SYNOPSIS
The 26S proteasome is a 2,500,000-dalton protease complex that degrades polyubiquitylated proteins by a mechanism that requires ATP hydrolysis. It also degrades short non-ubiquitylated peptides and certain unstructured proteins by an energy-independent mechanism that requires bound ATP to maintain its component subcomplexes, the 20S proteasome and PA700, in a functionally assembled state. Proteolysis of both types of substrates requires PA700-induced opening of reversible gates at substrate access pores of the 20S proteasome. Here we demonstrate that the rate of peptide substrate hydrolysis, a functional monitor of gate opening, is regulated variably by multiple effectors. ATPγS and other nonhydrolyzable ATP analogs increased peptide substrate hydrolysis by intact 26S proteasome. Thus, nucleotides that maintained 26S proteasome structure but did not support ATP hydrolysis or the degradation of polyubiquitylated proteins promoted enhanced rates of peptide hydrolysis. Polyubiquitin and a peptoid that binds selectively to a single ATPase subunit of PA700 also increased rates of peptide hydrolysis but had disparate effects on rates of ATP hydrolysis. The effect of polyubiquitin was specific for ubiquitin-ubiquitin linkages that supported proteolysis of protein substrates. These results indicate that gating of the 26S proteasome is not a simple two-state process but can be variably modulated. Our results suggest that modulated gating of the proteasome may be an important element of the mechanism of proteolysis of polyubiquitylated proteins.
Keywords: ubiquitin, PA700, AAA proteins, proteolysis, proteasome
INTRODUCTION
The 20S proteasome is the proteolytic component of a modular protease system responsible for most protein degradation in eukaryotic cells [6;16]. This 700,000-dalton complex is composed of four axially-stacked heteroheptameric rings [1;5]. Three subunits on each of the two compositionally-identical inner β rings contain N-terminal threonine residues that serve as catalytic nucleophiles for peptide bond hydrolysis [20]. These residues line a central lumen formed by the two inner rings and, thus, are sequestered from potential substrates by a shell of proteasome subunits. To reach these catalytic sites, substrates must traverse pores circumscribed by subunits of the outer α rings [19;21]. In addition to their narrow caliber that restricts substrates to short peptides or unfolded proteins, these pores are reversibly gated by N-terminal extensions of several α ring subunits [19;23]. Isolated 20S proteasomes normally assume gating conformations featuring occluded pores and therefore display low rates of proteolysis regardless of substrate size or structure. An open-pore conformation is achieved upon binding of any of several regulatory proteins to the proteasomes’s α rings [15;36;40].
The proteasome-regulator complex whose cellular function is best understood is the 2,500,000-dalton 26S proteasome comprising the 20S proteasome and a 700,000-dalton, 20-subunit regulator called PA700, or 19S regulator [18;36;39]. Six PA700 subunits, termed Rpt1–6, are distinct but homologous gene products of the AAA (ATPases Associated with various cellular Activities) protein family [3;9;11;18]. These subunits form a heterohexameric ring that abuts directly to the heteroheptameric α rings of the 20S proteasome [7;14]. PA700 binding to the 20S proteasome depends on ATP binding by Walker motifs of the AAA subunits [28;37]. The functional consequence of PA700 binding to 20S proteasome is conformational relief of the occlusion that blocks 20S proteasome pores, an effect manifested as increased rates of hydrolysis of peptides due to increased substrate access to the catalytic sites [19;23;30]. Remarkably, isolated C-terminal peptides of two AAA subunits (Rpt2 and Rpt5) are each sufficient to bind to and activate the 20S proteasome by a mechanism that repositions the blocking gates of the α subunit pores and presumably mimics the action of intact PA700 [17;37]. In contrast to PA700, however, proteasome binding and activation by Rpt peptides are independent of ATP. This result is expected for the conforrmationally unrestrained peptides, and suggests that ATP binding by PA700 promotes a conformation of the subcomplex that correctly orients the C-termini of component Rpt subunits for interaction with the cognate binding sites on the 20S proteasome.
Although gate opening is necessary and sufficient for the 26S proteasome’s ATP hydrolysis-independent degradation of short peptides and certain unstructured proteins, it is necessary but insufficient for the ATP-hydrolysis dependent degradation of protease’s major physiological substrates, which include highly structured proteins modified by polyubiquitin chains. Thus, in addition to opening substrate access pores, PA700 selectively binds polyubiquitin via several “receptor” subunits, unfolds the structured attached protein, and enzymatically removes the polyubiquitin chain to facilitate substrate translocation through the narrow open pore [39]. These additional substrate processing events likely account for the energy requirement of proteolysis and are powered by Rpt subunit-catalyzed hydrolysis of ATP.
While studying the effects of C-terminal Rpt subunit peptides on proteasome activation, we noted that peptides from different Rpt subunits produced additive, and under specific conditions, synergistic effects on proteolysis [17]. These results suggested that proteasome gating could be variably modulated. The purpose of this work was to determine whether substrate gating could be variably modulated in intact 26S proteasome. Our results reveal that rates of peptide hydrolysis by the 26S proteasome are altered in response to several conditions and elements involved in the degradation of polyubiquitylated proteins and provide insight to the mechanisms of 26S proteasome function.
EXPERIMENTAL
Materials
K48-linked tetra-ubiquitin and ubiquitin aldehyde were purchased from Boston Biochem. Bovine ubiquitin and ATP were purchased from Sigma Chemical Company. ATPγS was obtained from Sigma and Calbiochem. Rip-1 was a generous gift of Dr. Thomas Kodakek (UT Southwestern Medical Center, Dallas).
Protein purification
26S proteasome, 20S proteasome, PA700, and PA28 were purified from bovine red blood cells, as described previously [8;17;29]. Most experiments were conducted with 26S proteasome in which > 80% of the complex contained two PA700 caps. Human Rad23B and corresponding Ubl (residues 1–80) and Uba (residues 191–402) domains were expressed as GST-tagged recombinant proteins in E. coli and purified to homogeneity by affinity chromatography on glutathione-Sepharose [4;32]. GST was removed from the purified proteins by thrombin cleavage prior to use. Recombinant α-synuclein was purified as described previously [17;27].
Preparation of polyubiquitin and polyubiquitylated proteins
K48-linked polyubiquitin chains were prepared by incubation of purified ubiquitin (1.6 mM) with recombinant His-E1 (0.1 μM) and His-E2–25K (18 μM) conjugating enzymes, creatine phosphate (10 mM), inorganic pyrophosphatase (0.6 U/ml) and creatine kinase (0.6 U/ml) for 16 hrs at 37°C in buffer containing 50 mM Tris-HCl, pH 8.0, 2.5 mM ATP, 5 mM MgCl2, and 0.5 mM DTT. After incubation, conjugating enzymes were removed by affinity chromatography on Ni-beads and a high-molecular weight fraction (> 116,000 daltons) of polyubiquitin chains of heterogeneous length was isolated by gel-filtration chromatography. The fraction was concentrated and dialyzed extensively against buffer containing 20 mM Tris-HCl, pH 7.6, and 100 mM NaCl.
Polyubiquitylated His6-Sic was prepared as described previously [33]. In brief, His6-Sic containing a PY motif fused to the C-terminus was expressed in E. coli and purified by affinity chromatography. Ubiquitylation was conducted by incubation of Sic (10 μM) with purified recombinant E1 (0.12 μM), Ubc4 (3 μM), Rsp5 (0.53 μM), and bovine ubiquitin (230 μM), for 6 hrs at 37°C. A high molecular weight fraction of polyubiquitylated-Sic (> 160,000 daltons) was isolated by repeated rounds of glycerol density gradient centrifugation. After isolation and concentration, the proteins were dialyzed into buffer containing 20 mM Tris-HCl, pH 7.6, and 100 mM NaCl.
Proteasome activity
Proteasome activity was measured by determining rates enzymatic production of 7-amino-4-methylcourmarin (AMC) from peptide substrates Suc-Leu-Leu-Val-Tyr-AMC, Suc-Leu-Leu-Glu-AMC, and CBZ-Val-Leu-Arg-AMC, as described previously [17]. Standard assay conditions included 50 mM Tris-HCl, pH 7.8, 5 mM β-mercaptoethanol, and 50 μM substrate in a volume of 200 μl. Incubations were carried out at 37°C for 10 or 20 mins (depending on the experiment) in a Biotek PL600 fluorescence plate reader with filters at 380 nmex/460 nmem. AMC fluorescence was monitored once per minute during the assay and analyzed with kinetic software. Activity is expressed as arbitrary fluorescent units produced per minute. Routine control assays included reactions without enzyme. Other details of individual experiments are provided in appropriate figure legends. In some experiments semi-quantitative measures of proteasome activity were obtained by overlay of peptide substrates in situ on proteins separated in native polyacrylamide gels, as described previously [13;17]. After incubation at 37°C for 10 mins, AMC at the position of the protease responsible for its production was visualized by UV light.
Hydrolysis of polyubiquitylated-Sic by the 26S proteasome and PA700-dependent activation of 20S proteasome as a consequence of 26S proteasome assembly were determined as described previously [28].
ATPase activity
ATPase activity was determined by measuring production of [32P]-phosphate from [γ– 32P]ATP [9].
RESULTS
ATPγS increases peptidase activity of the 26S proteasome
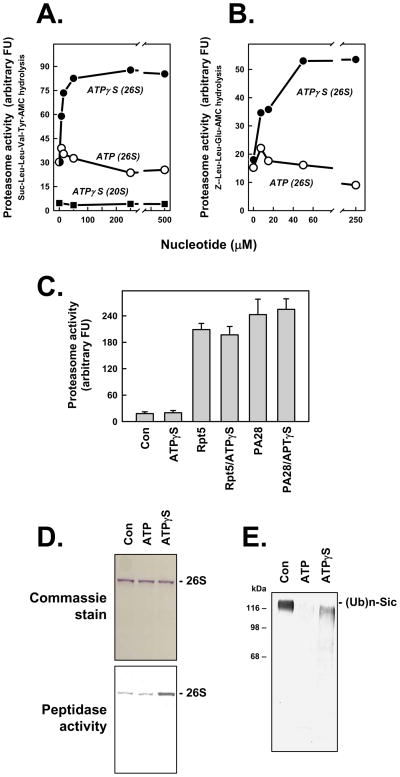
The 26S proteasome degrades peptide substrates at rates more than 20-fold faster than does the 20S proteasome. This effect reflects increased substrate access to the catalytic sites of the proteasome as a consequence of PA700-induced gate opening. To further explore the relationship between gate opening and ATP-dependent proteasome activation, we incubated the 26S proteasome with increasing concentrations of the non-hydrolyzable analog, ATPγS. ATPγS increased 26S proteasome activity by 3–5 fold over that achieved by comparable concentrations of ATP (Figure 1). Similar effects were observed with each of three different peptides hydrolyzed by distinct catalytic centers of the proteasome (Figures 1A, 1B, and data not shown). AMPPNP, another non-hydrolzyable ATP analog that maintained 26S proteasome structure, produced comparable results (data not shown). The stimulatory effect of ATPγS was specific for the 26S proteasome because ATPγS had no effect on the low basal rates of peptide hydrolysis by the 20S proteasome. Moreover, ATPγS had no effect on ATP-independent activation of 20S proteasome by the proteasome regulator PA28, or by a 10-residue peptide representing the C-terminus of the Rpt5 subunit of PA700 (Figures 1A and 1C) [17;29]. Thus, enhanced 26S proteasome activity by ATPγS appears to depend on ATPγS binding to one or more of the AAA subunits of PA700. Finally, ATPγS-stimulated proteasome activity did not reflect enhanced 26S proteasome stability because no differential effect of ATP and ATPγS on this property was detected by various sensitive indicators of this feature including native PAGE and density gradient centrifugation; in fact, the former method demonstrated directly that intact 26S proteasome had higher peptidase activity in the presence of ATPγS than in the presence of ATP (Figure 1D). In contrast to its stimulatory effect on peptidase activity, ATPγS inhibited the degradation of polyubiquitylated substrates (Figure 1E). This effect is consistent with the established requirement for ATP hydrolysis in the degradation of these substrates and reflects the need for energy consumption processing of such complex substrates. As described previously, ATPγS supports some deubiquitylation of polyubiquitylated substrates (Figure 1E) [28].
Figure 1. ATPγS stimulates peptidase activity of the 26S proteasome.
Panels A and B. 26S proteasome (0.7 nM) or 20S proteasome (2 nM) was assayed for peptidase activity using indicated substrates. Assays for 26S proteasome contained 5 μM ATP and indicated additional concentrations of either ATP or ATPγS. Assays for 20S proteasome contained indicated concentations of ATPγS. Data points represent mean values of triplicate assays. Similar effects were obtained in four independent experiments. Panel C. 20S proteasome (2 nM) was assayed for peptidase activity using Suc-Leu-Leu-Val-TryAMC substrate and indicated effectors: ATPγS (500 μM); Rpt5 (10 residue peptide of the C-terminus of Rpt5 PA700 subunit, 400 μM); Rpt5 (400 μM) and ATPγS (500 μM); PA28 (200 nM); PA28 (200 nM) and ATPγS (500 μM). Data represent mean values of quadruplicate assays (+/− SD). Similar results were obtained in three independent experiments. Panel D. 26S proteasome was preincubated with 10 μM (Con), 400 μM ATP (ATP) or 10 μM ATP and 400 μM ATPγS (ATPγS) and then subjected to native PAGE. One half of the get was stained with Commassie blue (upper panel) and the one half of the gel was incubated with Suc-Leu-Leu-Val-Tyr-AMC for 15 mins at 37°C and exposed to UV light to reveal AMC product (lower panel); the lower image has been converted to a positive exposure. Panel E. Polyubiquitylated-Sic was incubated in the absence (Con) or presence of 26S proteasome with 200 μM of either ATP or ATPγS, as described in Experimental Procedures. Samples were subjected to western blotting using anti-His antibody.
Polyubiquitin increases peptidase activity of the 26S proteasome
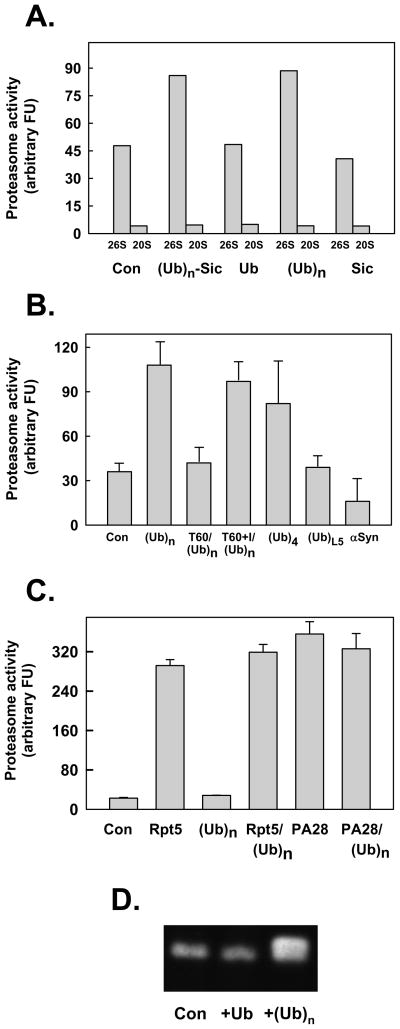
The stimulatory effect of ATPγS on peptide hydrolysis by 26S proteasome suggests that conformational changes of AAA subunits upon ATP binding induce a state of gate opening that optimizes peptide substrate transit and that ATP hydrolysis alters this conformation to one that attenuates transit. This model led us to question whether other factors that potentially alter 26S proteasome conformation could also influence gating. An obvious candidate for such a factor is a polyubiquitylated protein that interacts with polyubiquitin chain-binding subunits of PA700. To explore this possibility, we tested the effect of a polyubiquitylated substrate on the rate of peptide substrate hydrolysis by 26S proteasome. Polyubiquitylated-Sic, a model substrate degraded by the 26S proteasome, stimulated rates of hydrolysis of peptide substrates by 2–4 fold in the presence of ATP (Figure 2A). To determine whether this effect was mechanistically linked to the concomitant degradation of the protein substrate, we examined the individual effects of Sic and polyubiquitin. Non-ubiquitylated Sic, a protein refractory to degradation by the 26S proteasome, and α-synuclein a natively disordered protein readily degraded by the 26S proteasome in an ubiquitin-independent manner, failed to stimulate peptide hydrolysis (Figure 2A and 2B). In fact, the latter protein inhibited peptide hydrolysis, probably by substrate competition. In contrast, unanchored polyubiquitin stimulated peptide hydrolysis as well as, and in many experiments better than, its protein-conjugated counterpart. This effect was achieved by both high molecular weight K48-linked chains of heterogeneous length and tetra-ubiquitin, but by neither a linear fusion of five ubiquitin moieties nor mono-ubiquitin (Figure 2B). Preincubation of polyubiquitin with 26S proteasome for periods sufficient for nearly complete chain disassembly by deubiquitylating subunits of PA700, abrogated stimulation of peptidase activity in subsequent assays; this abrogation of stimulated peptidase activity was blocked by inhibitors of deubiquitylation (Figure 2B). As with the stimulatory effect of ATPγS, the stimulatory effect of polyubiquitin was specific for the 26S proteasome because it was not observed on basal activity of 20S proteasome or on 20S proteasome activity stimulated by PA28 or the C-terminal peptide of Rpt5 (Figure 2C). Thus, enhanced gate opening was not achieved by an unexpected interaction between polyubiquitin and the 20S proteasome, but instead appeared to require polyubiquitin binding to PA700. In fact, 26S proteasome preincubated with polyubiquitin migrated slower than 26S proteasome without polyubiquitin during PAGE, and has higher peptidase activity in a substrate overlay assay (Figure 2D). This results indicates that the polyubiquitin-bound form of 26S proteasome features increased gating. Finally, the effect of polyubiquitin was not mediated indirectly via promotion of assembly (and therefore activation) of any dissociated 20S and PA700 subcomplexes in the 26S proteasome preparations. In experiments to test this possibility directly, we found that polyubiquitin had no effect on the rate or extent of 26S proteasome assembly from purified 20S and PA700 subcomplexes (Figure 2D and data not shown).
Figure 2. Polyubiquitin stimulates peptidase activity of the 26S proteasome.
Panel A. 26S proteasome (0.7 nM) or 20S proteasome (2 nM) were assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Assays contained either no other additions (Con) or: polyubiquitylated-Sic (Ubn-Sic, 350 nM); monoubiquitin (Ub, 1 mM); K48-linked polyubiquitin (350 nM), or; Sic (50 nM). Panel B. 26S proteasome (0.7 nM) was assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Assays contained either no other additions (Con) or: K48-linked polyubiquitin (Ubn, 340 nM); K48- linked tetraubiquitn (Ub4, 400 nM); linear tandom head-to-tail fusion of five ubiquitins (UbL5, 400 nM); and α synuclein (α– syn, 200 nM). In two other assays, 26S proteasome (0.7 nM) and K48-linked polyubiquitin (Ubn, 350 nM) were preincubated for 60 min in the absence (T60) or presence (T60+I) of ubiquitin aldehyde (10 μM) and o-phenanthroline (5 mM) prior to assay. Panel C. 20S proteasome (2 nM) was assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Assays contained either no other additions (Con) or: Rpt5 (10 residue peptide of the C-terminus of Rpt5 PA700 subunit, 400 μM); K48-linked polyubiquitin (350 nM); 400 μM Rpt5 peptide and 350 nM K48-linked polyubiquitin; PA28 (200 nM); PA28 (200 nM) and K48-linked polyubiquitin (350 nM). For Panels A-C, data are expressed as mean values of triplicate assays (+/− SD); similar results were obtained in at least three independent experiments. Panel D. Purified 26S proteasome was preincubated briefly with ubiquitin (Ub), K48-linked polyubiquitin chains (Ubn), or buffer (Con), and then subjected to native PAGE. After electrophoresis, the gel was incubated with Suc-Leu-Leu-Val-Tyr-AMC for 15 mins at 37°C and exposed to UV light to reveal AMC product.
Polyubiquitin delivered to the proteasome by reversibly associated proteins stimulates proteasome activity
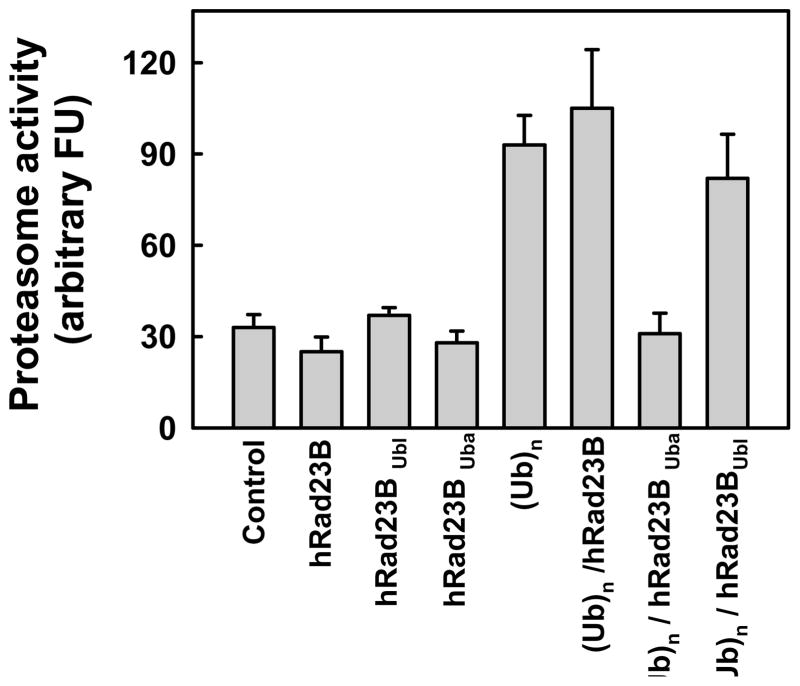
Polyubiquitylated proteins reach the 26S proteasome by multiple mechanisms including by direct binding of polyubiquitin chains to specific constituent subunits of PA700 and by extra-proteasomal polyubiquitin binding proteins that reversibly associate with PA700 [12;34]. Therefore, we determined whether the route by which polyubiquitin is delivered to the proteasome influenced the effect of polyubiquitin on gating. We utilized human Rad23B (hRad23B), a polyubiquitin chain binding protein responsible for proteasome-mediated degradation of certain cellular proteins. hRad23B contains an N-terminal Ubl domain that binds to PA700 and C-terminal Uba domains that bind polyubiquitin. In the absence of polyubiquitin, neither full-length hRad23B nor its separate Ubl or Uba domains significantly affected 26S proteasome-catalyzed peptide hydrolysis (Figure 3). Thus, ubiquitin-like proteins that bind to PA700 do not mimic the effect of polyubiquitin on proteasome gating. In contrast, a complex of hRad23B bound to polyubiquitin stimulated peptidase activity in a manner similar to that of polyubiquitin alone. Polyubiquitin bound to the isolated Uba domains had poor stimulatory activity, indicating that the Ubl domain of hRad23 was required for an interaction of the complex with proteasome. Finally, polyubiquitin stimulation of peptidase activity was not affected by the isolated Ubl domain (Figure 3), suggesting that polyubiquitin and the Ubl domain do not compete for the same binding site on the proteasome. In sum, these results indicate that polyubiquitin enhances proteasome gating by a mechanism independent of its mode of delivery to the proteasome.
Figure 3. Relative effects of polyubiquitin and Rad23 on peptidase activity of the 26S proteasome.
26S proteasome (0.7 nM) was assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Assays contained either no other additions (Con) or: hRad23B (300 nM); Ubl domain of hRad23B (300 nM); Uba domain of hRad23B; K48-linked polyubiquitin (Ubn, 300 nM); (Ub)n bound to hRad23B (300 nM); Ubn bound to the Uba domain of hRad23B (300 nM); Ubn (400 nM) and the Ubl domain of hRad23B (300 nM). Data represent mean values of quadruplicate assays (+/− SD). Similar results were obtained in two independent experiments.
Relative roles of ATPγS and polyubiquitin in modulation of 26S proteasome gating
To explore mechanistic relationships between ATPγS- and polyubiquitin–stimulated peptidase activity, we tested the combined effects of these agents. Neither significantly affected peptidase activity of the proteasome in the presence of maximally effective concentrations of the other (Figure 4A). In contrast, their stimulatory effects were approximately additive at submaximal concentrations of each (Figured 4B and 4C). These results suggest that ATPγS and polyubiquitin promote their effects via mechanisms that ultimately increase gating to similar degrees.
Figure 4. Combined effects of ATPγS and polyubiquitin on peptidase activity of the 26S proteasome.
26S proteasome (0.7 nM) was assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate. Panel A. Assays contained either no other additions (Con) or: ATPγS (400 μM); K48-linked polyubiquitin (510 nM), or; ATPγS (400 μM) and K48-linked polyubiquitin (510 nM). Panel B. Assays contained 20 μM ATPγS and indicated concentrations of K48-linked polyubiquitin. Panel C. Assays contained 100 nM K48-linked polyubiquitin and indicated concentrations of ATPγS. Data are expressed as mean values of triplicate assays (+/− SD). Similar results were obtain in three independent experiments for Panel A and two independent experiments for Panels B and C.
Functional relationships between ATPase activity and stimulation of peptidase activity
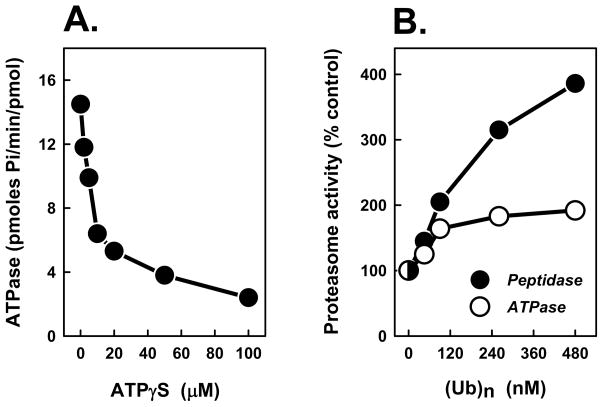
ATPγS inhibited 26S proteasome ATPase activity over the same concentration range in which it stimulated peptidase activity (Figure 5A). This effect likely reflects competition of ATPγS for ATP. We speculated that cycles of ATP binding and hydrolysis might promote altering conformations of optimized and suboptimized gating, respectively, that allow correspondingly increased and attenuated degrees of substrate transit. Therefore, we tested whether polyubiquitin-stimulated peptidase activity also was associated with altered ATPase activity. In contrast to ATPγS, polyubiquitin had a modest but reproducible stimulatory effect on 26S proteasome ATPase activity. Thus, two agents that stimulate 26S proteasome activity similarly have different effects on ATPase activity, indicating that the enhanced peptidase activity is not strictly correlated with altered rates of ATP hydrolysis.
Figure 5. Differential effects of ATPγS and polyubiquitin on ATPase activity of the 26S proteasome.
Panel A. 26S proteasome was assayed for ATPase activity in the presence of the indicated concentrations of ATPγS. Panel B. 26S proteasome was assayed for peptidase activity using Suc-Leu-Leu-Val-Tyr-AMC as substrate (closed circles) or ATPase activity (open circles) in the presence of indicated concentrations of K48-linked polyubiquitin. For each assay, activity in the absence of polyubiquitin was assigned a value of 100% and other activities are expressed as a percentage of that value. Each value represents the mean of triplicate assays. Similar results were obtained in at least three independent experiments.
Rip-1, an Rpt4-binding peptoid, activates 26S proteasome peptidase activity without altering ATPase activity
Recently, Rip-1 was identified as a peptoid (N-substituted oligoglycine) that binds specifically to the Rpt4 subunit of 26S proteasome [24–26]. Like ATPγS, Rip-1 stimulates 26S peptidase activity but inhibits degradation of polyubiquitylated proteins (Table 1, data not shown, and references [25;26]). To determine whether Rip-1 mimicked other features of ATPγS on proteasome function, we examined its effects on ATPase activity of purified 26S proteasome and on PA700 binding to and activation of the 20S proteasome. At Rip-1 concentrations that stimulated 26S proteasome peptidase activity by over 2.5-fold, ATPase activity was unaffected (Table 1). Moreover, unlike ATPγS, Rip-1 did not promote 26S proteasome assembly (and concomitant 20S proteasome activation) from component subcomplexes. Thus, although Rip-1 mimicked the stimulatory effects of ATPγS and polyubiquitin on 26S proteasome peptidase activity, it was distinct from each with respect to its effects on other aspects of proteasome function. These results indicate that proteasomal gating can be mediated by multiple mechanisms.
Table 1.
Relative effects of Rip-1 and ATPγS on proteasome functions.
| Assay | Control | + Rip-1 | + ATPγS |
|---|---|---|---|
| 26S proteasome ATPase (pmol Pi/min/pmol) | 12.2 +/− 2.5 | 14.5 +/− 4.3 | 3.6 +/− 0.8 |
| 26S proteasome activity (arbitrary FU) | 35.4 +/− 2.4 | 72.7 +/− 9.9 | 116 +/− 15 |
| PA700-dependent activation of 20S proteasome activity (arbitrary FU) | 14.2 +/− 4.7 | 13.1 +/− 0.9 | 198 +/− 13 |
The peptoid, Rip-1 was tested for effects on indicated functions of purified proteasomes as described in Experimental Procedures. Results are expressed as mean values (+/− SD) of triplicate (ATPase) or quadruplicate (26S proteasome activity and PA700-dependent activation of 20S proteasome activity) assays. Similar results were obtained in at least two independent experiments. For this experiment, Rip-1 and ATPγS were used at 20 μM concentrations, but no qualitative differences of their effects occurred over a wide range of concentrations.
DISCUSSION
The results presented here support a model in which the substrate access portal of the 26S proteasome can be variably modulated in a manner manifested as variable rates of peptide substrate hydrolysis. The 26S proteasome degrades short peptides by a mechanism that requires neither substrate modification by ubiquitin nor ATP hydrolysis [28;37]. Therefore, the degradation of these peptides reflects their ability to diffuse through the substrate access pores opened as a consequence of binding of C-termini of certain Rpt subunits of PA700 to cognate sites on the outer rings of the 20S proteasome. Although the binding of PA700 to the 20S proteasome required for gating depends on ATP binding to Rpt subunits, isolated C-terminal peptides of these subunits bind to and activate the proteasome in an ATP-independent fashion [17;37]. Thus, ATP-dependent activation of the proteasome probably involves an ATP binding-induced conformation of PA700 that orients C-terminal peptides of Rpt subunits for competent proteasome interaction. Recently we showed that the stimulatory effects of C-terminal peptides of different activating Rpt subunits were additive suggesting that proteasomal gating was not a simple two-state process (i.e. open and closed), but instead could be modulated in a graded fashion. Although the binding of individual isolated Rpt peptides to the proteasome is an artificial condition that may not mimic conditions achieved by intact PA700 (where all activating subunits probably bind simultaneously), these results demonstrate the capacity for variable gating of the proteasome. The current results reveal that variable gating is also a feature of the intact 26S proteasome and identify several factors that affect the process.
Non-hydrolyzable ATP analogs stimulated the peptidase activity of intact 26S proteasome. Thus, peptidase activity of 26S proteasome can be enhanced above that achieved by gating promoted upon complex assembly. One model consistent with this finding is that the ATP-bound state of 26S proteasome features a conformation optimized for substrate transit and ATP hydrolysis returns this conformation to a suboptimized state (Figure 6). Because altered conformations of Rpt subunits of PA700 are likely transmitted to the α proteasome subunits with which they are in contact, substrate transit could be affected by altered positions of the N-terminal α subunit gates or by other features of subunit conformation that affect the caliber of the access pore (Figure 6). A recent electron microscopy study of 26S proteasome indicated that PA700 promotes radial displacement of α subunits of the proteasome [7]. Thus, is it reasonable to imagine that effectors of the conformation of Rpt subunits can promote additional changes in the geometry of the substrate access pore. Such effects may be important for processing of polyubiquitylated substrates, whose degradation, in contrast to that of peptide substrates, requires ATP hydrolysis. Thus, toggling conformations of the 26S proteasome caused by rounds of ATP binding and hydrolysis could be a means of coordinating temporal alterations in processive substrate transit with other processes required for proteolysis such as substrate unfolding and deubiquitylation (Figure 6).
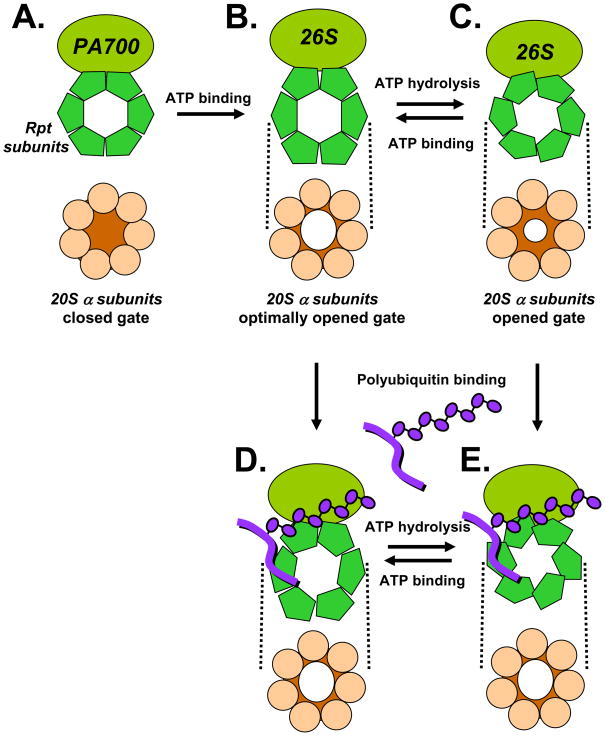
Figure 6. Model for variable modulation of gating of the 26S proteasome by ATP and polyubiquitin.
A. The 20S proteasome has low catalytic activity because substrate entry pores in the center of outer α rings are occluded by closed gates. B. Binding of ATP to Rpt subunits of PA700 induces conformational changes that orient C-termini of these subunits for binding to α subunits of 20S proteasome, resulting in 26S proteasome formation. PA700 binding alters the conformation of proteasome α subunits, resulting in opening of substrate access pores. C. Subsequent hydrolysis of ATP alters the conformation of Rpt subunits and transmits these changes to bound α subunits, promoting a suboptimized open gate. Rounds of ATP binding and hydrolysis cycle conformations between maximally open (ATP bound) and suboptimally open conformations. Polyubiquitin binds to PA700 directly (D and E) or via a reversibly associated polyubiquitin binding protein such as Rad23 (not shown). Polyubiquitin-induced alteration of PA700 conformation is transmitted to a subunits of the proteasome. In the ATP bound state, polyubiquitin binding produces no additional effect on gating (D), but in the suboptimal state increases gating the optimal state (E). Figures depict end-on views of 20S proteasome and Rpt subunits of PA700. Dashed lines indicate axial binding of these rings to one another in a plane perpendicular to the page.
In addition to nucleotides, peptide hydrolysis by the 26S proteasome also was stimulated by polyubiquitin. Neither monoubiqutin nor the Ubl domain of a protein that interacts with PA700 produced this effect. Polyubiquitin composed of a linear head-to-tail fusion of five ubiquitin monomers, which in separate experiments we have found to be a poor signal for proteolysis of attached protein substrates (X. Li and G.N. DeMartino, unpublished data), also failed to stimulate peptidase activity. Although most of our experiments were conducted with unanchored polyubiquitin chains composed of K48-linkages, we achieved similar results with proteolysis-susceptible polyubiquitylated-Sic, which was produced by a method that resulted in chains with a high percentage of K63 linkages. Thus, for the cases examined here, peptidase activity was stimulated only by forms of ubiquitin that supported protein degradation. While our work was under review, Bech-Otschir et al reported similar effects of polyubiquitin on stimulation of 26S proteasome peptidase activity [2]. In contrast to our results however, they failed to observe comparable effects of non-hydrolyzable ATP analogs. The basis for this discrepancy is unclear. Finally, polyubiquitin presented to the proteasome via a polyubiquitin chain-binding protein such as hRad23B stimulated peptidase activity as well as polyubiquitin itself. These results indicate either that polyubiquitin per se was responsible for this effect upon transfer from hRad23B to the proteasome or that polyubiquitin conferred proteasome stimulatory capacity to hRad23B when bound. Because the isolated Ubl domain of Rad23 did not interfere with polyubiquitin stimulation, polyubiquitin and hRad23B appear to interact at different sites on the proteasome.
The similar effects of ATPγS, polyubiquitin, and Rip-1 on increased 26S proteasome peptidase activity did not correlate with a correspondingly uniform effect on ATPase activity, which was inhibited, stimulated, and unaffected by these agents, respectively. Such results suggest that these various agents increased gating by different mechanisms. Additional evidence for the different actions of ATPγS on one hand and polyubiquitin and Rip-1 on the other comes from their different abilities to promote 26S assembly and activation from component subcomplexes, which ATPγS promoted, but polyubiquitin and Rip-1 did not (Table 1). ATPγS likely binds to Walker nucleotide binding motifs of Rpt subunits, whereas polyubiquitin binds to the 26S proteasome at several sites including subunits Rpn13 and Rpn10 [10;22;35;38]. Although Rip-1 binds specifically to Rpt4, it probably does not do so via nucleotide binding sites [24]. Thus, despite the distinct interactions of these different agents with the proteasome, each could produce functionally similar effects by promoting conformational changes in PA700 that are translated into enhanced opening of substrate access pores.
The discussion provided above assumes that rates of peptide hydrolysis are faithful monitors of proteasome gating. In the limited cases where proteasome activation has been correlated with detailed structural information, this assumption seems justified [15;31;40]. However, peptide hydrolysis could be affected by mechanisms independent of altered gating per se, such as allosteric effects that alter function of the proteasome’s catalytic sites. Our preliminary kinetic data give no evidence for such mechanisms as the basis of effects described here, but additional work will be required to confirm this assumption rigorously.
Acknowledgments
We thank David Thompson, Tsui-Ling Chang, and Kerry Wooding for help with specific experiments, Brajesh Kumar and Thomas Gillette for reagents and helpful advice, and Thomas Kodadek for supplying Rip-1.
FUNDING
This work was supported by grants from the National Institutes of Health (R01 DK 46181) and the Welch Foundation (I-500) to GND.
Reference List
- 1.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 2.Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhutter HG, Dahlmann B, Kloetzel PM. Nat Struct Mol Biol. 2009;16:219–225. doi: 10.1038/nsmb.1547. [DOI] [PubMed] [Google Scholar]
- 3.Beyer A. Protein Science. 1997;6:2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Madura K. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coux O, Tanaka K, Goldberg AL. Ann Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 6.Craiu A, Gaczynska M, Akapian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 7.da Fonseca PC, Morris EP. J Biol Chem. 2008 doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMartino GN. Methods Enzymol. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- 9.DeMartino GN, Moomaw CR, Zagnitko OP, Proske RJ, Ma CP, Afendis SJ, Swaffield JC, Slaughter CA. J Biol Chem. 1994;269:20878–20884. [PubMed] [Google Scholar]
- 10.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 11.Dubiel W, Ferrell K, Pratt G, Rechsteiner M. J Biol Chem. 1992;267:22699–22702. [PubMed] [Google Scholar]
- 12.Elasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 13.Elsasser S, Schmidt M, Finley D. Meth Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- 14.Ferrell K, Wilkinson CRM, Dubiel W, Gordon C. Trends Biochem Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- 15.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Gillette TG, DeMartino GN. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glickman MH, Rubin DM, Fried VA, Fischer JE, Finley D. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MN, Finley D. Nat Struct Biol. 2001;11:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 20.Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 21.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 22.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 24.Lim HS, Archer CT, Kim YC, Hutchens T, Kodadek T. Chem Commun (Camb) 2008:1064–1066. doi: 10.1039/b717861a. [DOI] [PubMed] [Google Scholar]
- 25.Lim HS, Archer CT, Kodadek T. J Am Chem Soc. 2007;129:7750–7751. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim HS, Cai D, Archer CT, Kodadek T. J Am Chem Soc. 2007;129:12936–12937. doi: 10.1021/ja075469+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CW, Giasson BI, Lewis KA, Lee VM, DeMartino GN, Thomas PJ. J Biol Chem. 2005;280:22670–22678. doi: 10.1074/jbc.M501508200. [DOI] [PubMed] [Google Scholar]
- 28.Liu CW, Li X, Thompson D, Wooding K, Chang T, Tang Z, Yu H, Thomas PJ, DeMartino GN. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma CP, Slaughter CA, DeMartino GN. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 30.Ma CP, Vu JH, Proske RJ, Slaughter CA, DeMartino GN. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 31.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rassi S, Pickart CM. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 33.Saeki Y, Isono E, Toh-e A. Meth Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M, Hanna J, Elsasser S, Finley D. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 35.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DM, Kafri G, Cheng Y, Ng D, Wala T, Goldberg AL. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 38.van Nocker S, Deveraux Q, Rechsteiner M, Vierstra RD. Proc Natl Acad Sci (USA) 1996;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voges D, Zwickl P, Baumeister W. Ann Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 40.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Nature. 2001;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]