determined the number of RPN, which we use as a proxy for transcript abundance (Mortazavi et al, 2008). When reads mapped to multiple 3′ UTRs, we split the reads evenly between the UTRs. We used alignments to 3′ UTRs as most validated microRNA targeting (and siRNA ‘off-targeting') has been identified in this region.
Alternative measures of transcript abundance included array fluorescence and number of genes targeted (Supplementary Figure 1). Array fluorescence lacks the dynamic range of RNA-seq. That is, probes called as ‘present' with low fluorescence frequently seem to be absent from the transcriptome, leaving the problem of determining a proper threshold for minimizing false positives and false negatives (Mortazavi et al, 2008). The raw number of genes targeted ignores the transcript abundance and thus may sometimes be a poor proxy for target concentration, especially for those small RNAs with few targets. In general, however, these measures are all highly correlated (Supplementary Figure 1). We also used the sum of all individual target sites on all target transcripts, as opposed to just the total number of target transcripts, and came to equivalent conclusions. To determine the total ‘predicted target concentration,' we sum the abundance for all the predicted targets.
Quantification of target downregulation
We used microRNA and siRNA transfection experiments followed by microarray assay of gene expression to determine small RNA-mediated downregulation. We first performed target predictions using heptamer seed matches in the longest 3′ UTR of Refseq annotated genes to determine a set of predicted targets. We also performed target prediction using several other methods: (1) TargetScan-conserved targets (Lewis et al, 2005; Grimson et al, 2007; Linsley et al, 2007); (2) heptamer seed matches that are conserved in three of four distantly related organisms (mouse, rat, chicken, and dog) (Khan et al, 2009); (3) heptamer seed match to Aceview 3′ UTRs (Thierry-Mieg and Thierry-Mieg, 2006); and (4) the miRanda algorithm (John et al, 2004). All target prediction methods yielded similar results (Supplementary Figure 2).
We examined three measures of downregulation:
(1) Average downregulation (log expression change) of predicted targets.
(2) Area between the predicted target cumulative distribution and the background cumulative distribution. Example cumulative distribution functions for targets and background are shown in Figure 1C and D. The signed area between these curves yields an estimate for how much downregulation an mi/siRNA is able to exert.
(3) Estimate of the total number of molecules degraded as a percentage of initial number of predicted target molecules. The values for x(T=1) and x(0) are estimated as in the kinetic model and the measure we examine is (x(T=1)−x(0))/x(0).
These three methods are highly correlated and yield similar results in our analysis (Supplementary Figure 1).
Representative set of independent microRNA experiments
There are several data sets that have multiple small RNAs with identical seed sequences. In some cases, the seeds are the same because the entire small RNA is the same. In other cases, the seed is identical but other nucleotides in the small RNA are different. When the entire small RNA is identical, we only include replicate experiments if they were performed by separate laboratories. When only the seed is identical, we take two members of the seed class. When multiple time points are available and we only use one, we take the time point at which the targets are most downregulated on average (Supplementary Figure 11).
Downregulation of primary siRNA targets
We provide evidence suggesting that siRNAs with fewer off-target molecules are more effective at downregulating their direct/primary targets (Figure 3B). There are several large sets of microarrays where many siRNAs (each separately transfected) are targeted to a single gene. Each of these experiments shows correlation between target abundance and downregulation; however, only one shows significant correlation (siMAPK14; P<0.02), whereas the others show nearly significant correlation. To elucidate the significance of correlation, we combined all the experiments into a single data set. However, different primary targets are knocked down to different extents on average. To normalize for this effect, we subtracted the mean and divided by s.d. for each set of siRNAs targeting a single gene. That is, for each panel of ‘raw data' in Supplementary Figure 9, we subtract the mean of the y axis and divide by the s.d. of the y axis. We do not change the x axis. The resulting transformed data are shown in Figure 3B and Supplementary Figure 9. The transformed data show highly significant correlation.
Note that the siRNAs targeted to MAPK14 and SOD1 have a single nucleotide mismatch to the target in HeLa cells. Although MAPK14 has a perfect match siRNA, we do not use it as all other siRNAs have a single nucleotide mismatch. Inclusion of the perfect match siRNA does not alter conclusions.
Supplementary Material
Supplementary Text and Figures
Plots showing the target log expression ratio and target abundance for each gene in each transfection experiment. The data for each plot can be found in Supplementary Table 3
Information regarding the small RNA transfection experiments, including the accession, publication reference, seed, sense strand, etc.
Quantification of the target abundance and amount of down-regulation for each transfection experiment
The target log expression ratio and target abundance for each transfection experiment
The shared targets of microRNA pairs, as shown in Figure 2. The log expression ratio and target abundance is given for each shared target
Acknowledgments
We thank Javier Apfeld, Nick Stroustrup, Grégoire Altan-Bonnet, Walter Fontana, and Aly Khan for useful discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson E, Birmingham A, Baskerville S, Reynolds A, Maksimova E, Leake D, Fedorov Y, Karpilow J, Khvorova A (2008) Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA 14: 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M, Neilson J, Sharp P (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A, John B, Gaul U, Tuschl T, Sander C, Marks D (2003) MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K, Grimson A, Jan C, Lewis B, Johnston W, Lim L, Burge C, Bartel D (2005) The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya S, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore P (2004) Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 11: 599. [DOI] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V (2008) mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods 5: 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim L, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson A, Linsley P, Chen C, Lowe S, Cleary M, Hannon G (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS (2006) Widespread siRNA ‘off-target' transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Betel D, Miller M, Sander C, Leslie C, Marks D (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotech 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger A, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, Wagner J, Rees E, Korn B, Frenzel C, Sachse C, Sönnichsen B, Guo J, Schelter J, Burchard J, Linsley P et al. (2007) Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods 4: 337–344 [DOI] [PubMed] [Google Scholar]
- Lewis B, Burge C, Bartel D (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lewis B, Shih I-H, Jones-Rhoades M, Bartel D, Burge C (2003) Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lim L, Lau N, Garrett-Engele P, Grimson A, Schelter J, Castle J, Bartel D, Linsley P, Johnson J (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Linsley P, Schelter J, Burchard J, Kibukawa M, Martin M, Bartz S, Johnson J, Cummins J, Raymond C, Dai H, Chau N, Cleary M, Jackson A, Carleton M, Lim L (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M (2008) Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. BioTechniques 45: 81–94 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ramsköld D, Wang E, Burge C, Sandberg R (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD (2006) Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genetics 2: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell R, Cohen S (2005) Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM (2003) Identification of Drosophila microRNA targets. PLoS Biol 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Li H, Rossi J (2010) Sequence context outside the target region influences the effectiveness of miR-223 target sites in the RhoB 3′UTR. Nucleic Acids Res 38: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J (2006) AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7: S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text and Figures
Plots showing the target log expression ratio and target abundance for each gene in each transfection experiment. The data for each plot can be found in Supplementary Table 3
Information regarding the small RNA transfection experiments, including the accession, publication reference, seed, sense strand, etc.
Quantification of the target abundance and amount of down-regulation for each transfection experiment
The target log expression ratio and target abundance for each transfection experiment
The shared targets of microRNA pairs, as shown in Figure 2. The log expression ratio and target abundance is given for each shared target