Abstract
The papillomaviruses (PVs) are a family of viruses infecting several mammalian and nonmammalian species that cause cervical cancer in humans. The evolutionary history of the PVs as it associated with a wide range of host species is not well understood. Incongruities between the phylogenetic trees of various viral genes as well as between these genes and the host phylogenies suggest historical viral recombination as well as violations of strict virus–host cospeciation. The extent of recombination events among PVs is uncertain, however, and there is little evidence to support a theory of PV spread via recent host transfers. We have investigated incongruence between PV genes and hence, the possibility of recombination, using Bayesian phylogenetic methods. We find significant evidence for phylogenetic incongruence among the six PV genes E1, E2, E6, E7, L1, and L2, indicating substantial recombination. Analysis of E1 and L1 phylogenies suggests ancestral recombination events. We also describe a new method for examining alternative host–parasite association mechanisms by applying importance sampling to Bayesian divergence time estimation. This new approach is not restricted by a fixed viral tree topology or knowledge of viral divergence times, multiple parasite taxa per host may be included, and it can distinguish between prior divergence of the virus before host speciation and host transfer of the virus following speciation. Using this method, we find prior divergence of PV lineages associated with the ancestral mammalian host resulting in at least 6 PV lineages prior to speciation of this host. These PV lineages have then followed paths of prior divergence and cospeciation to eventually become associated with the extant host species. Only one significant instance of host transfer is supported, the transfer of the ancestral L1 gene between a Primate and Hystricognathi host based on the divergence times between the υ human type 41 and porcupine PVs.
Keywords: papillomavirus, phylogenetic incongruence, importance sampling, host transfer, relaxed clock models, Bayesian analysis
Introduction
Papillomaviruses (PVs) are small, nonenveloped viruses that cause lesions on the surface of mucosal and cutaneous tissue. The lesions typically regress spontaneously although persistent infection of a subset of these viruses can lead to the formation of malignant tumors (Durst et al. 1983; Boshart et al. 1984; zur Hausen 1989, 2000; Ho et al. 1995; Campo 2002; Ferenczy and Franco 2002; Pfister 2003; Schiffman et al. 2005; Doorbar 2006).
PV contains a circular double-stranded DNA genome approximately 8 kb in size commonly consisting of up to 8 genes, which are classed as either “early” (genes E1–E2, E4–E7) or “late” (L1 and L2) based on their temporal expression (Danos et al. 1982). E1 and E2 encode proteins important in the viral replication process and are present in all PV genomes; the function of E4 is not yet fully understood but is thought to contribute to replication efficiency and virus release. The remaining early genes (E5, E6, and E7) are known as the “transforming genes”; at least one of these genes is observed in all PV genomes. The transforming genes play an important role in disrupting cellular processes and thus allowing cell cycle progression in order to facilitate viral genome amplification (Doorbar 2005). The late genes, L1 and L2, are referred to as “structural genes” due to their role in the formation of the virus particle. Both late genes are also universally present among PV genomes.
One of the most striking features about the PVs is their association with a wide range of hosts. Well established as parasites of two avian species and numerous species from the various mammalian orders, the host range was recently extended to include reptiles (Herbst et al. 2009). The evolutionary history of this family of viruses, and details of how the various host associations arose, remains a source of controversy. The benign nature of most PV infections coupled with the high host specificity indicates a long association of host and virus, supporting the idea of “host-linked evolution” characterized by simultaneous cospeciation of host and virus (Chan et al. 1992; Ong et al. 1993; Bernard 1994; Tachezy, Duson, et al. 2002; Tachezy, Rector, et al. 2002). The association of PVs with birds, turtles, and mammals dictates that, under a cospeciation mechanism, PVs have been evolving with their hosts for over 300 million years.
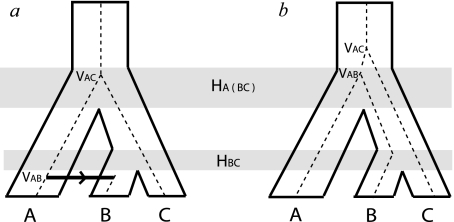
Cospeciation of viruses and hosts should result in agreement between the host and virus phylogenetic tree topologies. As an increasing number of PV sequences has enabled the generation of more detailed phylogenetic trees, however, discordances between the evolutionary histories of the PVs and that of the associated host species have become apparent (Chan et al. 1992, 1995; Chan, Bernard, et al. 1997; Chan, Ostrow, et al. 1997; Garcia-Vallve et al. 2005; Bravo and Alonso 2007; Gottschling, Kohler, et al. 2007; Gottschling, Stamatakis, et al. 2007). These incongruities, or conflicts, between host and virus phylogenies may be explained by one of two mechanisms as shown in figure 1. The first mechanism is host transfer, where either the whole or part of a viral genome adapted to one host species infects another, resulting in a stable lineage in the new host. The second mechanism involves prior divergence, the radiation of a single viral lineage into multiple viral lineages within a single ancestral host. Subsequent speciation of the host may result in differential segregation of the viral lineages among the resultant host species (incomplete lineage sorting), extinction of some of these viral lineages among descendent hosts, or simply a failure to detect the multiple virus lineages in the descendent species, so that the occurrence of prior divergence is not readily discernible from the sampled taxa. Either of these two mechanisms can produce a virus phylogeny that fails to match the host phylogeny, as shown in figure 1.
FIG. 1.
Viral diversification mechanisms that may result in incongruent topologies between virus (dashed) and host (solid). (a) A virus lineage associated with the ancestral host ABC cospeciates with its host at the initial speciation event. The descendent virus lineage in host BC is not observed in host B—this may be due to extinction of the virus in B, failure of the ancestral lineage virus to associate with B following speciation of host BC (“incomplete lineage sorting”), or a failure to detect the virus. The virus lineage that has been detected in B is the result of a host transfer of the virus lineage from A. This host transfer event can be identified by comparing the host and viral divergence times as VAB < HA(BC) in contrast to the previous node where VAC = HA(BC) consistent with cospeciation of the viruses in A and C with their hosts. (b) The virus lineage associated with ancestral host ABC diverges prior to the first host speciation event resulting in two virus lineages that can then segregate along the descendant hosts independently of each other. The viruses associated with hosts A and B are descended from one lineage, whereas the virus in host C is descended from the other lineage. Once again, these events are reflected in divergence times: VAB = HA(BC) as the virus cospeciated with its host, however, VAC > HA(BC) indicating the divergence of virus C prior to speciation of host ABC.
Evidence of prior divergence is abundant among PVs. Over a hundred PV types are currently known in humans, most of which cluster into three main groups—the α, β, and γ human papillomaviruses (HPVs)—which may reflect viral divergences to occupy different biological niches (Chan et al. 1995). Monkeys are also found to be infected by PVs from the α and β genera (Chan, Bernard, et al. 1997; Chan, Ostrow, et al. 1997; Gottschling, Kohler, et al. 2007) implying the occurrence of these viral radiations prior to primate host speciations. Multiple PV types have also been detected in various nonprimate species such as cows, dogs, and horses.
In contrast, evidence for host transfer of extant PVs is extremely rare. The bovine papillomavirus (BPV1) and BPV2 remain the only types to have been isolated from heterogeneous hosts, all of which were ungulates (Otten et al. 1993; Bloch et al. 1994; Antonsson and Hansson 2002; Chambers et al. 2003; Bogaert et al. 2008). Unlike normal PV infection of the epithelium, these zoonotic BPVs can produce only nonproductive fibroblastic tumors or sarcoids in horses and donkeys. Among rabbits, cottontail rabbit PV produces productive infections in its natural host but produces only poorly productive infections that can progress to cancers in domestic rabbits (Rous and Beard 1935). The difficulty of successful host transfer of PVs is not surprising given the slow evolutionary rate, and thus long adaptation times, of DNA viruses.
Despite seemingly greater evidence in favor of prior divergence over host transfer, differences in opinion exist as to which diversification mechanisms, or combination of such mechanisms, best explain the phylogenetic incongruities between PVs and their hosts. Gottschling, Stamatakis, et al. (2007) proposed that multiple host transfer events of the β PVs might have occurred between primate species. Chan, Bernard, et al. (1997), however, conjectured in favor of prior divergence of the α PVs between their associated primate species based on shorter evolutionary distances between intraspecies PVs compared with interspecies PVs.
Distinguishing between ancestral diversification mechanisms is not a trivial task. Methods developed to resolve host and parasite phylogenetic incongruities are predominantly based around finding the optimal set of events that can reconcile the observed evolutionary relationships between parasites with that of the hosts (Brooks 1981; Page 1994; Charleston 1998). These methods, however, do not consider the relative timings of species along the trees and thus may generate historically impossible solutions involving transfer between noncontemporaneous species. Statistical methods (Huelsenbeck et al. 1997; Legendre et al. 2002) provide a different approach by evaluating differences in various phylogenetic parameters of the host and virus trees but are limited by their inability to handle data sets with multiple parasites per host. Both of these approaches can be compromised by uncertainties in either the parasite or host phylogenies. In general, none of these methods provide confident inferences about the nature of alternative diversification mechanisms when cospeciation is rejected.
One possible way to distinguish between these two different mechanisms is to examine the relationship between host and virus divergence times, as shown in figure 1; for prior divergence, the divergence of the viruses “precedes” that of the host, whereas with host transfers the temporal order is reversed. Unfortunately, unlike the faster evolving RNA viruses, substitution rates for PV cannot be obtained by sampling viral sequences in real time, making it difficult to date the viral divergences. Estimates of PV substitution rates on the order of 10−8 nucleotide substitutions per site per year have been obtained by assuming correspondence between viral and host divergence times (Van Ranst et al. 1995; Tachezy, Duson, et al. 2002; Rector et al. 2007), although this analysis may be susceptible to saturation effects as well as assuming the cospeciation mechanism that we are interested in testing. The study of PV evolutionary history is further complicated by the observation of phylogenetic incongruence “between” genes (Bravo and Alonso 2004; Garcia-Vallve et al. 2005; Narechania et al. 2005) and “within” genes (Angulo and Carvajal-Rodriguez 2007; Carvajal-Rodriguez 2008), suggesting that even the various genetic elements of PV do not share a common phylogenetic relationship.
In this paper, we present the results of our analysis of the evolutionary history of PVs. We first examined the validity of combining PV genes for phylogenetic analysis of host association mechanisms by testing the phylogenetic incongruence between the PV genes E1, E2, E6, E7, L1, L2 using human and nonhuman PV sequences. We applied Bayesian phylogenetic methods to incongruence testing and report significant topological incongruence between all six genes studied, indicating that the phylogenetic history of each PV gene must be analyzed separately. To examine the ancestral diversification mechanisms of the PVs, we have devised a new method of characterizing such events when viral divergence times are not available and the phylogenetic relationships are uncertain. In this method, we applied importance-sampling techniques from stochastic simulation theory to Bayesian phylogenetic estimation. Biased distributions of divergence times are specified for the internal nodes of the PV tree in order to evaluate the likelihood of viral divergence due to prior divergence or host transfer at these nodes. We observe multiple incongruities between the host phylogeny and both the L1 and E1 PV gene phylogenies; most of these incongruities are explained by prior divergence of multiple PV lineages associated with the ancestor of all extant mammals but significant support is observed for host transfer events between ancestral Primates and Hystricognathi (porcupine). This method has general application potential in resolving host–parasite phylogenetic incongruities when parasite divergence times are unavailable and when topological uncertainties exist.
Materials and Methods
Amino acid and nucleotide sequences of the PV genes E1, E2, E6, E7, L1, and L2 were obtained from Genbank (Benson et al. 2005). Our data set consisted of the entire genome sequence of 108 different PVs (supplementary table 1, Supplementary Material online). E1, E2, L1, and L2 are present in all 108 PVs, whereas E6 and E7 are only present in 102 of these PVs. The protein sequences were aligned individually using MUSCLE (Edgar 2004). Nucleotide alignments were then constructed from the amino acid alignments using Pal2nal (Suyama et al. 2006). Gapped positions in the resulting nucleotide alignments were removed.
All phylogenetic inference of the PV genes was performed using the Bayesian methods implemented in BEAST (Drummond and Rambaut 2007). In contrast to methods such as maximum likelihood and neighbor joining, which determine evolutionary parameters based on the analysis of an optimal phylogenetic tree, Bayesian phylogenetics use a Markov chain Monte Carlo (MCMC) algorithm to average over tree topologies, branch lengths, and evolutionary parameters for a given set of sequences, and phylogenetic inferences are made from the distributions of the observed parameters (Drummond et al. 2002; for reviews, see Huelsenbeck et al. 2001; Yang 2006). In this way, we can often make confident predictions about some aspects of the evolutionary process (e.g., the substitution rate) in the absence of confidence about other aspects (e.g., the true phylogenetic tree). In Bayesian methods, the support for a hypothesis is represented by the “posterior probability,” the fraction of the samples that conform to the particular hypothesis, whereas the ability of the model to represent the data is represented by the (generally harmonic) average of the sampled log likelihoods generated by the Markov chain. The BEAST program allows incorporation of complex evolutionary models involving rate heterogeneity across sites and lineages. For each analysis, we used the HKY + Γ(5) + Inv evolutionary model, with each codon position partitioned and branch rates selected from a relaxed clock lognormal distribution (Drummond et al. 2006). We used the relaxed clock model following rejection of the molecular clock assumption. A Yule model of speciation was specified for the tree prior. In each analysis, the initial tree was generated randomly.
Gene Incongruence Test
To determine if the six genes had the same evolutionary history and could be combined in further phylogenetic analysis, we investigated the phylogenies of the genes in pairs. By employing a Bayesian approach, we could determine the evidence for incongruence between the two genes in the absence of confident phylogenetic trees for either. For each gene pair, we performed two separate BEAST analyses, each sampling over two separate phylogenetic trees, one for each gene. The question was whether constraining the sampling process to only consider pairs of trees with identical topologies would produce a significantly worse fit to the observed data, quantified as change in the total log likelihood, indicating differences in tree topologies and evidence for incongruence (similar to the approach of Huelsenbeck and Bull [1996] but applied in a Bayesian framework). In the “constrained topology” analysis, the two gene alignments were linked together by specifying the same evolutionary parameters and tree topology for each gene. Branch lengths, however, were varied independently for the two trees. For the “unconstrained topologies” analysis, the two gene alignments had the same evolutionary parameters but individual tree topologies and branch rates were specified for each gene. Each analysis was repeated again to ensure convergence. For each gene pairing, we calculated the marginal log likelihoods, estimated by calculating the harmonic means of the log likelihoods at each state, for both the topologically constrained and the unconstrained MCMC chains. Tests of incongruence between the genes E1, E2, L1, and L2 were performed using a data set of 108 PV sequences; tests of incongruence which included either E6 or E7 were performed using a data set of 102 PV sequences as the following PV types lack one or both of these transforming genes: PePV, PsPV, TtPV2, BPV3, BPV9, BPV10.
Importance Sampling of Diversification Mechanisms
The observed phylogenetic incongruities between viral genes and hosts suggest a more complicated history than simple virus–host cospeciation. Piecing together this evolutionary history is difficult. One important clue is evidence for either prior divergence or host transfer events, which can be determined based on the relative timing of the virus and host divergences. We can determine the evidence for prior divergence or host transfers based on the probability that a divergence between virus lineages occurred prior to or after the divergence between the corresponding hosts. Unfortunately, dating these events for the virus genes has generally been based on estimated substitution rates that are derived assuming virus–host cospeciation, the process that we are evaluating. As an alternative, we designed and implemented a new method to investigate divergence mechanisms among PVs that applies the concept of importance sampling in stochastic simulations to Bayesian phylogenetic inference.
As Bayesian phylogenetics incorporates the sampling of tree topologies and evolutionary parameters for a set of sequences, we could, in principle, perform such a procedure to investigate how often the sampled viral divergence times corresponded to the known host divergence times. The problem is that without some timing information included in the simulation, the viral divergence time is equally likely to be at any time in the past; the probability that the viral divergence times correspond to the rather narrow interval of the host speciation time is extremely remote, as is the probability that the viral divergence time occurred after the origin of life or within the lifetime of the universe. We must include some timing information in the MCMC analysis in order for the sampling procedure to be constrained to realistic timings. Yet, we do not wish to put in fixed constraints that imply assumptions about cospeciation that presuppose the relationships that we are interested in investigating. We make the assumption that cospeciation is common, and therefore bias our MCMC sampling in favor of large number of cospeciation events, without assuming that any “specific” divergence corresponds to cospeciation. We do this by imposing a penalty term in the log-likelihood calculation for each node where cospeciation is violated, that is, when times are sampled outside the host speciation range, resulting in enhanced sampling of trees and timings where cospeciation is common but avoiding the imposition of any fixed constraints. This corresponds to what would be called “importance sampling” in stochastic simulations (for a review, see, for instance, Srinivasan 2002). Importance sampling provides a way of ensuring the sampled parameters are relevant so that meaningful inferences can be made from the resulting distributions.
In importance sampling, it is standard to correct for the effect of the bias in order to determine what the underlying distribution would have been in the absence of the bias. Unfortunately, this would result in the same situation we had prior to the imposition of the biases; the calculation, appropriately corrected, would again be dominated by the vast space of possible trees where cospeciation occurs at some random time in the past. An alternative is to consider violations of cospeciation at individual nodes simply as a measure of the evidence against cospeciation, given an overall bias toward cospeciation. Because of our general assumption of cospeciation, our observed violations represent a conservative estimate. We only need to translate the magnitude of these violations into statistical significance. This can be done through parametric bootstrapping, where we can access the probability that a similar or greater violation would be observed if cospeciation had in fact occurred at that node. We do this by constructing synthetic data modeled on the PV tree where all the nodes under investigation have been adjusted to conform to cospeciation. The nature of the mechanism will be revealed by the timing of viral divergence relative to that of host divergence—viral divergence prior to host speciation indicates prior divergence, whereas viral divergence after host speciation indicates host transfer.
From the PV phylogeny, we identified nodes which formed the most recent common ancestor to PV lineages from different hosts and for which corresponding host divergence times were available. Although a priori knowledge of the viral phylogeny (or the posterior distribution of trees) is required in order to identify nodes at which biased distributions can be applied, uncertainties in the viral phylogeny do not pose a problem as long as the nodes of interest can be confidently identified by their high posterior probabilities. Almost all nodes identified from the E1 and L1 gene trees had posterior probabilities ranging from 0.77 to 1.00; the majority of these posterior probabilities were greater than 0.98. The only exception involved the human–monkey split for which posterior probabilities of 0.6 were observed in both gene trees. We ran multiple independent MCMC chains for phylogenetic estimation and obtained consistent results at this node and thus we proceeded with the biased sampling at this node in spite of the lower posterior. Host divergence times were obtained from molecular estimates along a Mammalian supertree (Bininda-Emonds et al. 2007). For the nodes so identified, we modified the BEAST source code to provide a modified uniform distribution prior on the age of the node. In this distribution, the probability of node ages outside the host speciation range, which formed the upper and lower bounds of the standard uniform distribution, was nonzero but lower than the probability of divergence times within the host speciation range. Both noncospeciation priors were assigned the same probability so there was no bias for prior divergence over host transfer or vice versa. This bias was implemented by imposing a (nonnormalized) prior of 0.05 for noncospeciation relative to speciation by adding a log-likelihood penalty of ln(0.05) = −1.301 for each node that violated cospeciation. It is important to note that the amount of bias against host transfer and prior divergences does not imply an assumption about the likelihood of these two possible scenarios relative to the null model of virus–host cospeciation. We are simply measuring how much the resulting trees violate this null model for a given level of bias and then, using parametric bootstrapping, determine the probability that such violations would be observed, for the same level of bias, were the null model to be correct. In order to see if the magnitude of the bias affected the results, we repeated the analysis with a log-likelihood penalty of ln(0.005) = −2.301.
We performed this modified BEAST analysis on the E1 and L1 genes from the PV data set, with the same model specifications as before and the biased prior distributions specified for the ages of certain nodes. Parrot PV (PePV), which shows the greatest evolutionary distance to all mammalian PVs, was specified as an outgroup. For each of these nodes, we were interested in the proportion of the sampled states in which the node age agreed with the associated host speciation time, the proportion in which the node ages predate host speciation (in agreement with prior divergence) and the proportion in which the node ages postdate host speciation (in agreement with host transfer). Each BEAST analysis was run for 30,000,000 generations with states sampled every 1,000 generations.
In order to calculate P values for the results of this biased BEAST analysis, we simulated PV data sets according to a tree where all viral divergences (for the nodes of interest) occurred via cospeciation. These times were randomly sampled from the host speciation times, assuming a uniform distribution. Using a consensus maximum a posteriori (MAP) tree from the above BEAST analysis, we specified times randomly sampled from the corresponding host speciation times and reestimated the times of the remaining internal nodes using r8s (Sanderson 2003) and the nonparametric rate smoothing algorithm, which allowed for rate heterogeneity between branches. We repeated the process using different sets of sampled times resulting in ten trees with different divergence times of the internal nodes. To convert the branch lengths from units of time to units of distance, we sampled rates for each branch from the distribution of branch rates obtained in the above BEAST analysis. Sequences were simulated along the resulting trees using Evolver from the PAML package (Yang 1997, 2007). Each codon position was simulated separately using the mean values of substitution parameters κ and α obtained from the partitioned BEAST analysis. Ten data sets were simulated for each tree, resulting in 100 simulated data set in total. The biased BEAST analysis was then performed for each simulated data set.
Results
Testing Gene Incongruence
We used BEAST (Drummond and Rambaut 2007), a program for Bayesian phylogenetic inference, to evaluate the evidence that the genes, taken two at a time, have incompatible evolutionary histories. In contrast to other approaches, this method does not consider a specific tree topology for each gene but rather averages over a wide range of tree topologies either constrained or not constrained to be the same for both genes. We then compare the log likelihoods obtained with and without this constraint. The marginal log likelihoods for each of the topologically constrained and unconstrained MCMC chains are shown in supplementary table 2 (Supplementary Material online). (Log-likelihood values for different gene pairs are not directly comparable as different correction constants had to be applied to calculate the marginal log likelihood for each gene pair.) In all 15 gene pairings, we find higher mean log likelihoods for chains run with independent topologies for each gene than when both genes are constrained to the same topology at each state in the chain. As shown in supplementary table 2 (Supplementary Material online), in each case, we found the difference to be significantly greater than the variation in log likelihood observed in each analysis. The large absolute values of the log-likelihood differences (ranging from 21.10 to 264.26) as well as the magnitude relative to the size of the variations both argue strongly for incongruent phylogenies. Significant differences in the log likelihoods, in favor of unconstrained topologies, were also observed when the analysis was repeated with separate evolutionary parameters for each gene partition. This provides evidence that all six genes have distinct phylogenetic topologies.
No evidence of recombination has been observed in experimental work or molecular epidemiological data. This lack led researchers to propose that the observed tree incongruence might be the result of convergent evolution (Narechania et al. 2005); convergent evolution at the amino acid level has recently been shown to result in incongruent trees at the nucleotide level in extreme situations (Castoe et al. 2009). In order to test this possibility, we repeated the analysis restricting the data set to third codon positions that would be unexpected to show this type of effect, as the influence of selective pressure acting on the amino acid sequence at these positions would be greatly reduced. We observed similar measures of phylogenetic incongruity, suggesting that the observed incongruity is the result of viral recombination rather than convergent evolution.
We obtained consensus MAP trees for each gene and investigated topological differences in an attempt to pinpoint the incongruent branches. The MAP trees for E2, E6, E7, and L2 displayed more uncertainty in the reconstruction than were observed for E1 and L1, especially for the deeper branches. This is likely due to the short alignment lengths of these four genes, following the removal of locations with gaps or with uncertain alignments. We therefore restricted further topological comparisons to the E1 and L1 genes.
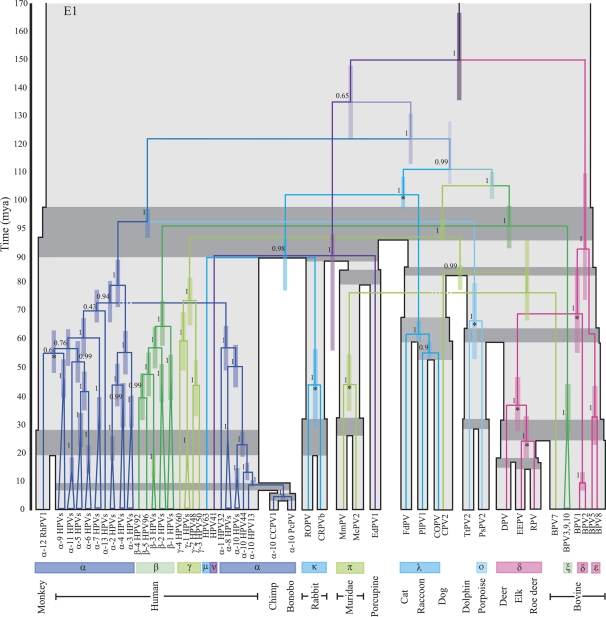
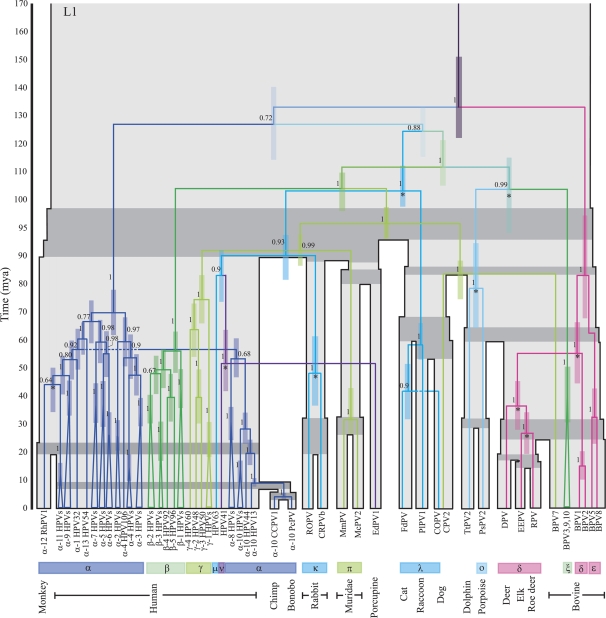
As is shown in figure 2 certain groupings of taxa cluster together with high posterior probabilities in both the E1 and L1 gene trees. In particular, the classification of the majority of PVs by genera (16 genera labeled from α–π; de Villiers et al. 2004), which is achieved based on a region of the L1 gene, is maintained in both gene trees. These classifications largely correspond with previous groupings of PVs based on tissue tropism and biological manifestation (Chan et al. 1995). A noticeable difference in the topologies of the E1 and L1 genes concerns the position of the ν HPV (HPV41)-porcupine (EdPV1) clade. There is also a rearrangement in the position of the Cetacean (porpoise and dolphin) PVs and in the position of the π Muridae PVs. Further topological incongruities are observed within the α, β, γ PVs of E1 and L1 including the characteristic L1 gene split of the high-risk α HPVs and different associations of the rhesus monkey PV with the high-risk α PVs.
FIG. 2.
The E1 and L1 gene trees each shown on top of the associated host tree (grey), which is scaled according to the times of the host divergences (Ma). The timings of the PV splits correspond with the mean times sampled from the biased sampling analysis of each gene; the 95% CIs of viral divergence times at each node are represented by the colored bars. The host speciation times of related host taxa are highlighted in dark grey. Posterior probabilities of internal PV branches are indicated beside the branches. PV nodes labeled with a * indicate divergences for which cospeciation violations were found to be statistically significant. Labels below the tree indicate 1) the names of the PV taxa—“α-2 HPVs” groups together all HPVs included in our analysis from species 2 of the α genus, 2) the genus classifications of the PV taxa, 3) the host species from which the virus was isolated. PV clades are colored according to genus classifications; for simplicity, some genera that consistently group together in both gene trees have been assigned the same color.
Figure 3 shows a splits network generated by combining the E1 and L1 MAP topologies in SplitsTree (Huson and Bryant 2006). A split is defined as the partition of taxa obtained following removal of any branch in the tree. SplitsTree obtains all the splits for the E1 and L1 MAP trees and creates a network consisting of edges for each split observed in the two trees. Regions of the gene trees which are congruent are represented by single edges in the network and are “tree-like” in appearance; however, if two taxa (or sets of taxa) are connected to each other in different ways in the two gene trees this is represented in the network by a set of parallel edges. Such regions in the network therefore display where incongruities between the evolutionary histories of the two genes lie. The network shows several incongruent regions that may be due to multiple recombination events in the evolutionary history of the viruses. Most of these incongruities are located at the base of the network and involve viral lineages that descend to a wide range of hosts. Further incongruities are observed within the primate α PV clades and among the β-HPVs and γ-HPVs. One incongruity is observed nearer the tips of the network; this involves the λ Carnivora PVs. The overall compatibility of splits near the tips of the network suggests against recent recombination events between the PV types in our data set, consistent with the lack of experimental observations.
FIG. 3.
A splits network generated from the E1 and L1 consensus trees using SplitsTree. Sets of parallel edges in the network indicate locations of incongruence and potential recombination.
Host–Virus Tree Incongruence
In addition to observing incongruities between the evolutionary histories of different PV genes, we also observe incongruities between the derived E1 and L1 gene virus trees and the known host tree. High posterior probabilities are observed at almost all the internal branches in each gene tree. The HPVs, which have been heavily sampled, fail to form a monophyletic clade in either the E1 or the L1 gene tree. Instead there are three consistently distinct main HPV clades, which correspond to the α, β, and γ genera of PVs. HPV63 and HPV41, which have been assigned to the μ and ν genera of PVs, respectively, did not cluster with any of the three main HPV clades. Of the three nonhuman primate PVs included in our analysis, chimpanzee and bonobo cluster within the low-risk α HPVs while rhesus monkey PV clusters separately within the high-risk α HPVs. Multiple PV types isolated from cattle also fail to form a monophyletic clade and instead are separated into δ, ϵ, ξ, and BPV7 clades located in different regions of the virus trees. The two canine PVs (COPV, CPV2) appear to be distantly related to each other and do not cluster together in either tree.
The lack of monophyly among PVs at the hosts’ species level further extends at the order and superorder levels. Our data set contains viruses isolated from the mammalian orders of Rodentia (Muridae and porcupine), Primates (human, chimpanzee, bonobo, and monkey), Lagomorpha (rabbit), Carnivora (cat, dog, and raccoon), Cetacea (porpoise and dolphin), and Artiodactyla (bovine, elk, deer, and roe deer). Rodentia, Primates, and Lagomorpha fall under the superorder Euarchontoglires, whereas the remaining orders fall under the superorder Laurasiatheria. Among the Rodentia, the Muridae PVs and the porcupine PVs are in different parts of the tree, with the porcupine EdPV1 clustering with ν HPV. The PV trees do not show an early divergence of sequences from Euarchontoglires and Laurasiatheria but instead we see Euarchontoglire-derived PVs clustering with Laurasiatheria-derived PVs at several points in the gene trees.
As mentioned above, the E1 and L1 gene trees reflect different evolutionary histories of the PV genes. These differences extend to the grouping of PVs from different host species. The Cetacean PVs cluster with Primate α PVs in the E1 tree but with the ξ BPVs in the L1 tree. The ν HPV-EdPV1 clade occupies different position in the two trees, and although this clade associates with human and rabbit PVs in the L1 tree, the Glire PVs (rabbits, porcupine) do not form a monophyletic group.
Importance Sampling of Diversification Mechanisms
As described in the Materials and Methods section, we applied importance-sampling techniques drawn from statistical simulation theory to Bayesian phylogenetics in order to determine whether the virus divergence time matched, preceded, or followed the host divergence time, indicating cospeciation, prior divergence, or host transfers, respectively. This analysis involved biasing the Bayesian sampling of viral divergence times toward cospeciation, finding those nodes in which the statistical support against cospeciation was sufficient to overcome the applied bias, and then using parametric bootstrapping, that is, applying the same calculations to simulated data, in order to evaluate the probability of getting such a strong support if the null model of cospeciation were to have occurred.
We performed this analysis independently on the E1 and L1 gene data sets because of the observed phylogenetic incompatibility of these genes. Divergence time constraints were placed on 24 nodes of the E1 and L1 trees, as shown in supplementary table 3 (Supplementary Material online), in accordance with the corresponding host divergence times (Bininda-Emonds et al. 2007). A log-likelihood penalty of ln(0.05) was set for the sampling of times outside of the specified host speciation range at each node. We repeated the analysis with a stronger bias of ln(0.005). The results described below refer to the results obtained with the weaker bias, unless stated.
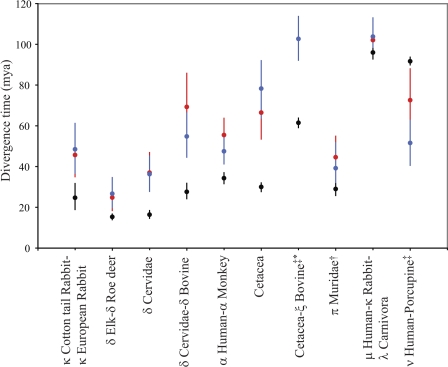
Supplementary table 4 (Supplementary Material online) shows the statistical support for prior divergence or host transfer for both E1 and L1 genes. The resulting divergence timings with confidence intervals (CIs) are shown in figure 2. Figure 4 shows the ancestral PV splits for which violation of the host speciation times in one or both of the data sets was found to be statistically significant, with CIs for the viral divergence times compared with the uncertainty in the host divergence times. Node times sampled after the host speciation times suggest host transfer events. Node times sampled prior to the host speciation time are taken as indication of prior divergence possibly followed by incomplete lineage sorting, extinction, or lack of detection.
FIG. 4.
Divergence times for the host (black), E1 (red), and L1 (blue) genes. CIs for the host and viral divergence times are indicated with error bars; unseen error bars represent CIs smaller than the size of the symbols. Viral divergence times further back than host divergence times (e.g., human–monkey) represent prior divergence, whereas viral divergence times more recent than host divergence times (e.g., human–porcupine) represent likely host transfer events. †Statistically significant violation of host divergence time observed for E1 only. ‡Statistically significant violation of host divergence time observed for L1 only. *Node present in L1 gene tree only.
Most nodes do not reject the cospeciation process, suggesting that the data are consistent with our assumption of the generality of cospeciation. Statistically significant support for prior divergence at the ancestral PV nodes of human–monkey, dolphin–porpoise, domestic–cottontail rabbits, elk–roe deer, Cervinae (deer)–Capreolinae (elk, roe deer), and Cervidae–δ BPV was observed for both genes. As seen in figure 4, there is generally good agreement between the timing of these prior divergence events in both gene trees, arguing against recombination at these points. In addition, there is strong support for prior divergence of the E1 genes of the Muridae (harvest and multimammate mouse) PVs, whereas for the L1 genes, the divergence times sampled for this node largely agree with the host speciation times. For the L1 genes, prior divergence at the ancestral node of the Cetacean–ξ BPV was also found to be statistically significant; these two groups of PVs do not share an immediate common ancestor in the E1 gene tree.
The only host transfer event found to be statistically significant with the weaker penalty was the post-host speciation divergence of the ν HPV-porcupine (EdPV1) L1 genes. The proposed host transfer of the E1 genes of these PV species was not found to be significant (P ∼ 0.30). For the E1 gene, we observe divergence times of 55.38–88.14 million years ago (Ma) compared with 40.70–62.82 Ma for the L1 gene, however, the position of this node differs in both gene trees.
Results obtained with the stronger cospeciation bias of ln(0.005) were in general similar, as shown in supplementary table 4 (Supplementary Material online). All the nodes where cospeciation was rejected with the weaker bias produced similar results with the stronger bias, with the exception of the prior divergence of the Cottontail and European rabbit divergence of the E1 gene, which was strongly supported with the weaker bias (P < 0.01) but not quite supported with the stronger bias (P < 0.06). A number of nodes seemed to reject cospeciation with the stronger bias based on minimal posterior probabilities, generally corresponding to phylogenetic trees that were far from the MAP tree. When analyzing the simulated data sets using the higher bias, the MCMC chain does not appear to sample these topologies. It is therefore possible that there is inadequate mixing of the MCMC sampling procedure at this higher bias.
When sampling PV divergence times we allowed for rate heterogeneity across branches following rejection of a molecular clock from both likelihood ratio tests and Bayesian analysis for both genes. Branch rates were sampled from a lognormal distribution; the mean evolutionary rate estimated for the E1 genes was 7.1 × 10−9 (standard deviation [SD] = 3.1 × 10−10) nucleotide substitutions per site per year and 9.7 × 10−9 (SD = 5.2 × 10−10) nucleotide substitutions per site per year for the L1 genes. In order to provide a more accurate estimate of the rate, we performed the BEAST analysis again for each gene, specifying constraints only for those nodes that did not show significant violations of cospeciation, for which we applied the standard uniform prior distribution of divergence times. The resulting mean rates obtained were 7.1 × 10−9 (SD = 1.5 × 10−10) nucleotide substitutions per site per year for the E1 genes and 9.6 × 10−9 (SD = 2.1 × 10−10) nucleotide substitutions per site per year for the L1 genes, which are in good agreement with our previous estimates. Branch-specific evolutionary rates are similar at the top and bottom of our trees suggesting against saturation having an affect on our analysis.
Discussion
Previous studies of phylogenetic incongruence among PV genes have largely reported incongruence between the early genes and the late genes (Bravo and Alonso 2004; Garcia-Vallve et al. 2005; Narechania et al. 2005). The most recent analysis of PV gene incongruence, however, did not find significant incongruence between the amino acid sequences of the E1, E2, and L1 genes and therefore claimed that the protein products of these three genes may be combined in phylogenetic analyses (Gottschling, Stamatakis, et al. 2007). Similar conclusions could not be drawn at the nucleotide level, however, and it must be noted that only a small representative set of PVs was used in the study.
We first investigated topological incongruence between various mammalian PV genes. Our test involved a larger data set of PVs than has been previously considered, including many nonhuman PV sequences and did not make the assumptions of previous work that we can confidently determine the correct single topology for each gene or that there is an overall tree representing the “true” phylogeny. Instead, we employed Bayesian methods to integrate over uncertainties in the specific topology of each gene and examined the effect on the likelihood when each pair of genes is constrained to assume identical topologies. We find significant topological incongruence between the E1, E2, E6, E7, L1, and L2 genes. These incongruities were observed at multiple branches between the gene phylogenies and involved rearrangements between multiple sets of taxa. We propose that multiple recombination events may have occurred among the E1 and/or L1 genes of PV lineages that coexisted in ancestral mammalian species. Multiple recombination events within the α, β, and γ primate PV genera may also be inferred. This has important implications for the interpretation of phylogenetic analyses of PVs as analyses performed on combined data sets may be compromised. The topological incongruities also indicate the role of recombination in the evolutionary history of the PVs.
The application of recombination detection programs to PV sequences has highlighted potential intragene recombination events within several PV genes. Varsani et al. (2006) located 4 statistically significant recombination events in the L2 gene, 2 statistically significant recombination events in the L1 gene, and 1 statistically significant recombination event in the E1 gene from a data set consisting of 105 sequences. In a separate analysis of the α HPVs, significant recombination signal was detected in the L1 and L2 genes and also in the E6 and E7 genes (Angulo and Carvajal-Rodriguez 2007; Carvajal-Rodriguez 2008). The location of recombination breakpoints in these genes remains undetermined. In our analysis, we treated each gene individually and specifically neglected these potential intragene recombination events.
By employing a novel importance-weighting scheme, we were able to identify viral divergences where the evidence indicates a process other than cospeciation—either prior viral divergence preceding the host divergence or host transfers following the host divergence. There are uncertainties in the analysis, but we can characterize the overall picture. There was a wide diversification of PVs among mammals starting from around 150 Ma. Starting with an early divergence of the Cervidae-δ BPV-ε BPV lineage from that of the other mammalian PVs, by the time of the Euarchontoglires–Laurasiatheria divergence approximately 96 Ma both genes had well-defined α, β, Cervidae-δ BPV-ε BPV, ξ Bovine, Cetacean, and λ (Carnivora excluding CPV2) lineages. In addition, L1 had two lineages containing the γ HPV-mouse and BPV7-CPV2 types, whereas for E1, these types were divided into γ HPV and mouse-BPV7-CPV2 lineages. The μ HPV-ν HPV-EdPV1-κ rabbit clade present at this time in L1 was divided into μ HPV-κ rabbit and ν HPV-EdPV1 clades in E1.
The L1 gene exhibits a cospeciation divergence between the μ HPV-ν HPV-EdPV1 and κ rabbit PV lineages, followed by a divergence between μ HPV and ν HPV-EdPV1 and a host transfer event between humans (ν HPV) and porcupine (EdPV1). The E1 gene follows a different trajectory, with the ν HPV-EdPV1 lineage diverging from the rest of the PV species quite early; the μ HPV–κ rabbit PV lineage diverges from the λ clade sometime later but still prior to the split between Euarchontoglires and Laurasiatheria.
Varsani et al. (2006) analyzed various PV sequences using a suite of recombination detection methods and identified ν HPV as a putative recombinant sequence with the canine PV (COPV) from the λ genus as one of the parent sequences. Their analysis highlighted the E1 gene of ν HPV as the location of recombination. Our gene trees do not concur with their finding as ν HPV is quite distantly related to the λ Carnivora PVs in the E1 gene tree. The consistent grouping of ν HPV and the porcupine PV in our trees suggests that ν HPV is unlikely to be a recombinant genome but the variable position of this clade in the different gene trees may indicate a recombination event in the ancestral viral lineage. One possible explanation involves a recombination event occurring between an unknown ancestral PV lineage and the ancestral lineage of μ HPV resulting in the ancestral lineage of ν HPV and the porcupine EdPV1. Subsequent to the recombination event the PV lineage diverged with the Primate and Rodent hosts but coinfection in either host approximately 50 Ma resulted in transfer of the late region of the genome from one species to the other, although we cannot deduce the direction of this transfer. A simpler scenario would be that the two human PV subtypes diverged from an ancestral Primate PV lineage, and subsequently, there was a host transfer of the ancestral ν HPV lineage to an ancestral Hystricognathi species resulting in the porcupine PV; although our E1 gene analysis does not support the divergence of ν HPV and μ HPV and failed to support a host transfer event involving ν HPV and the porcupine PV, a proportion of the divergence times sampled outside of the host speciation do overlap with those of L1 gene. In order to observe cospeciation of the ν HPV and porcupine PV L1 gene sequences, we would have to apply a substitution rate of approximately 6.7 × 10−9 nucleotide substitutions per site per year, which falls below the distribution of rates applied across the different branches of the L1 tree and supports the rejection of cospeciation at this node.
For the L1 gene, we do observe cospeciation between the γ HPV and π Muridae PV lineages, as well as between the dog (CPV2)–BPV7 divergence. For the E1 gene, we observe early divergence of γ HPVs from the π Muridae-CPV2–BPV7 lineage, followed by cospeciation divergences in the latter. Despite the lack of statistical support for rejecting cospeciation at these various nodes, the observed E1 topology is inconsistent with the host topology, as we would expect the Euarchontoglires (humans and Muridae) and Laurasiatheria (dogs and cows) to cluster separately. It is important to note that we can only detect host transfers and prior divergences when these events occur sufficiently far from the divergence between the hosts. It may be that the prior divergence or host transfer events occurred within the estimated time for the host divergence event or that there is insufficient data to make a reliable identification of the process of the virus divergence. It is also possible that the topology of the viral trees is erroneous, although the posterior probability for the derived topology is high.
A similar situation was observed in the λ clade of the Carnivora PVs: the E1 gene tree topology of this clade is congruent with the host topology and there is no evidence of host transfer or prior divergence of the cat, dog (COPV), and raccoon PVs. In the L1 gene tree, however, the cat PV is more closely related to the dog PV than the raccoon PV with insufficient statistical support in favor of host transfer at the cat–dog PV node. Again, these events might have occurred within the estimated time for the host divergence event or represent our limited statistical power.
Topological differences between the E1 and L1 genes did not result in conflicting divergence times for the majority of viral nodes, which lead us to conclude that the different phylogenies observed for the PV genes arise from recombination events among the ancestral viral lineages. The Cetacean PVs provide an interesting example of this. Both Cetacean PVs were extracted from genital warts; in the E1 gene tree, they form a clade sister to the α PVs, which are the only other clade comprising of genital PVs. In the L1 gene tree, the Cetacean PVs form a clade sister to the ξ BPVs and thus for these PVs the L1 gene tree appears to reflect the host phylogeny, whereas the E1 gene tree reflects the biological properties of the virus. Our sampled divergence times reveal cospeciation of the α Primate PV-Cetacean PV divergence of the E1 gene and prior divergence of the ξ BPV-Cetacean PV L1 genes. In addition, the sampled divergence times for the E1 α Primate–Cetacean node are similar with those of the L1 ξ bovine–Cetacean node. Our results appear to suggest that the ancestral PV lineage that was passed on to the two Cetacean animals may be a recombinant of the ancestral α Primate PV lineage and the ancestral ξ BPV lineage. A recombination event was identified in the L2 gene of porpoise PsPV1 in the analysis of Varsani et al. (2006); however, an ancestral PV of the β genus of HPVs was proposed as one of the parental sequences of the recombination despite the inclusion of ξ BPVs in the analysis. Two other Cetacean PVs—TtPV1 and TtPV3—both obtained from the bottlenose dolphin are also thought to be the descendants of a recombinant PV derived from PsPV and TtPV2 (Rector et al. 2008). There is therefore accumulating evidence of recombinant PV lineages specifically among the Cetacean species.
The relative rarity of zoonotic transmissions of PVs in our analysis is in agreement with the practical difficulties associated with such events. PVs may only gain entry to the basal cells of epithelial tissue via epithelial wounds therefore zoonotic transmissions would require direct contact between the different host species at the very least. Details of the molecular mechanism of PV recognition by host cells are not known but the slow evolutionary rate of PVs is likely to hinder rapid adaptation to a new host. Current evidence indicates that PV gene expression outside of the natural host fails to result in productive infection.
The absence of PV lineages from various extant hosts may be explained by incomplete lineage sorting of the virus among the descendant host species (the virus was not vertically transmitted to all descendant hosts), extinction of virus lineages along particular hosts or a failure to detect these viruses in nonhuman species. There has been an increased sampling effort among other mammalian species, particularly in zoo animals (Antonsson and Hansson 2002; Antonsson and McMillan 2006), however, there has been little success in detecting new PV types, and for many species no PV infection is found. It may be that many nonhuman PV lineages have become extinct but it is difficult to explain this pattern of extinction given the extent of PV diversification observed among humans. Our analysis shows the HPV radiations began prior to the existence of humans—the divergence of last common ancestor of the α PVs is estimated to have occurred 70–80 Ma, that of the β PVs is estimated at around 55–65 Ma, and that of the γ PVs is estimated at around 75 Ma in our analysis. According to these timings, all three genera existed prior to the divergence of the ancestral Primate species, the α and γ PVs may even have existed prior to the divergence of the Euarchonta, which include the Dermoptera (e.g., flying lemurs) and Scandentia (e.g., tree shrews) orders as well as the Primates. No PVs have been isolated from the Dermoptera or the Scandentia, however, and the observed PV diversity among nonhuman Primates is currently much less than that observed among humans. If similar radiations had occurred in other mammalian orders then the Papillomaviridae family has the potential to be substantially larger than previously imagined under a strictly cospeciating mechanism of PV divergence.
Previous estimates of the rate of evolution of PVs have been obtained from PV sequences between closely related hosts under the assumption of cospeciation of host and virus. For feline PVs, an initial estimate of 7.3–9.6 × 10−9 nucleotide substitutions per site per year (Tachezy, Duson, et al. 2002) was later revised to an overall rate of 1.95 × 10−8 (95% CI: 1.32 × 10−8, 2.47 × 10−8) nucleotide substitutions per site per year for the viral-coding genome and with evolutionary rates for individual genes ranging from 1.44 × 10−8 (for E7) to 2.39 × 10−8 (for E6; Rector et al. 2007). A rate of 1.2 × 10−8 nucleotide substitutions per site per year was estimated from Artiodactyla PV sequences (Van Ranst et al. 1995). The Bayesian approach used to investigate cospeciation involves estimation of the evolutionary rates along each branch. The mean rate from the resulting distribution of branch rates therefore allows us to supply estimates of the overall average rate of PV evolution, as well as an estimation of how much this rate varies along various branches of the phylogenetic tree. We found different rates for the E1 genes and the L1 genes. The former are found to evolve slower than the latter with mean evolutionary rates of 7.10 × 10−9 (SD = 1.49 × 10−9) nucleotide substitutions per site per year and 9.57 × 10−9 (SD = 2.08 × 10−9) nucleotide substitutions per site per year, respectively. Previous estimates for these two genes found evolutionary rates of 1.76 × 10−8 (95% CI: 1.2 × 10−8, 2.31 × 10−8) and 1.84 × 10−8 (95% CI: 1.27 × 10−8, 2.35 × 10−8), respectively; however, this analysis was restricted to feline PVs (Rector et al. 2007). Our lower evolutionary rates correlate with our observations of prior divergence of PV lineages, whereas previous estimates have assumed strict correspondence with host divergence times among a small set of closely related PVs. The E1 gene codes for a protein that initiates replication, whereas the L1 gene codes for the viral capsid protein. We may expect the L1 gene to have a higher evolutionary rate than the E1 gene, as the capsid proteins must maintain diversity in order to evade recognition by the host immune system.
In performing this analysis, we are introducing a new general method to investigate diversification mechanisms of viruses and other parasites. Previous methods have generally relied on host–parasite tree reconciliations, which involve counting events necessary to explain discrepancies between the calculated host and parasite trees. These methods suffer from the problems of unknown host and parasite phylogenies, the need to assign relative weights to the different diversification events and the existence of equally parsimonious but different solutions. The incorporation of host transfer events adds additional uncertainty to proposed solutions as temporal information on divergence events is not known but host transfer can only occur between contemporaneous hosts. Other methods have assumed that timing could be derived by assuming strict cospeciation events at some part of the tree, assuming what is being examined elsewhere. Our method considers cospeciation to represent the “null hypothesis” and tests for violations of cospeciation by sampling viral divergence times that are biased for the host speciation times. We are therefore able to analyze the different evolutionary scenarios without requiring explicit knowledge of the viral divergence times. The bias toward cospeciation means that only those divergences that strongly conflict with the cospeciation times will be identified. By utilizing Bayesian phylogenetic analysis, we are able to examine host–parasite associations without restricting the parasite phylogeny to one topology whereas also incorporating evolutionary information present in the data set to evaluate temporal congruence. The only assumption made in our method is that host tree and the associated divergence times are correct, which is necessary in order for the method to produce results. Explicit consideration of evolutionary events along each lineage is circumvented making the biased sampling method more suitable for complex data sets with high parasite-to-host ratios than are alternative methods.
The derived timings of the distant viral divergences can be compromised by saturation. We observed no correlation between the branch-specific substitution rates and the depth of the branch on the phylogenetic tree, providing no evidence for such saturation effects. More conclusive evidence of the lack of such saturation would require a better characterization of the timing of these deeper nodes, something that is not available given the current sequence data and available host speciation information.
The choice of the bias is important. If the bias toward cospeciation is not sufficiently strong, the MCMC sampling will be dominated by irrelevant timescales, and the posterior probabilities of both real and synthetic data will include negligible cospeciation posteriors, resulting in lack of statistical power. Conversely, when the bias is too strong the MCMC mixing times become inconveniently long; this is especially a problem when there is evidence rejecting cospeciation based on minimal posteriors, as occurred with the higher bias used in this paper. It is best to be suspicious of results rejecting cospeciation unless there are substantial posteriors on multiple MCMC threads, as in the results reported here.
The calculations described here are computationally intensive, as the MCMC analysis must be repeated for each of the parametric bootstrap simulations. The statistical power of this method is also reduced by the conservative nature of the assumption of the general predominance of cospeciation. Further work is necessary to better characterize the statistical power of this method, including how this depends upon both the sequence data and the applied bias. Comparison of viral speciation times with that of their hosts will always be conservative, however, as prior radiation and host transfer events that occur within the uncertainty of the host speciation time cannot be detected with this method.
Our analysis presents the first attempt to characterize alternative host association mechanisms of the PVs and therefore resolve PV–host phylogenetic incongruities. The current PV genome data set extends that included in our analysis, however, we restricted ourselves to those sequences that could be confidently placed on the PV tree. As more PV types are identified, we are hopeful that biased sampling of divergence times for a larger data set of PVs, and with greater coverage of the host tree, will generate an even clearer picture of PV evolutionary history.
Supplementary Material
Supplementary tables 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We would like to thank Mario dos Reis for the valuable discussions and assistance provided, Andrew Rambaut for advice on modifying the BEAST source code and the Associate Editor for his insightful comments, concerns, and remarks. This research was supported by funding from the Medical Research Council (UK).
References
- Angulo M, Carvajal-Rodriguez A. Evidence of recombination within human alpha-papillomavirus. Virol J. 2007;4:33. doi: 10.1186/1743-422X-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Hansson BG. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, McMillan NA. Papillomavirus in healthy skin of Australian animals. J Gen Virol. 2006;87:3195–3200. doi: 10.1099/vir.0.82195-0. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H-U. Coevolution of papillomaviruses with human populations. Trends Microbiol. 1994;2:140–143. doi: 10.1016/0966-842x(94)90602-5. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Bloch N, Breen M, Spradbrow PB. Genomic sequences of bovine papillomaviruses in formalin-fixed sarcoids from Australian horses revealed by polymerase chain reaction. Vet Microbiol. 1994;41:163–172. doi: 10.1016/0378-1135(94)90145-7. [DOI] [PubMed] [Google Scholar]
- Bogaert L, Martens A, Van Poucke M, Ducatelle R, De Cock H, Dewulf J, De Baere C, Peelman L, Gasthuys F. High prevalence of bovine papillomaviral DNA in the normal skin of equine sarcoid-affected and healthy horses. Vet Microbiol. 2008;129:58–68. doi: 10.1016/j.vetmic.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Alonso A. Mucosal human papillomaviruses encode four different E5 proteins whose chemistry and phylogeny correlate with malignant or benign growth. J Virol. 2004;78:13613–13626. doi: 10.1128/JVI.78.24.13613-13626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Alonso A. Phylogeny and evolution of papillomaviruses based on the E1 and E2 proteins. Virus Genes. 2007;34:249–262. doi: 10.1007/s11262-006-0017-4. [DOI] [PubMed] [Google Scholar]
- Brooks DR. Hennig's parasitological method: a proposed solution. Syst Zool. 1981;30:229–249. [Google Scholar]
- Campo MS. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Carvajal-Rodriguez A. Detecting recombination and diversifying selection in human alpha-papillomavirus. Infect Genet Evol. 2008;8:689–692. doi: 10.1016/j.meegid.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, Naylor G, Jiang ZJ, Parkinson CL, Pollock DD. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers G, Ellsmore VA, O'Brien PM, Reid SW, Love S, Campo MS, Nasir L. Association of bovine papillomavirus with the equine sarcoid. J Gen Virol. 2003;84:1055–1062. doi: 10.1099/vir.0.18947-0. [DOI] [PubMed] [Google Scholar]
- Chan S-Y, Bernard H-U, Ong C-K, Chan S-P, Hofmann B, Delius H. Phylogenetic analysis of 48 papillomavirus types and 28 subtypes and variants: a showcase for the molecular evolution of DNA viruses. J Virol. 1992;66:5714–5725. doi: 10.1128/jvi.66.10.5714-5725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S-Y, Bernard H-U, Ratterree M, Birkebak TA, Faras AJ, Ostrow RS. Genomic diversity and evolution of papillomaviruses in rhesus monkeys. J Virol. 1997;71:4938–4943. doi: 10.1128/jvi.71.7.4938-4943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S-Y, Delius H, Halpern AL, Bernard H-U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S-Y, Ostrow RS, Faras AJ, Bernard H-U. Genital papillomaviruses (PVs) and epidermodysplasia verruciformis PVs occur in the same monkey species: implications for PV evolution. Virology. 1997;228:213–217. doi: 10.1006/viro.1996.8400. [DOI] [PubMed] [Google Scholar]
- Charleston MA. Jungles: a new solution to the host/parasite phylogeny reconciliation problem. Math Biosci. 1998;149:191–223. doi: 10.1016/s0025-5564(97)10012-8. [DOI] [PubMed] [Google Scholar]
- Danos O, Katinka M, Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among papovaviridae. EMBO J. 1982;1:231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32(1 Suppl):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond). 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczy A, Franco E. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 2002;3:11–16. doi: 10.1016/s1470-2045(01)00617-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve S, Alonso A, Bravo IG. Papillomaviruses: different genes have different histories. Trends Microbiol. 2005;13:514–521. doi: 10.1016/j.tim.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Kohler A, Stockfleth E, Nindl I. Phylogenetic analysis of beta-papillomaviruses as inferred from nucleotide and amino acid sequence data. Mol Phylogenet Evol. 2007;42:213–222. doi: 10.1016/j.ympev.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Stamatakis A, Nindl I, Stockfleth E, Alonso A, Bravo IG. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol Biol Evol. 2007;24:1242–1258. doi: 10.1093/molbev/msm039. [DOI] [PubMed] [Google Scholar]
- Herbst LH, Lenz J, Van Doorslaer K, Chen Z, Stacy BA, Wellehan JF, Jr., Manire CA, Burk RD. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology. 2009;383:131–135. doi: 10.1016/j.virol.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Ho GY, Burk RD, Klein S, Kadish AS, Chang CJ, Palan P, Basu J, Tachezy R, Lewis R, Romney S. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–1371. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Bull JJ. A likelihood ratio test to detect conflicting phylogenetic signal. Syst Biol. 1996;45:92–98. [Google Scholar]
- Huelsenbeck JP, Rannala B, Yang Z. Statistical tests of host-parasite cospeciation. Evolution. 1997;51:410–419. doi: 10.1111/j.1558-5646.1997.tb02428.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Legendre P, Desdevises Y, Bazin E. A statistical test for host-parasite coevolution. Syst Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- Narechania A, Chen Z, DeSalle R, Burk RD. Phylogenetic incongruence among oncogenic genital alpha human papillomaviruses. J Virol. 2005;79:15503–15510. doi: 10.1128/JVI.79.24.15503-15510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CK, Chan SY, Campo MS, Fujinaga K, Mavromara-Nazos P, Labropoulou V, Pfister H, Tay SK, ter Meulen J, Villa LL. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67:6424–6431. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten N, von Tscharner C, Lazary S, Antczak DF, Gerber H. DNA of bovine papillomavirus type 1 and 2 in equine sarcoids: PCR detection and direct sequencing. Arch Virol. 1993;132:121–131. doi: 10.1007/BF01309847. [DOI] [PubMed] [Google Scholar]
- Page RDM. Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics. 1994;10:155–173. [Google Scholar]
- Pfister H. Chapter 8: Human papillomavirus and skin cancer. J Natl Cancer Inst Monogr. 2003;31:52–56. doi: 10.1093/oxfordjournals.jncimonographs.a003483. [DOI] [PubMed] [Google Scholar]
- Rector A, Lemey P, Tachezy R, et al. (15 co-authors) Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 2007;8:R57. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A, Stevens H, Lacave G, et al. (13 co-authors) Genomic characterization of novel dolphin papillomaviruses provides indications for recombination within the Papillomaviridae. Virology. 2008;378:151–161. doi: 10.1016/j.virol.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Rous P, Beard JW. The progression to carcinoma of virus-induced rabbit papillomas (Shope) J Exp Med. 1935;62:523–554. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Herrero R, Desalle R, et al. (18 co-authors) The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Srinivasan R. Importance sampling: applications in communications and detection. Berlin (Germany): Springer-Verlag; 2002. [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy R, Duson G, Rector A, Jenson AB, Sundberg JP, Van Ranst M. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology. 2002;301:313–321. doi: 10.1006/viro.2002.1566. [DOI] [PubMed] [Google Scholar]
- Tachezy R, Rector A, Havelkova M, Wollants E, Fiten P, Opdenakker G, Jenson B, Sundberg J, Van Ranst M. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002;2:19. doi: 10.1186/1471-2180-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ranst M, Kaplan JB, Sundberg JP, Burk RD. Molecular evolution of the human papillomaviruses. In: Gibbs AJ, Calisher CH, Garcia-Arenal F, editors. Molecular basis of virus evolution. Cambridge: Cambridge University Press; 1995. pp. 455–476. [Google Scholar]
- Varsani A, van der Walt E, Heath L, Rybicki EP, Williamson AL, Martin DP. Evidence of ancient papillomavirus recombination. J Gen Virol. 2006;87:2527–2531. doi: 10.1099/vir.0.81917-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Computational molecular evolution. Oxford: Oxford University Press; 2006. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses as carcinomaviruses. Adv Viral Oncol. 1989;8:26. [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.