Abstract
In this review, we describe the history of amniote sex determination as a classic example of Darwinian evolution. We suggest that evolutionary changes in sex determination provide a foundation for understanding important aspects of chromosome and genome organization that otherwise appear haphazard in their origins and contents. Species with genotypic sex determination often possess heteromorphic sex chromosomes, whereas species with environmental sex determination lack them. Through a series of mutations followed by selection at key genes, sex-determining mechanisms have turned over many times throughout the amniote lineage. As a consequence, amniote genomes have undergone gains or losses of sex chromosomes. We review the genomic and ecological contexts in which either temperature-dependent or genotypic sex determination has evolved. Once genotypic sex determination emerges in a lineage, viviparity and heteromorphic sex chromosomes become more likely to evolve. For example, in extinct marine reptiles, genotypic sex determination apparently led to viviparity, which in turn facilitated their pelagic radiation. Sex chromosomes comprise genome regions that differ from autosomes in recombination rate, mutation rate, levels of polymorphism, and the presence of sex-determining and sexually antagonistic genes. In short, many aspects of amniote genome complexity, life history, and adaptive radiation appear contingent on evolutionary changes in sex-determining mechanisms.
Key Words: Amniotes, Archosauromorpha, Hemizygosity, Heterogamety, Pseudoautosomal region, Sexual antagonism, Viviparity
Sex-Determining Mechanisms Change Over Time
Sex-determining mechanisms appear to have changed several times over at least the last 300 million years of amniote history [Reisz, 1997; van Tuinen and Hadly, 2004]. In this review, we describe the relevance of these changes to genome structure and dynamics. For example, genotypically sex-determined species typically have sex chromosomes and temperature-dependent sex-determined species lack them. Here, we describe contexts that divide species with different sex-determining mechanisms and the genomic consequences of that division. At present, 3 reviews have characterized changes in the amniote lineage from genotypic sex determination (GSD) to temperature-dependent sex determination (TSD) and from TSD to GSD [Janzen and Krenz, 2004; Organ and Janes, 2008; Pokorna and Kratochvil, 2009]. Phylogenetic study of sex-determining mechanisms is challenged by questions regarding commonalities and hence homoplasies among those mechanisms. Since the molecular targets affected by temperature in TSD organisms have not yet been identified, researchers have been hesitant to conclude that all incidences of TSD indicate the same molecular action. Similar concerns are attached to the study of GSD. The molecular interactions involved with GSD are also poorly described in different amniote groups.
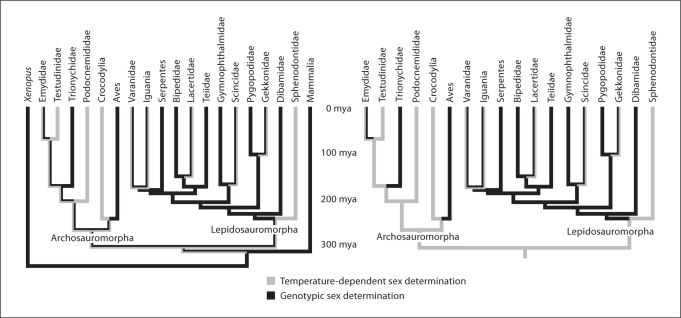
Species may have common organizations of sex chromosomes (i.e., Z and W sex chromosomes (female heterogamety) as seen in birds or X and Y sex chromosomes (male heterogamety) as seen in most mammals). Yet, these chromosomes may have little or nothing in common. For example, Z and W sex chromosomes are found in both snakes and birds, but the 2 pairs of sex chromosomes share no sequence similarity or recent evolutionary history [Matsubara et al., 2006]. Recent phylogenetic studies explain this lack of homology as a consequence of independent origins of female heterogamety in snakes and birds [Organ and Janes, 2008]; but how many independent origins of sex chromosomes and sex-determining mechanisms have occurred? A large research community is addressing this question with a multi-pronged approach that includes phylogenetic reconstruction, comparative mapping and population genetics, and analyses of expression of sex-determining and sex-differentiating genes. Synthesis of these approaches will enable researchers to answer important questions about a system that appears, in part, to have been conserved over long spans of time and also to have evolved repeatedly and reversibly between 2 basic modes (GSD including female and male heterogamety and TSD) multiple times (fig. 1).
Fig. 1.
The evolution of sex-determining mechanisms in reptiles. Clades were included to highlight the major divisions of reptiles. Both trees are identical except the tree on the left has outgroups Mammalia and Xenopus whereas the tree on the right used no outgroup in ancestral state reconstruction. Choice of outgroup affects number and kind of changes in reptilian sex-determining mechanisms as reconstructed by BayesTraits (http://www.evolution.rdg.ac.uk/BayesTraits.html). Note that several species-rich groups within Aves and Serpentes are entirely genotypically sex-determined with a ZZ/ZW sex chromosome system while many lepidosauromorph groups show a variety of sex-determining mechanisms.
Inferences of ancestral sex-determining mechanisms (SDM) in reptiles have not resulted in clear answers regarding ancestral conditions and direction of evolutionary shift between SDMs because of uncertain estimations of the true reptile tree, the lability of SDM, and the selection of outgroups in the analysis (fig. 1). For example, the Sphenodontia (the group to which extant and extinct tuataras belong) diverged from the lineage of snakes and lizards early, about 230 mya, but now the clade is almost entirely extinct [Miller et al., 2006]. The number of extant species is low and the phylogenetic branch leading to them is long, especially for characterization of a trait that readily changes. Therefore, extant tuataras have an unreasonably and probably inappropriately large influence on phylogenetic reconstructions of SDM in amniotes. Additionally, the phylogenetic placement of turtles has been contentious [Near et al., 2005; Shedlock et al., 2007], although for the case illustrated in figure 1, the reconstructions of SDM do not change appreciably if the clade Reptilia is more heavily sampled taxonomically or if turtles are moved from Archosauromorpha to the base of the tree.
Why Do Sex-Determining Mechanisms Change?
Sex-determining mechanisms appear to change in response to environmental changes and/or intragenomic degeneration. For example, extant pelagic amniotes such as sea snakes and cetaceans universally exhibit viviparity as well as GSD, and fossil evidence demonstrates viviparity in several extinct marine reptiles [Caldwell and Lee, 2001]. For these reasons, extinct marine reptiles like ichthyosaurs, mosasaurs, and sauropterygians most likely also exhibited GSD [Organ et al., 2009]. Temperature-dependent sex determination would likely have precluded the ability of these species to be viviparous as maternal body temperatures, especially in deep ocean environments, would be too constant to incubate both male and female offspring in response to temperature. Extant marine reptiles such as sea turtles are not obligatorily pelagic. Their body plan allows them to exit the water and deposit gas-exchanging eggs on nesting beaches. However, the body plan of mosasaurs and other extinct pelagic reptiles did not permit transport of eggs to nesting beaches and amniotic eggs would not effectively exchange gases underwater. Therefore, we have hypothesized that GSD enabled viviparity and viviparity, in turn, facilitated adaptive radiation of pelagic amniotes [Organ et al., 2009].
In addition to changes in habitat such as the transition from land to water, there are also environmental shifts in which local population sex ratios and resource availability could cause changes in intersexual differences in fitness. According to the Charnov-Bull hypothesis, TSD could allow parents finer control over offspring sex ratios in environments where intersexual differences in fitness make TSD adaptive [Charnov and Bull, 1977]. Empirical support for this model was provided by Warner and Shine [2008] in the form of enhanced fitness of males incubated at the natural temperature most likely to yield male offspring over fitness of males produced at other temperatures in jacky dragons, Amphibolurus muricatus, a TSD lizard. The same pattern was reported for females incubated at the optimal female-producing temperature and females incubated at other temperatures. These studies suggest that SDMs change in response to environmental variation.
Intragenomic changes that may influence SDM involve the origin and degeneration of sex chromosomes. Sexually antagonistic genes are genes that are beneficial to one sex but detrimental to the other sex [Arnqvist and Rowe, 2005]. The adaptive significance of sex chromosomes appears to be that they allow the sequestration of sexually antagonistic loci so that they are passed from parents either only to sons or daughters, depending on which sex carries the sex-specific chromosome [Charlesworth et al., 2005]. Once sequestered on a nascent sex chromosome, sexually antagonistic genes can arise through the inhibition of recombination. Hemizygous regions – regions present in only one copy in the heterogametic sex – by necessity lack recombination. Such regions can arise by exaptation of a previously existing gene to a novel and sexually antagonistic function or by translocation of a sexually antagonistic locus to a nascent sex chromosome [Graves, 2001]. The cessation of recombination that comes with hemizygosity leads to the accumulation of deleterious mutations in the absence of gene conversion or copy correction [Rice, 1987; Skaletsky et al., 2003; Graves, 2006] ultimately resulting in a deleterious mutation load that leads to the dissolution of a sex chromosome. This conclusion has been invoked to explain the absence of a Y chromosome in the mole vole, Ellobius lutescens, and other mammals [Just et al., 1995]. It is possible that the demise of a sex chromosome due to purged excess of deleterious mutations leads to the loss of GSD and an opportunity for SDM shift [Graves, 2006; Mank and Avise, 2009].
Consequences of SDM Shift
Characteristics of Species with Environmental Sex Determination
Species in which offspring sex is primarily determined by incubatory environment may find themselves more vulnerable to environmental changes than their GSD counterparts. If clutch sex ratios are determined by nest temperatures and nest temperatures are in turn determined by climate, then sex ratios could change as a result of rapid climate change. For example, nest temperatures are significantly changed by invasive plants that overshadow nesting areas of Nile crocodiles, Crocodylus niloticus, a TSD reptile [Leslie and Spotila, 2001]. Also, many TSD species, such as tuatara, Sphenodon guntheri, nest in regions where global warming is expected to affect nesting temperature within the next century [Mitchell et al., 2008]. For TSD species at risk of negative effects of climate change, options are available. These species must choose among several challenging options: they could migrate to an area that is more amenable to their SDM; they could alter their nesting phenology or SDM sensitivity to temperature; or they could undergo a genomic shift to GSD [Huey and Janzen, 2008; Schwanz and Janzen, 2008].
Genomic Connections between Sexual Antagonism and Sexual Selection via GSD
Once a lineage shifts to GSD and subsequently possesses sex chromosomes, sequence linked to the sex chromosomes exhibit population genetic dynamics that differ from those exhibited by sequence on autosomes [Ellegren, 2009]. Sex chromosomes consist of pseudoautosomal regions (PAR) that continue to recombine between pairs of homologous sex chromosomes and hemizygous sex-specific regions that carry sexually antagonistic loci and no longer recombine. Within a genome, sex-specific regions occur in different effective population sizes than autosomes. For example, in birds such as chicken, Gallus gallus, females carry a Z and a W sex chromosome that contain one or more PARs as well as Z- and W-specific sequence, whereas males carry 2 Z sex chromosomes. In an avian population with a balanced sex ratio, effective populations of chromosome types occur in a ratio of 4 autosomes, 3 Z sex chromosomes, and one W sex chromosome. In birds, for example, these ratios are known to affect a number of population genetic parameters affecting sex chromosomes, such as the level of standing genetic diversity. Sex-specific regions of sex chromosomes typically exhibit lower diversity than autosomes as reported in chicken [Berlin and Ellegren, 2004; Sundstrom et al., 2004], and pseudoautosomal regions have been shown to recombine more frequently than autosomes as reported in emus, Dromaius novaehollandiae, as well as humans, Homo sapiens [Lien et al., 2000; Janes et al., 2009]. In addition, sex linkage presents opportunities for sexual antagonism. For example, in chicken, genes that are detrimental to females but neutral to males are significantly overrepresented on the Z chromosome [Mank and Ellegren, 2009]. Mank et al. [2007] described a fast-Z effect in chicken in which Z-linked protein-coding genes evolve faster than autosomal genes. In the collared flycatcher, Ficedula albicollis, a number of plumage-defining genes have been mapped to avian Z chromosomes [Saetre et al., 2003]. Z-linkage of these genes and the evidence for faster rates of evolution on avian Z chromosomes suggest that runaway selection of male ornamentation is a direct result of linkage to a sex chromosome [Saetre et al., 2003, but see Mank et al., 2006]. In addition, 6 genes associated with sex-differential song memory acquisition in zebra finch, Taeniopygia guttata, have been mapped to the Z chromosome [Tomaszycki et al., 2009]. Opportunities for sexual antagonism and faster rates of evolution appear to be afforded by sex linkage. The connection between sex linkage and sexually selected plumage and song seen in birds is further supported by the observation that sexual dimorphism is weak to nonexistent in TSD species such as alligators and sea turtles which lack sex chromosomes. In summary, sex chromosomes enabled by GSD appear to have resulted in rapid evolution of sex differences and sexually selected traits.
Genotypic Sex Determination Increases Genome Complexity
In this review, we define genome complexity in terms of the number of characterized genome compartments, including autosomes, pseudoautosomal regions, and hemizygous regions. The causes and consequences of increased genome complexity in GSD, not TSD, species are described in figure 2. Some GSD species appear not to have detectable sex chromosomes, but this may be a consequence of inadequate cytological techniques. For example, Ezaz et al. [2005] and Martinez et al. [2008] recently reported previously unidentified sex chromosomes in bearded dragons, Pogona vitticeps, and Macquarie turtles, Emydura macquarii, respectively. As protocols improve, sex chromosomes are likely to be discovered in more GSD species that are currently described as lacking them. Species that lack sex chromosomes, regardless of SDM, likely have fewer opportunities for sexual antagonisms [Mank and Ellegren, 2009]. TSD species in particular exhibit no sex-specific regions but instead exhibit varying patterns of sex-determining response to incubation temperature. For example, in leopard geckos, Eublepharis macularius, a TSD lizard, the SDM of different matrilineal lines responded differently to incubation temperatures that reproducibly yield consistent offspring sex ratios. This experiment separated clutches of eggs into different experimental treatment groups, thereby controlling for maternal effects. Variable responses to incubation temperature of the SDMs of different matrilineal lines in this TSD lizard suggest a gene by environment interaction that is indicative of polygenic inheritance [Janes and Wayne, 2006]. In light of the finding that TSD in leopard geckos is polygenic, we suggest that TSD does not depend on the function of one sex-linked factor such as DMRT1 in birds or SRY in mammals [Sinclair et al., 1990; Smith et al., 2009] but rather on a rapidly evolving network of environmentally sensitive genes. In light of interspecific differences in sex-determining response to incubation temperature among TSD reptiles, we conclude that similar experimentation with other TSD reptiles should be conducted and is expected to support our characterization of the genetic underpinnings of TSD.
Fig. 2.
A flowchart describing possible changes in sex-determining mechanism (SDM) among amniotes. Novel sex-determining (SD) gene(s) can override temperature-dependent sex determination (TSD) or pre-existing genotypic sex determination (GSD). Once GSD evolves, genome complexity increases as illustrated here in the grey shaded box. This box illustrates genome characteristics that arise subsequent to the evolution of GSD by this model.
Conclusions within Reach
The presence or absence of sex chromosomes likely explains a great deal of the evolutionary innovations seen in animals that possess them and not seen in animals that lack them. Furthermore, within GSD species, evolution from presumably homomorphic (cryptic) sex chromosomes to partially and fully differentiated sex chromosomes adds another important level of complexity in genome evolution, but one that as yet cannot be adequately understood in amniotes given the lack of data on sex chromosomes in reptiles. There is evidence that suggests that tempo and mode of evolution change at different organizational scales of the genome. For instance, variation in the reptilian karyotype is consistent with a model of early rapid evolution and anagenesis, while variation in global genome size is best described by continuous gradual evolution [Organ et al., 2008]. These different evolutionary modes for different compartments and levels of the genome suggest that the dynamics at one level may be largely independent of dynamics at another.
Theoretical linkages between shifts in SDM and traits such as sexually dimorphic gene expression, male and female ornamentation, and speciation should be investigated in greater detail [Kirkpatrick and Hall, 2004]. In addition, further work is required on the mechanisms underlying and interconnections between GSD and TSD. For example, the discovery of the sex-reversing effect of incubation temperature in a lizard that exhibits sex chromosomes makes the distinction of GSD and TSD less clear [Quinn et al., 2007]. For this reason, further empirical support is required for characterization of TSD and GSD as either distinct mechanisms under different genetic controls or as a series of conserved sex-determining genes with different sensitivities to temperature and other environmental variables [Sarre et al., 2004]. One way to attack this question is through gene expression studies, and all would agree that current descriptions of cascades of sex-differentiating genes are incomplete [Janes et al., 2008a; Valenzuela, 2008]. For example, differences in expression profiles of sex-determining and sex-differentiating genes between TSD and GSD species should be studied via high-throughput sequencing of transcripts retrieved from embryonic and gonadal tissues of TSD and GSD species. High-throughput sequencing should identify novel sexually dimorphic expression patterns and missing elements in the chain of up-regulated and down-regulated elements that feed back upon each other [Janes et al., 2008b]. Knock-down and knock-out research models in TSD species would be extremely valuable but are likely to be technically challenging [Thomas and Capecchi, 1987]. A more feasible approach would be to recognize relevant sex-determining and sex-differentiating genes in a TSD species and prepare a knock-down chicken or some other closely related species that is more pliable at the bench [Sinclair, A.H., personal communication].
A clear and exhaustive description of sex determination will be possible only after more thorough investigation of the molecular basis of sex determination and differentiation. How similar are the SDMs of distantly related TSD species? What genomic or expression patterns can be discerned between pairs of sex chromosomes even if no sequence is shared? Does male or female heterogamety evolve multiple times in response to the same ecological pressure(s) or is each case of GSD unique in its derivation? In this review, we have characterized relationships between the environment and large-scale genome evolution. The environment influences sex-determining mechanisms that, in turn, influence genome complexity. In this way, sex determination offers one of the best examples of contingent and hierarchical evolution. In the wake of new genome resources [Janes et al., 2008b], answers to these questions and a more detailed history of this Darwinian model are within reach.
Acknowledgements
We thank Ettore Olmo for the opportunity to contribute to this issue. Our conversations with Dr. Olmo regarding this effort began with a symposium at the 2008 meeting of the Society for Integrative and Comparative Biology funded by 2 divisions within SICB: Developmental and Cell Biology and Evolutionary Developmental Biology. The National Science Foundation (IOB 0809547) also supported the symposium. Postdoctoral support for D.E.J. was provided by NSF grant MCB-0817687 to N. Valenzuela and S.V.E. and NIH #5F32GM072494. C.L.O. was supported by NIH postdoctoral fellowship #5F32GM075490. This manuscript benefited from comments from Christopher Balakrishnan, Ryan Kearney, Stephen Sarre, and Andrew Sinclair.
References
- Arnqvist G, Rowe L. Sexual Conflict. Princeton: Princeton University Press; 2005. [Google Scholar]
- Berlin S, Ellegren H. Chicken W: a genetically uniform chromosome in a highly variable genome. Proc Natl Acad Sci USA. 2004;101:15967–15969. doi: 10.1073/pnas.0405126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MW, Lee MSY. Live birth in Cretaceous marine lizards (mosasauroids) Proc Biol Sci. 2001;268:2397–2401. doi: 10.1098/rspb.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Charnov EL, Bull J. When is sex environmentally determined? Nature. 1977;266:829–830. doi: 10.1038/266828a0. [DOI] [PubMed] [Google Scholar]
- Ellegren H. The different levels of genetic diversity in sex chromosomes and autosomes. Trend Genet. 2009;25:278–284. doi: 10.1016/j.tig.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Quinn AE, Miura I, Sarre SD, Georges A, Graves JAM. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13:763–776. doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- Graves JAM. From brain determination to testis determination: evolution of the mammalian sex-determining gene. Reprod Fertil Dev. 2001;13:665–672. doi: 10.1071/rd01093. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Huey RB, Janzen FJ. Climate warming and environmental sex determination in tuatara: the last of the Sphenodontians? Proc Biol Sci. 2008;275:2181–2183. doi: 10.1098/rspb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes DE, Wayne ML. Evidence for a genotype × environment interaction in sex-determining response to incubation temperature in the leopard gecko, Eublepharis macularius. Herpetologica. 2006;62:56–62. [Google Scholar]
- Janes DE, Ezaz T, Graves JAM, Edwards SV. Characterization, chromosomal location, and genomic neighborhood of a ratite ortholog of a gene with gonadal expression in mammals. Integrat Comp Biol. 2008a;48:505–511. doi: 10.1093/icb/icn024. [DOI] [PubMed] [Google Scholar]
- Janes DE, Organ C, Valenzuela N. New resources inform study of genome size, content, and organization in nonavian reptiles. Integrat Comp Biol. 2008b;48:447–453. doi: 10.1093/icb/icn010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes DE, Ezaz T, Graves JAM, Edwards SV. Recombination and nucleotide diversity in the sex chromosomal pseudoautosomal region of the emu, Dromaius novaehollandiae. J Heredity. 2009;100:125–136. doi: 10.1093/jhered/esn065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen FJ, Krenz JG. Phylogenetics: which was first, TSD or GSD? In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington DC: Smithsonian Books; 2004. p. 121.p. 130. [Google Scholar]
- Just W, Rau W, Vogel W, Akhverdian M, Fredga K, et al. Absence of Sry in species of the vole Ellobius. Nat Genet. 1995;11:117–118. doi: 10.1038/ng1095-117. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Hall DW. Sexual selection and sex linkage. Evolution. 2004;58:683–691. doi: 10.1111/j.0014-3820.2004.tb00401.x. [DOI] [PubMed] [Google Scholar]
- Leslie AJ, Spotila JR. Alien plant threatens Nile crocodile (Crocodylus niloticus) breeding in Lake St. Lucia, South Africa. Biol Conserv. 2001;98:347–355. [Google Scholar]
- Lien S, Szyda J, Schechinger B, Rappold G, Arnheim N. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am J Hum Genet. 2000;66:557–566. doi: 10.1086/302754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Evolutionary Diversity and turn-over of sex determination in teleost fishes. Sex Dev. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution. 2009;63:1464–1472. doi: 10.1111/j.1558-5646.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hall DW, Kirkpatrick M, Avise JC. Sex chromosomes and male ornaments: a comparative evaluation in ray-finned fishes. Proc Biol Sci. 2006;273:233–236. doi: 10.1098/rspb.2005.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: Rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez PA, Ezaz T, Valenzuela N, Georges A, Graves JAM. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: A new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 2008;16:815–825. doi: 10.1007/s10577-008-1228-4. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, et al. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HC, Belov K, Daugherty CH. MHC class I genes in the tuatara (Sphenodon spp.): evolution of the MHC in an ancient reptilian order. Mol Biol Evo. 2006;23:949–956. doi: 10.1093/molbev/msj099. [DOI] [PubMed] [Google Scholar]
- Mitchell NJ, Kearney MR, Nelson NJ, Porter WP. Predicting the fate of a living fossil: how will global warming affect sex determination and hatching phenology in tuatara? Proc Biol Sci. 2008;275:2185–2193. doi: 10.1098/rspb.2008.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Meylan PA, Shaffer HB. Assessing concordance of fossil calibration points in molecular clock studies: An example using turtles. Am Naturalist. 2005;165:137–146. doi: 10.1086/427734. [DOI] [PubMed] [Google Scholar]
- Organ CL, Janes DE. Evolution of sex chromosomes in Sauropsida. Integr Comp Biol. 2008;48:512–519. doi: 10.1093/icb/icn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Moreno RG, Edwards SV. Three tiers of genome evolution in reptiles. Integr Comp Biol. 2008;48:494–504. doi: 10.1093/icb/icn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Janes DE, Meade A, Pagel M. Genotypic sex determination enabled adaptive radiations of extinct marine reptiles. Nature. 2009;461:389–392. doi: 10.1038/nature08350. [DOI] [PubMed] [Google Scholar]
- Pokorna M, Kratochvil L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linnean Soc. 2009;156:168–183. [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JAM. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411–411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Reisz RR. The origin and early evolutionary history of amniotes. Trend Ecol Evol. 1997;12:218–222. doi: 10.1016/s0169-5347(97)01060-4. [DOI] [PubMed] [Google Scholar]
- Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre GP, Borge T, Lindroos K, Haavie J, Sheldon BC, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc Biol Sci. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarre SD, Georges A, Quinn A. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. Bioessays. 2004;26:639–645. doi: 10.1002/bies.20050. [DOI] [PubMed] [Google Scholar]
- Schwanz LE, Janzen FJ. Climate change and temperature-dependent sex determination: can individual plasticity in nesting phenology prevent extreme sex ratios? Physiol Biochem Zool. 2008;81:826–834. doi: 10.1086/590220. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Botka CW, Zhao SY, Shetty J, Zhang TT, et al. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genorne. Proc Natl Acad Sci USA. 2007;104:2767–2772. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Fairlie PG, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Sundstrom H, Webster MT, Ellegren H. Reduced variation on the chicken Z chromosome. Genetics. 2004;167:377–385. doi: 10.1534/genetics.167.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Tomaszycki ML, Peabody C, Replogle K, Clayton DF, Tempelman RJ, Wade J. Sexual differentiation of the zebra finch song system: potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N. Evolution of the gene network underlying gonadogenesis in turtles with temperature-dependent and genotypic sex determination. Integr Comp Biol. 2008;48:476–485. doi: 10.1093/icb/icn031. [DOI] [PubMed] [Google Scholar]
- van Tuinen M, Hadly EA. Error in estimation of rate and time inferred from the early amniote fossil record and avian molecular clocks. J Mol Evol. 2004;59:267–276. doi: 10.1007/s00239-004-2624-9. [DOI] [PubMed] [Google Scholar]
- Warner DA, Shine R. The adaptive significance of temperature-dependent sex determination in a reptile. Nature. 2008;451:566–568. doi: 10.1038/nature06519. [DOI] [PubMed] [Google Scholar]