Abstract
The epidermal growth factor receptor (EGFR) is an important chemotherapeutic target for tyrosine kinase inhibitors and antibodies that block the extracellular domain of EGFR. Betulinic acid (BA) and curcumin inhibited bladder cancer cell growth and downregulated specificity protein (Sp) transcription factors, and this was accompanied by decreased expression of EGFR mRNA and protein levels. EGFR, a putative Sp-regulated gene, was also decreased in cells transfected with a cocktail (iSp) containing small inhibitory RNAs for Sp1, Sp3 and Sp4, and RNA interference with individual Sp knockdown indicated that EGFR expression was primarily regulated by Sp1 and Sp3. BA, curcumin and iSp also decreased phosphorylation of Akt in these cells and downregulation of EGFR by BA, curcumin and iSp was accompanied by induction of LC3 and autophagy which is consistent with recent studies showing that EGFR suppresses autophagic cell death. The results show that EGFR is an Sp-regulated gene in bladder cancer, and drugs such as BA and curcumin that repress Sp proteins also ablate EGFR expression. Thus, compounds such as curcumin and BA that downregulate Sp transcription factors represent a novel class of anticancer drugs that target EGFR in bladder cancer cells and tumors by inhibiting receptor expression.
Keywords: EGFR suppression, Sp transcription factors, curcumin, betulinic acid
INTRODUCTION
Sp transcription factors are critical for early embryonic development in mouse models; however, there is evidence that expression of Sp1 decreases with age in humans and laboratory animal models (1–6). Several different cancer cell lines overexpress Sp1, Sp3 and Sp4 proteins including breast cancer cell lines (7–11); however, in immortalized but not transformed MCF10A cells, expression of these proteins was significantly decreased (11). Similar differences were observed in human prostate tumors (xenografts) in athymic nude mice and mouse liver (9), and ongoing studies in mouse tissue/organs including proliferative gastrointestinal tissue and bone marrow confirm the low to non-detectable expression of Sp1, Sp3 and Sp4 in mature mice. Moreover, clinical studies show that overexpression of Sp1 in gastric and pancreatic cancer correlates with poor survival (12, 13) and pancreatic tumor aggressiveness (14). Differences in expression of Sp proteins in tumor vs. non-tumor tissue suggests that these transcription factors are potential targets for cancer chemotherapy, particularly since regulation of several pro-oncogenic genes including survivin, cyclin D1, VEGF, VEGFR1 and VEGFR2 are Sp-dependent (8–10). Anticancer drugs such as curcumin and betulinic acid (BA) act, in part, by decreasing expression of Sp1, Sp3 and Sp4 in bladder and prostate tumors, respectively, and the low toxicity of these compounds suggest that their effects on Sp proteins are specific for cancer cells and tumors (9, 10).
The epidermal growth factor receptor (EGFR) plays a critical role in cellular homeostasis (15, 16); however, EGFR and other ErbB members are frequently activated in many tumor types and this is due to several factors including activating mutations, gene amplifications, overexpression of the receptor and/or its cognate ligands and loss of inhibitory factors that regulate receptor activity (17–19). Enhanced EGFR activity in cancer cells and tumors is associated with increased growth, survival and angiogenesis of tumors and thereby contributes significantly to the phenotypic characteristics of cancer cells (17–19). Not surprisingly, EGFR has become a major target for cancer chemotherapy and development of two major classes of anti-EGFR agents, namely, monoclonal antibodies against the extracellular domain of EGFR and low molecular weight drugs that competitively inhibit ATP binding to the intracellular tyrosine kinase domain.
Bladder tumors also overexpress EGFR and ligands for this receptor (20, 21), and clinical applications of EGFR blocking agents such as the tyrosine kinase inhibitor gefitinib in combination with other drugs are underway or in development (22, 23). Regulation of EGFR expression in cancer and non-cancer cell lines is complex and cell context-dependent. For example, early growth response-1 (Egr-1) enhances basal and hypoxia-induced EGFR expression in human osteosarcoma U2OS and SaOS-2 and cervical cancer HeLa cells (24). However, in some cancer cell lines, EGFR expression is dependent on Sp1 (25, 26), and we hypothesized that the anticancer activity of curcumin and BA in bladder cancer cells may be due, in part, to downregulation of Sp proteins and EGFR. Our results show that BA and curcumin decrease EGFR expression in bladder cancer cells through downregulation of Sp1 and Sp3 transcription factors and this represents a novel pathway for targeting EGFR in cancer cells and tumors.
MATERIALS AND METHODS
Cell Lines
KU7 and 253JB-V human bladder cancer cells were provided by Dr. A. Kamat, (M.D. Anderson Cancer Center, Houston, TX). 253JB-V and KU7 cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 0.15% sodium bicarbonate, 0.011% sodium pyruvate, 0.24% HEPES and 10 ml/L of antibiotic/antimycotic cocktail solution (Sigma Aldrich, St. Louis, MO). Cells were grown in 150 cm2 culture plates in an air/CO2 (95:5) atmosphere at 37°C and passaged approximately every 3 days.
Antibodies, Chemicals and Other Materials
Sp1 (PEP2), Sp3 (D-20), Sp4 (V-20), VEGF (147), Survivin (FL-142), AKT (sc-8312), p-AKT (sc-7985-R) and EGFR (1005) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cleaved PARP (ASP 214), p-MAPK (197G2) and MAPK (137F5) antibody was purchased from Cell Signaling Technology (Danvers, MA) and SGLT antibody was purchased from Abcam Inc, (Cambridge, MA) (ab7970-1). LC3 antibody was purchased from MBL International Corporation, (Woburn, MA). Monoclonal β-actin antibody was purchased from Sigma-Aldrich. Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA). BA was purchased from Sigma-Aldrich, curcumin (98% pure) was purchased from Indofine Chemical Company, Inc. (Hillsborough, NJ) and gefitinib (>99% pure) was obtained from LC Laboratories (Woburn, MA). The GFP-LC3 plasmid was kindly provided by Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan).
Cell Proliferation Assays
Bladder cancer cells (3 × 104 cells per well) were seeded using DMEM: Ham’s F-12 medium containing 2.5% charcoal-stripped FBS in 12-well plates and left to attach for 24 h. Cells were then treated with either vehicle (DMSO) or the indicated concentrations of BA, curcumin and gefitinib. Fresh medium and test compounds were added every 24 h for curcumin, BA and gefitinib. Cells were then counted at the indicated times using a Coulter Z1 particle counter. Each experiment was done in triplicate and results are expressed as means ± SE for each determination. The concentration of epidermal growth factor (EGF) used to induce cell proliferation was 100 ng/ml.
Western Blot Assays
Bladder cancer cells were seeded in DMEM: Ham’s F-12 medium containing 2.5% charcoal-stripped FBS. Twenty-four h later, cells were treated with either vehicle (DMSO) or the indicated compounds for 48 h. Cells were collected using high-salt buffer (50 mmol/L HEPES, 0.5 mol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 10% glycerol, and 1% Triton-X-100, pH 7.5) and 10μL/mL of Protease Inhibitor Cocktail (Sigma Aldrich). The lysates were incubated on ice for 1 h with intermittent vortexing every 10 min, followed by centrifugation at 20,000 g for 10 min at 4°C. Lysates were then incubated for 3 min at 100°C before electrophoresis, and then separated on 10% SDS-PAGE 120 V for 3 to 4 h in 1X running buffer (25 mM tris-base, 192 mM glycine, and 0.1% SDS). Proteins were transferred onto polyvinylidene difluoride (PVDF) membranes by wet electroblotting in a buffer containing 25 mmol/L Tris, 192 mmol/L glycine, and 20% methanol for 1.5 h at 0.9 A at 4°C. The membranes were blocked for 30 min with 5% TBST-Blotto [10 mmol/L Tris-HCl, 150 mmol/L NaCl (pH 8.0), 0.05% Triton X-100, and 5% nonfat dry milk] and incubated in fresh 5% TBST-Blotto with 1:200–1:1000 primary antibody overnight with gentle shaking at 4°C. After washing with TBST for 10 min, the PVDF membrane was incubated with secondary antibody (1:5000) in 5% TBST-Blotto for 2 h by gentle shaking. The membrane was washed with TBST for 10 min, incubated with 6 mL of chemiluminescence (PerkinElmer Life Sciences, Waltham, MA) substrate for 1.0 min, and exposed to Kodak X-OMAT AR autoradiography film (American X-ray supply Inc, Jackson, CA). Quantification of the proteins was done using Image J software and the optical densities were plotted after normalization with lamin/β-actin.
siRNA Interference Assay
The two bladder cancer cell lines, 253JB-V and KU7 were seeded (1 × 105 per well) in 6-well plates in DMEM: Ham’s F-12 medium supplemented with 2.5% charcoal-stripped FBS without antibiotic and left to attach for one day. The triple Sp siRNA knockdown (iSp1, iSp3, iSp4 complex) along with iLamin as control was performed using Liopfectamine reagent according to the manufacturer’s instructions. Small inhibitory RNAs were prepared by Dharmacon RNA Technologies (Chicago, IL). The iRNA complexes used in this study are indicated as follows:
| LMN | 5′ - CUG GAC UUC CAG AAG AAC ATT |
| Sp1 | SMARTpool L-026959-00-0005 |
| Sp3 | 5′ - GCG GCA GGU GGA GCC UUC ACU TT |
| Sp4 | 5′ - GCA GUG ACA CAU UAG UGA GCT T |
Real-Time PCR
Total RNA was isolated using the RNeasy Protect Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNA was eluted with 30 μL of RNase-free water and stored at −80°C. RNA was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. cDNA was prepared from the 253JB-V and KU7 bladder cancer cell lines at different time intervals using a combination of oligodeoxythymidylic acid and dNTP mix (Applied Biosystems, Foster City, CA) and Superscript II. Each PCR was carried out in triplicate in a 20 μL volume using SYBR Green Master mix (Applied Biosystems) for 15 min at 95°C for initial denaturing, followed by 40 cycles of 95°C for 30 s and 60°C for 1 min in the ABI Prism 7700 sequence detection system (Applied Biosystems). The ABI Dissociation Curves software was used after a brief thermal protocol (95°C for 15 s and 60°C for 20 s, followed by a slow ramp to 95°C) to control for multiple species in each PCR amplification. The comparative CT method was used for relative quantitation of samples. Primers were purchased from Integrated DNA Technologies (Coralville, IA). The sequences of primers for EGFR were 5′ - TTT CGA TAC CCA GGA CCA AGC CAC AGC AGG - 3′ and 5′ - AAT ATT CTT GCT GGA TGC GTT TCT GTA - 3′. Values for each gene were normalized to expression levels of TATA-binding protein. The sequences of the primers used for TATA-binding protein were: 5′-TGC ACA GGA GCC AAG AGT GAA-3′ (sense) and 5′-CAC ATC ACA GCT CCC CAC CA-3′ (antisense).
Transfection and Luciferase Assays
The luciferase construct of EGFR containing five Sp1 binding sites (PER6-Luc) was a kindly provided by Dr. A.C Johnson (NCI-NIH, Bethesda, MD). Bladder cancer cells (1 × 105 per well) were plated in 12-well plates in DMEM: Ham’s F-12 medium supplemented with 2.5% charcoal-stripped FBS. After 16–24 h, various amounts of DNA (i.e., 0.4 μg pGL3; 0.04 μg β-galactosidase; and 0.4 μg PER6-Luc) were transfected using Lipofectamine reagent according to the manufacturer’s protocol and luciferase activity (normalized to β-galactosidase) was determined essentially as described.
Electrophoretic Mobility Shift Assay
Cells were rinsed in cold PBS buffer and harvested in reporter lysis buffer (Promega). After 15-min incubation on ice and 10-min centrifugation at 16,000 × g, 4°C, the pellet was resuspended in reporter lysis buffer supplemented with 0.5 mol/L KCl and incubated on ice for 60 min. The supernatant containing nuclear proteins was collected after centrifugation for 10 min at 16,000 × g, 4°C and quantified for protein concentrations by Bradford method. The GC-rich probe was prepared by annealing the two complementary polynucleotides: 5′-CTC GTC GGC CCC CGC CCC TCT-3′ and 5′-AGA GGG GCG GGG GCC GAC GAG-3′. The EGFR strand was 5′-AGC TTC GCG TCC GCC CGA GTC CCC GCC TCG CCG CCA ACG CCA-3′ and mutant EGFR 5′-AGC TTC GCG TCC GCC CGA GTC TTT GTC TCG CCG CCA ACG CCA-3′. The annealed probe was 5′-end–labeled using T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP (Perkin-Elmer). The labeled probe was purified with the Chroma Spin TE-10 column (BD Biosciences). The electrophoretic mobility shift assay reaction was carried out by incubating 10 μg of nuclear extract with binding buffer (25 mM HEPES, 1.5 mM EDTA, 1.0 mM DDT, 2 mM magnesium chloride, 10% glycerol, 100 mM KCl at pH 7.6) in the presence of 1 μg of poly(dI-dC) (Roche Molecular Biochemicals) with or without unlabeled competitor oligonucleotides, and 10 fmol of labeled probe. The mixture was incubated for 15 min on ice. Protein-DNA complexes were resolved by 5% native PAGE at 160 V at room temperature for 1.5 h. The gels were then dried and visualized by autoradiography.
Fluorescence Microscopy and GFP-LC3 Localization
Monolayers of cells were cultured for 24 h in 2-well coverglass chamber slides in medium containing 10% serum and treated as indicated. The GFP-LC3 plasmid was kindly provided by Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan). KU7 cell lines were transfected with 1 μg/well GFP-LC3 plasmid using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Slides were examined by fluorescence microscopy using Zeiss Stallion Dual Detector Imaging System (Carl Zeiss Microimaging Inc., Thornwood, NY). The intracellular distribution of GFP-LC3 was evaluated by monitoring GFP-LC3 and DIC images throughout the entire thickness of the cell by optical slices at 0.5 μM intervals using a C-Apochromat 63X, 1.2 NA water immersion lens. Digital images were acquired using Slide Book software (Intelligent Imaging Innovations, Denver, CO). The entire z-stack was subjected to fluorescence deconvolution to remove out of plane fluorescence. Cells were examined in more than five fields per slide on multiple slides and the number of punctae per cell was determined by microscopic analysis. The GFP-LC3 punctate dot structures in individual live cells were imaged and quantitated using a fluorescence microscope (Olympus IX70 inverted fluorescent light microscope system) equipped with a digital camera (Olympus DP70 digital camera system). The number of GFP-LC3 punctate dots per cells in GFP-LC3-positive cells was determined. A minimum of 15 cells per sample were counted per condition per experiment. Results (mean number of punctae per cell) are expressed as mean ± SD for combined data from the representative of 3 independent experiments.
Staining for Acridine Orange
253JB-V and KU7 bladder cancer cells were seeded in monolayers in 2.5% serum containing medium and at 70% confluence, cells were untreated or treated with 10 μM BA and 40 μM curcumin for various time points. At the appropriate time points, cells were incubated with 1 μg/ml acridine orange (Molecular Probes, Eugene, OR) in serum-free medium for 15 min. The acridine orange was removed and fluorescence images were obtained before and after removing the dye. The cytoplasm and nucleus of the stained cells fluoresced bright green, whereas the acidic autophagic vacuoles fluoresced bright red.
Statistical Analysis
Statistical significance of differences was determined by analysis of variance and student t-test, and the levels of probability were noted. All statistical tests were two-sided.
RESULTS
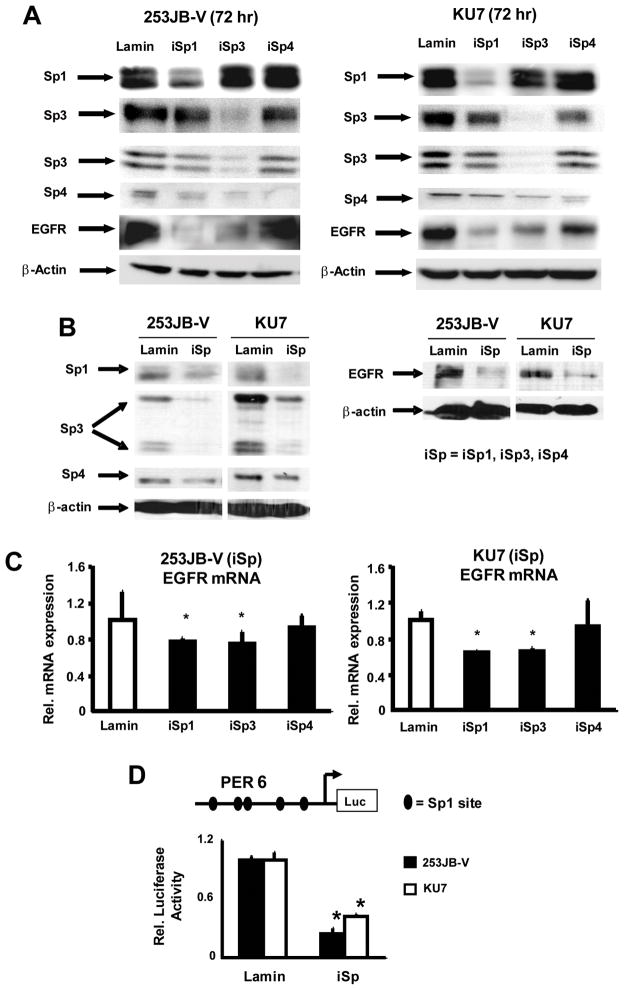
The epidermal growth factor receptor is overexpressed or constitutively active in bladder and other cancer cell lines and tumors, and expression of this gene/gene product can be regulated by multiple factors including Sp transcription factors (25, 26). The role of Sp proteins in regulating EGFR expression in bladder cancer cells was investigated by RNA interference using small inhibitory RNAs for Sp1 (iSp1), Sp3 (iSp3), and Sp4 (iSp4) or a cocktail of iSp1, iSp3 and iSp4 combined (iSp) which simultaneously decrease expression of all three transcription factors as previously described (10). Figure 1A illustrates knockdown of the individual Sp proteins in both bladder cancer cell lines and the results show that only iSp1 and iSp3 significantly decrease EGFR protein levels. Figure 1B demonstrates that transfection of 253JB-V and KU7 cells with the iSp cocktail decreased Sp1, Sp3 and Sp4 and EGFR protein levels confirming that expression of this receptor tyrosine kinase is Sp-dependent in bladder cancer cells and this is consistent with the multiple GC-rich Sp binding sites identified in the EGFR promoter (25, 26). However, results of RNA interference which knocks down individual Sp proteins show that EGFR is primarily regulated by Sp1 and Sp3 (Fig. 1A). Moreover, in a parallel experiment, we also show by RNA interference that knockdown of Sp1 or Sp3 but not Sp4 decreased EGFR mRNA levels (Fig. 1C). We also investigated the effects of Sp knockdown in 253JB-V and KU7 cells cotransfected with PER6-luc and iSp or non-specific iLamin (Fig. 1D). Knockdown of Sp proteins significantly decreased luciferase activity demonstrating that expression of the EGFR promoter was also Sp-dependent.
FIGURE 1.
Regulation of EGFR in bladder cancer cells is Sp-dependent. Downregulation of Sp1, Sp3, Sp4 and EGFR proteins in bladder cancer cells transfected with iLamin (control), small inhibitory RNAs for Sp1 (iSp1), Sp3 (iSp3), Sp4 (iSp4) (A) or iSp (combined iSp1, iSp3 and iSp4) (B). Cells were transfected with the appropriate oligonucleotide and after 72 h, whole cell lysates were obtained and analyzed by western blots as outlined in the Materials and Methods. Similar results were observed in replicate (2–3) experiments. C. Effects of iSp1, iSp3 and iSp4 on EGFR mRNA levels. 253JB-V and KU7 cells were transfected with iSp1, iSp3, iSp4 or iLamin (control) and mRNA was isolated from these cells and EGFR mRNA was determined by real-time PCR as described in the Materials and Methods. Results are expressed as means ± SE for 3 separate experiments and significantly (p < 0.05) decreased EGFR mRNA levels (relative to iLamin set at 1.0) are indicated (*). D. Effects of iSp on EGFR promoter expression. Cells were transfected with PER6 and iSp or iLamin (non-specific), and luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SE for 3 replicate determinations and significantly (P < 0.05) decreased activity after Sp knockdown is indicated (*).
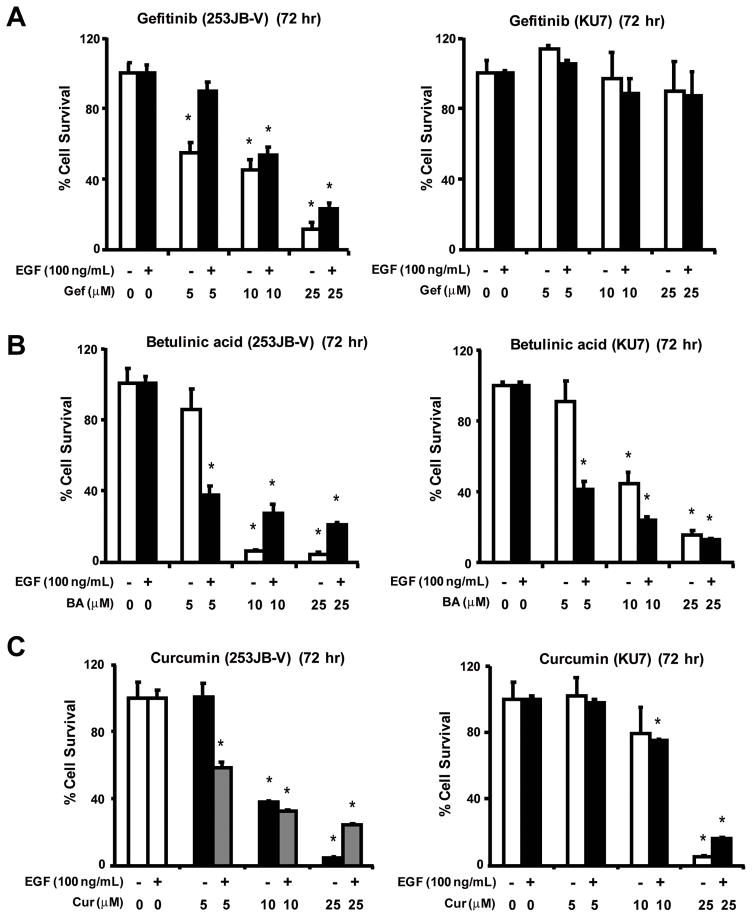
In previous studies, we reported that BA and curcumin inhibit growth and decrease Sp1, Sp3, Sp4 and Sp-dependent genes in prostate and bladder cancer cells, respectively (9, 10). Since EGFR plays an important role in the growth of bladder cancer cells, we hypothesized that the anticarcinogenic activity of BA and curcumin may be due, in part, to downregulation of EGFR and other Sp-dependent genes. Results illustrated in Figure 2 show that both BA and curcumin inhibited basal and EGF-induced growth of 253JB-V and KU7 bladder cancer cell growth, whereas gefitinib, the clinically used EGFR tyrosine kinase, inhibitor inhibited basal and EGF-induced proliferation of 253JB-V but not KU7 cells. The differential gefitinib-responsiveness of these bladder cancer cell lines has previously been reported (27).
FIGURE 2.
Inhibition of bladder cancer cell growth. 235JB-V and KU7 cells were treated with gefitinib (A), betulinic acid (B), or curcumin (C) for 72 h in the presence or absence of EGF (100 ng/ml), and the effects of the treatments on cell proliferation were determined as described in the Materials and Methods. Results are expressed as means ± SE for at least 3 replicate experiments per treatment group, and a significant (P < 0.05) decrease in cell proliferation is indicated by an asterisk.
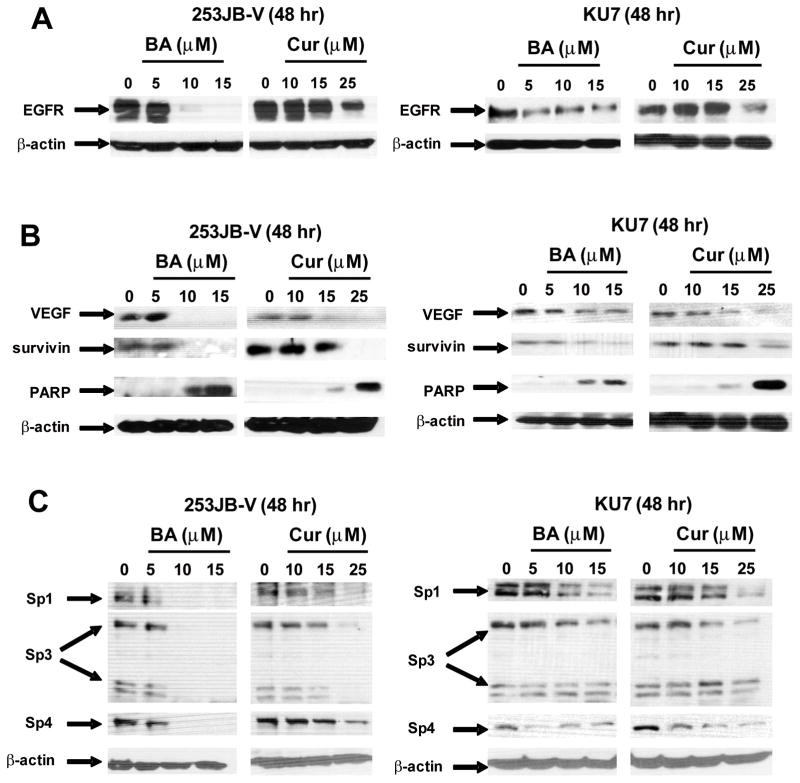
Results in Figures 3A and 3B show that after treatment of 253JB-V and KU7 cells with BA or curcumin for 48 h, there was a decrease in EGFR protein expression and this was accompanied by a parallel decrease in other Sp-dependent genes/proteins, namely VEGF and survivin, and also induction of cleaved PARP (Figs. 3A and 3B). Moreover, treatment of 253JB-V and KU7 cells with BA or curcumin for 48 h also decreased expression of Sp1, Sp3 and Sp4 proteins (Figs. 3C). These results confirm that like curcumin (10), BA also decreased Sp proteins and Sp-dependent proteins in bladder cancer cells and show for the first time that both BA and curcumin decrease expression of EGFR protein, an important target for bladder cancer chemotherapy (17–19). In contrast, the tyrosine kinase inhibitor gefitinib did not affect EGFR or Sp protein expression in these cell lines (data not shown) and decreased growth of 253JB-V but not KU7 cells corresponding to the reported gefitinib-responsiveness and -nonresponsiveness of these cell lines (27).
FIGURE 3.
BA and curcumin decrease EGFR and Sp proteins and Sp-dependent genes. Compound-induced repression of EGFR (A) and other Sp-dependent proteins (B) in 253JB-V and KU7 cells. Cells were treated with DMSO or different concentrations of the compounds for 48 h, and whole cell lysates were analyzed by western blots as described in the Materials and Methods. C. Compound-induced repression of Sp1, Sp3 and Sp4 proteins in 253JB-V and KU7 cells. Protein expression was determined as outlined above in (A). Results (A–D) were observed in replicate experiments (at least 3).
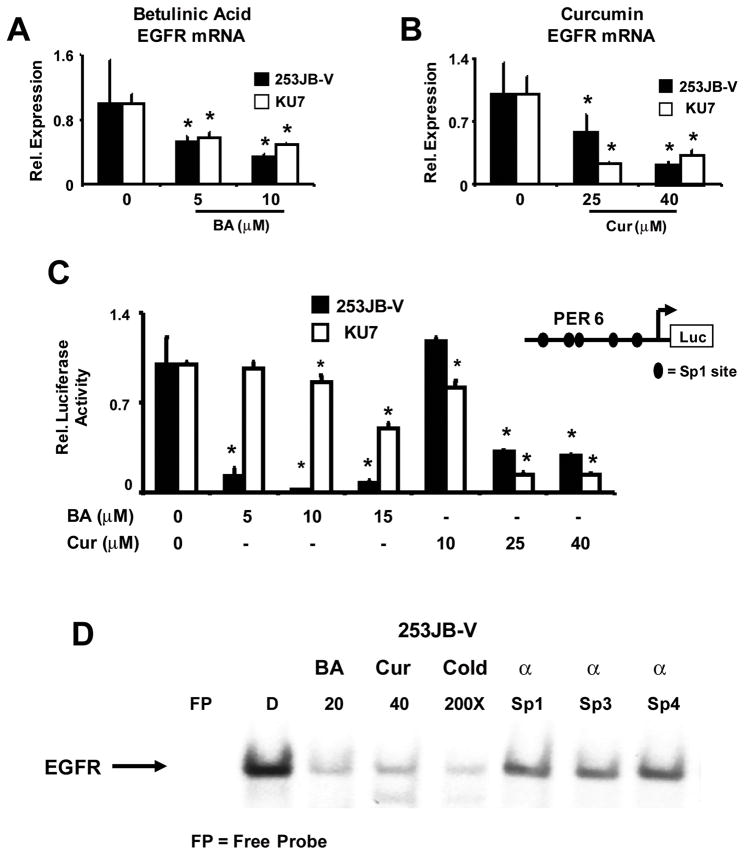
Treatment of 253JB-V and KU7 cells with 5 or 10 μM BA for 24 h also significantly decreased EGFR mRNA levels (Fig. 4A). Moreover, treatment with 25 or 40 μM curcumin also significantly decreased EGFR mRNA levels in both bladder cancer cell lines (Fig. 4B). BA- and curcumin-dependent inhibition of EGFR transcription was further investigated in bladder cancer cells transfected with PER6-luc, a construct which contains the −771 to −16 region of the EGFR promoter and many of the proximal GC-rich sites are located in this region of the promoter (Fig. 4C). Both compounds significantly decreased luciferase activity in 253JB-V and KU7 cells and these results confirm that curcumin and BA inhibit EGFR transcription. The effects of BA on downregulation of luciferase activity in KU7 required relatively high concentrations, suggesting that for BA additional cis-elements may also be important. In contrast, gefitinib did not affect EGFR transcription in these cell lines (data not shown). The EGFR promoter contains GC-rich Sp binding sites and we used the EGFR oligonucleotide containing the −112 to −77 GC-rich EGFR sequence in EMSA assays to investigate the effects of BA and curcumin on Sp protein binding to the EGFR promoter (Fig. 4D). Incubation of nuclear extracts from 253JB-V cells with the GC-rich oligonucleotide formed a retarded band complex, whereas decreased binding was observed using extracts from cells treated with 20 μM BA or 40 μM curcumin. Competition with unlabeled EGFR or a consensus GC-rich oligonucleotide decreased retarded band formation, and antibody experiments showed immunodepletion with Sp1 and Sp3 antibodies, and similar results were noted with Sp4 antibodies. Supershifted bands were not observed. Similar results were obtained using nuclear extracts from KU7 cells, thus confirming that BA- and curcumin-dependent downregulation of Sp1, Sp3 and Sp4 proteins decreases Sp binding to the GC-rich region of the EGFR promoter.
FIGURE 4.
BA and curcumin decrease EGFR expression in bladder cancer cells. BA (A) and curcumin (B) decrease EGFR mRNA levels. Cells were treated with DMSO or different concentrations of BA or curcumin for 24 h, and EGFR mRNA levels were determined by RT-PCR as described in the Materials and Methods. Results are expressed as means ± SE for 3 replicate determinations for each treatment group and significantly (P < 0.05) decreased expression (relative to DMSO) is indicated (*). C. Decreased EGFR promoter activity. Cells were transfected with PER6 and treated with DMSO, BA or curcumin, and luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SE for 3 replicate determinations and significant (P < 0.05) downregulation relative to DMSO is indicated (*). D. Gel shift assay. Nuclear extracts from cells treated with DMSO, BA or curcumin were incubated with a 32P-labeled GC-rich probe derived from the EGFR promoter, and gel mobility shift assays were determined as described in the Materials and Methods. Competition assays used 100-fold less of unlabeled oligonucleotide.
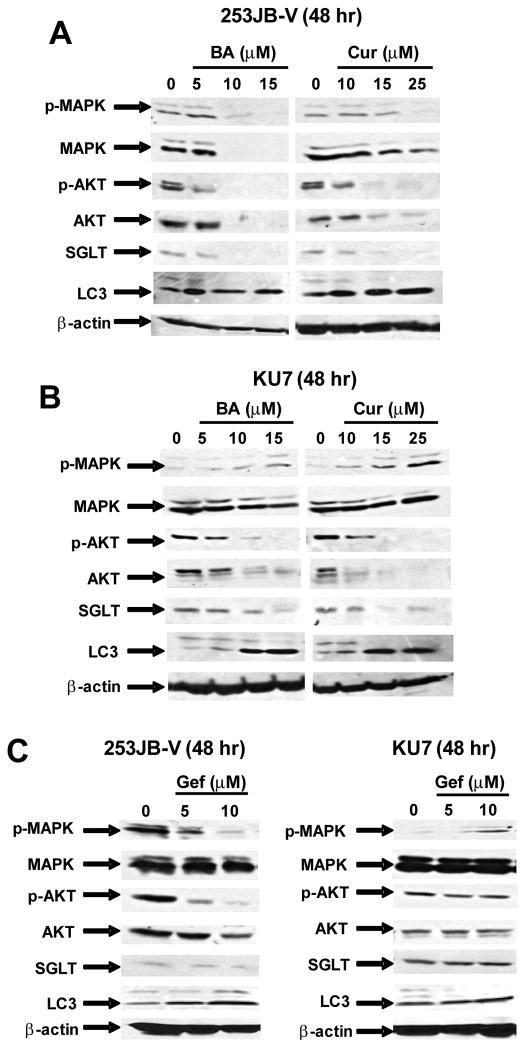
EGFR regulates multiple genes and pathways through activation of downstream kinases such as PI3-K and MAPK. Figure 5A summarizes the effects of BA and curcumin on Akt/phospho-Akt and MAPK/phospho-MAPK expression in 253JB-V cells. Cells treated with 10 or 15 μM BA and 25 μM curcumin decreased constitutive phospho-MAPK expression; however, the same concentrations of BA also decreased levels of MAPK protein, whereas curcumin had minimal effects on MAPK protein levels. Both BA and curcumin also decreased phospho-Akt in 253JB-V cells and this was accompanied by decreased Akt protein. In KU7 cells, BA and curcumin increased levels of phospho-MAPK but did not affect MAPK protein and both compounds decreased phospho-Akt and Akt protein expression (Fig. 5B). Thus, in the gefitinib-resistant cells, BA and curcumin inhibited the PI3-K and not the MAPK signaling pathways.
FIGURE 5.
Modulation of putative EGFR-dependent responses. Effects of BA and curcumin on EGFR-dependent effects in 253JB-V (A) and KU7 (B) cells compared to effects of gefitinib (C) in both cell lines. Cells were treated with DMSO or different concentrations of BA, curcumin or gefitinib for 48 h, and whole cell lysates were analyzed by western blot analysis as described in the Materials and Methods. Similar results were observed in replicate (2) experiments.
EGFR also enhances cancer cell survival by inhibition of autophagic cell death in breast cancer cells through stabilization of the sodium/glucose cotransporter 1 (SGLT1) and this response is independent of the kinase activity of this receptor (28). Results in Figures 5A and 5B also demonstrate that after treatment of 253JB-V and KU7 cells with BA or curcumin, there was a decrease in SGLT1 protein expression in both cell lines. Moreover, this was also accompanied by induction of LC3 which is a protein biomarker of autophagy (29). Thus, knockdown of EGFR in bladder cancer cells after treatment with curcumin or BA inhibited both EGFR kinase-dependent (PI3-K) and kinase-independent (SGLT downregulation and LC3 induction) survival pathways. Gefitinib also decreases expression of phospho-Akt and phospho-MAPK in 253JB-V cells (Fig. 5C) which is consistent with inhibition of EGFR tyrosine kinase activity by this compound. However, in KU7 cells, gefitinib did not affect expression of phospho-Akt and induced phospho-MAPK which is consistent with previous studies showing that this cell line is gefitinib-resistant (21). Gefitinib did not affect SGLT or LC3 protein expression in either cell line which is in contrast to the effects of BA and curcumin (Figs. 5A and 5B).
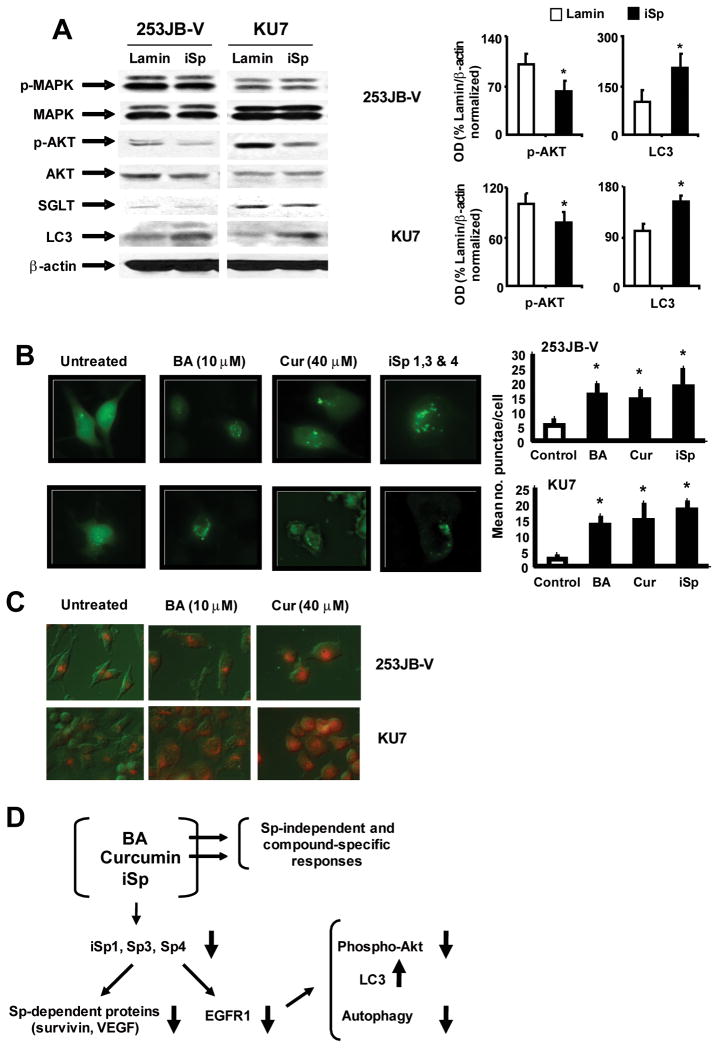
Knockdown of Sp proteins by RNA interference in KU7 and 253JB-V cells decreased EGFR expression (Fig. 1); however, the effects of Sp knockdown on EGFR kinase-dependent and -independent pathways has not previously been investigated. Results in Figure 6A show that in 253JB-V and KU7 cells transfected with iSp, phospho-MAPK, MAPK or Akt expression was not changed, whereas phospho-Akt protein levels were decreased and this was similar to the effects of curcumin and BA on phospho-Akt in these cell lines, suggesting that this response is EGFR-dependent (Figs. 5A and 5B). SGLT expression was not affected, whereas LC3 was induced in 253JB-V and KU7 cells transfected with iSp (Fig. 6A). Induction of LC3, a protein biomarker for autophagy was observed after ablation of EGFR by treatment of 253JB-V and KU7 cells with curcumin and BA (Figs. 5A and 5B) or transfection with iSp (Fig. 6A), and this was observed in a recent study showing that downregulation of EGFR (by RNA interference) induced autophagy in prostate and breast cancer cells (28). This was further confirmed in 253JB-V and KU7 cells transfected with the GFP-LC3 construct. In untreated cells, there was a diffuse pattern of green fluorescence throughout the cells (Fig. 6B); however, after treatment with 10 μM BA and 40 μM curcumin or transfection with iSp, a punctate fluorescent staining was observed and this is characteristic of cells undergoing autophagy (30). Quantitation of the number of punctae per cell showed that BA, curcumin and iSp significantly increased punctae formation in both cell lines.
FIGURE 6.
Sp protein knockdown decreases EGFR-dependent PI3K signaling and induces autophagy. A. Effects of iSp on kinases and autophagy-related proteins and quantitation of p-Akt and LC3. Whole cell lysates from bladder cancer cells transfected with iSp were analyzed by western blots as described in the Materials and Methods. Quantitation of phospho-Akt and LC3 proteins in cells transfected with iSp was determined as a % of Lamin/β-actin protein ratios from 3 replicate western blot analyses as indicated in and described in the Materials and Methods. Results are expressed as means ± SE and significant (P < 0.05) increases or decreases in the iSp transfected compared to the iLamin transfected groups are indicated (*). B. Induction of punctuate green fluorescence in 253JB-V and KU7 cells transfected with GFP-LC3. Cells cotransfected with iSp (or iLamin as control) and GFP-LC3 were treated with DMSO, BA or curcumin for 18 h and the fluorescence associated with GFP-LC3 was determined as described in the Materials and Methods. Punctae per cell were quantitated and results are expressed as means ± SE for 3 replicate determinations. Significant (P < 0.05) induction of punctae is indicated (*). C. Induction of autophagy. Induced acridine orange staining in 253JB-V and KU7 cells. Cells were treated with DMSO (untreated), 10 μM BA, or 40 μM curcumin for 18 h, and detection of autophagic vacuoles by acridine orange staining was determined as described in the Materials and Methods. Observations were typical of replicate experiments. D. Model for the effects of BA, curcumin and iSp in bladder cancer cells.
Further confirmation that BA, curcumin and Sp knockdown by RNA interference induced autophagy in 253JB-V and KU7 cells is illustrated in Figure 6C. Compared to DMSO (untreated controls), BA, curcumin and iSp induced acridine orange staining which is consistent with formation of acidic autophagic vacuoles (autophagolysomes) which are characteristically observed in autophagic cells (31). These results clearly demonstrate that BA- and curcumin-dependent downregulation of EGFR and this results in the loss of EGFR-dependent kinase activity (decreased phospho-Akt) and upregulation of LC3 and autophagy which are also EGFR-regulated (suppressed) responses (Fig. 6D) and these effects are also observed after knockdown of Sp1, Sp3 and Sp4 by RNA interference. BA and curcumin also affect other genes and responses in bladder and other cancer cell lines that are due to loss of Sp proteins and to Sp-independent compound-specific responses (Fig. 6D).
DISCUSSION
EGFR and ErbB2/HER2 are frequently overexpressed or constitutively activated in multiple tumor types. Monoclonal antibodies against EGFR (Cetuximab) and ErbB2 (Trastuzumab) are used alone or in combination for cancer chemotherapy, and applications of other antibodies are also being investigated [reviewed in (18, 19)]. Tyrosine kinase inhibitors for EGFR and ErbB2 have been identified and these include gefitinib, erlotinib, lapatinib and others which are used as single agents or in combination with other drugs for treatment of multiple cancers (17–19). Initial studies of kinase inhibitors in non-small cell lung cancer (NSCLC) patients observed minimal treatment benefits (32–35) and administration of gefitinib after radiotherapy did not improve survival of NSCLC patients (36). In contrast, treatment of NSCLC patients with tyrosine kinase inhibitors was highly successful for subsets of patients expressing EGFR kinase domain mutations (36–39) and similar results were observed for gefitinib in lung cancer cell lines (39). However, in bladder tumors and cancer cell lines, differential gefitinib responsiveness is not dependent on these kinase-domain mutants since wild-type EGFR expression is predominant in bladder tumors (40).
Initial studies confirmed the differential responsiveness of 253JB-V and KU7 cells to gefitinib (15 μM) which inhibited basal and EGF-induced proliferation of 253JB-V but not KU7 cells (Fig. 2). In contrast, BA and curcumin inhibited basal and EGF-induced growth of both cancer cell lines, and 253JB-V cells were slightly more sensitive to these compounds. Both bladder cancer cell lines expressed EGFR and are responsive to the mitogenic effects of EGF and we hypothesized that the anticarcinogenic activity of BA and curcumin may be due, in part, to downregulation of EGFR. This hypothesis was also based on the fact that both BA and curcumin decrease Sp1, Sp3 and Sp4 proteins in prostate and bladder cancer cells, respectively (9, 10), and EGFR expression in some cancer cell lines is also dependent on these transcription factors (25, 26). Sp2 expression in cancer cells is highly variable (data not shown) and this transcription factor does not bind GC-rich sequences and regulate prototypical Sp1(3/4)-dependent genes. Figure 1 illustrates that simultaneous knockdown of Sp1, Sp3 and Sp4 in 253JB-V and KU7 cells also decreases EGFR protein expression and EGFR promoter activity which is consistent with previous studies showing that GC-rich Sp binding sites were important for basal expression of EGFR (25, 26). However, in contrast to previous results with VEGF, VEGFR1 and VEGFR2, EGFR expression was regulated by Sp1 and Sp3 but not Sp4, demonstrating differential effects of these transcription factors on gene regulation. Figure 3A illustrates that EGFR protein was also decreased in 253JB-V and KU7 cells after treatment with BA or curcumin. Moreover, both compounds also decreased Sp1, Sp3 and Sp4 expression in 253JB-V and KU7 cells and this was accompanied by decreased expression of other Sp-dependent genes (survivin and VEGF) and induction of cleaved PARP (Figs. 3B and 3C). BA and curcumin also decreased EGFR mRNA levels and luciferase activity in 253JB-V and KU7 cells transfected with the EGFR promoter construct PER6 (Fig. 4), whereas the tyrosine kinase inhibitor gefitinib did not affect any of these responses (data not shown). These results, coupled with decreased binding of nuclear extracts from bladder cancer (treated with BA or curcumin) to GC-rich sequences from the EGFR promoter (Fig. 4D) indicate that the effects of BA and curcumin on EGFR are also Sp-dependent in bladder cancer cells and this correlates with the RNA interference studies summarized in Figure 1.
EGFR is an important receptor tyrosine kinase that regulates multiple kinase pathways (15–17) and a comparison of the effects of gefitinib, BA and curcumin in gefitinib-responsive 253JB-V indicates that all three agents decreased EGFR-dependent phosphorylation of MAPK and Akt (Figs. 5A and 5C). In contrast, BA and curcumin but not gefitinib decreased phospho-Akt levels in KU7 cells, and BA and curcumin increased phospho-MAPK expression in KU7 cells, whereas minimal effects were observed for gefitinib. These responses, coupled with the downregulation of Akt protein (KU7 and 253JB-V cells) by BA and curcumin and MAPK protein by BA only in 253JB-V cells, may be associated with effects of these compounds that are independent of their downregulation of Sp or EGFR proteins (Fig. 6D) and are currently being investigated.
A recent study in prostate and breast cancer cell lines investigated EGFR kinase-dependent and -independent responses by directly decreasing EGFR by RNA interference or by overexpression of wild-type and kinase domain mutant EGFR expression plasmids (28). One of their important observations was identification of a kinase-independent function of EGFR in which the wild-type and mutant (kinase domain) EGFR stabilized SGLT1 to prevent autophagic cell death and EGFR knockdown resulted in decreased SGLT1 expression and enhanced accumulation of LC3. Formation of the cleaved form of LC3 is critical for autophagosome formation and is a positive marker for autophagolysosomes (29). Both BA and curcumin induced LC3 accumulation and downregulated SGLT1 in 253JB-V and KU7 cells (Figs. 5A and 5B) and knockdown of EGFR by transfection with iSp also induced LC3 accumulation but did not decrease SGLT1 expression (Fig. 6A). Not surprisingly, gefitinib did not affect expression of either SGLT1 or LC3 (Fig. 5C). BA, curcumin and iSp transfection decreased EGFR expression (Figs. 1A and 3A) and therefore, their differences with respect to expression of SGLT1 in bladder cancer cells may be EGFR-independent and associated with other activities of BA and curcumin (Fig. 6D). The induction of autophagy by BA, curcumin and iSp was also confirmed by their induction of acridine orange staining (Figs. 6B and 6C) and induction of punctuate perinuclear green fluorescence in KU7 and 253JB-V cells transfected with GFP-LC3 (Fig. 6B). Both of these staining/fluorescent responses are characteristic of autophagy (30).
In summary, results of this study demonstrate that BA- and curcumin-dependent repression of Sp1, Sp3 and Sp4 proteins in bladder cancer cells also decreased expression of the Sp-dependent gene EGFR in both gefitinib-responsive 253JB-V and gefitinib-nonresponsive KU7 cells. Thus, BA and curcumin represent a novel type of EGFR inhibitor that block EGFR and EGFR-mediated responses through repression of Sp transcription factors and this results in inhibition of EGFR-dependent kinases and activation of autophagic cell death which is repressed by EGFR (kinase-independent) in cancer cell lines (28). Previous studies have reported that curcumin inhibited EGFR signaling in cancer cell lines (41–43), and one report showed that BA enhanced EGFR signaling in a relatively BA-resistant melanoma cell line (44). The curcumin-dependent effects in breast and colon cancer cells were accompanied by downregulation of EGFR, and it is possible that decreased expression of Sp proteins may contribute to this response. In PC3 prostate cancer cells, curcumin alone or in combination with phenylethyl isothiocyanate inhibited epidermal growth factor-induced signaling but did not downregulate EGFR and it was concluded that inhibition was associated with attenuation of IkBα and Akt phosphorylation. The role (if any) of Sp downregulation on these responses in PC3 cells is unclear. A chemotherapeutic advantage of compounds, such as BA and curcumin that downregulate Sp proteins, is that they also induce pro-apoptotic, antiproliferative and antiangiogenic activities through downregulation of other Sp-dependent genes such as survivin, cyclin D1 and VEGF/VEGFR1 (7–10) (Figs. 3A and 3B) and modulate other Sp-independent responses (10) (Fig. 6D). Relative contributions of Sp-dependent and Sp-independent pathways to the overall anticarcinogenic activity of curcumin and BA and other drugs that repress Sp proteins and Sp-regulated proteins such as EGFR will also vary with tumor type. The concentrations of BA used in this and other studies are comparable; however, low bioavailability of curcumin in vivo is a continuing concern, and it is possible that newer methods of drug delivery may overcome this problem. Currently, we are investigating the mechanisms of action and clinical applications of drugs such as BA and curcumin alone and in combination with other cytotoxic compounds used for clinical treatment of bladder cancer.
Acknowledgments
Funding: This work was supported by the National Institutes of Health [R01CA108718] and Texas A&M AgriLife.
Advice from Dr. Kathy Vanderlaag is gratefully appreciated.
Abbreviations
- BA
betulinic acid
- EGFR
epidermal growth factor receptor
- iSp
cocktail containing small inhibitory RNAs for Sp1, Sp3 and Sp4
- Sp
specificity protein transcription factor
References
- 1.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 2.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–60. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 3.Ammendola R, Mesuraca M, Russo T, Cimino F. Sp1 DNA binding efficiency is highly reduced in nuclear extracts from aged rat tissues. J Biol Chem. 1992;267:17944–8. [PubMed] [Google Scholar]
- 4.Adrian GS, Seto E, Fischbach KS, et al. YY1 and Sp1 transcription factors bind the human transferrin gene in an age-related manner. J Gerontol A Biol Sci Med Sci. 1996;51:B66–B75. doi: 10.1093/gerona/51a.1.b66. [DOI] [PubMed] [Google Scholar]
- 5.Oh JE, Han JA, Hwang ES. Downregulation of transcription factor, Sp1, during cellular senescence. Biochem Biophys Res Commun. 2007;353:86–91. doi: 10.1016/j.bbrc.2006.11.118. [DOI] [PubMed] [Google Scholar]
- 6.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Abdelrahim M, Baker CH, Abbruzzese JL, Safe S. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J Natl Cancer Inst. 2006;98:855–68. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- 8.Abdelrahim M, Baker CH, Abbruzzese JL, et al. Regulation of vascular endothelial growth factor receptor-1 (VEGFR1) expression by specificity proteins 1, 3 and 4 in pancreatic cancer cells. Cancer Res. 2007;67:3286–94. doi: 10.1158/0008-5472.CAN-06-3831. [DOI] [PubMed] [Google Scholar]
- 9.Chintharlapalli S, Papineni S, Ramaiah SK, Safe S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007;67:2816–23. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- 10.Chadalapaka G, Jutooru I, Chintharlapalli S, et al. Curcumin decreases specificity protein expression in bladder cancer cells. Cancer Res. 2008;68:5345–54. doi: 10.1158/0008-5472.CAN-07-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein (Sp) transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Wei D, Huang S, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–80. [PubMed] [Google Scholar]
- 13.Yao JC, Wang L, Wei D, et al. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109–17. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 14.Jiang NY, Woda BA, Banner BF, Whalen GF, Dresser KA, Lu D. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:1648–52. doi: 10.1158/1055-9965.EPI-07-2791. [DOI] [PubMed] [Google Scholar]
- 15.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–43. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 16.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–57. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37 (Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 18.Baselga J. The EGFR as a target for anticancer therapy--focus on cetuximab. Eur J Cancer. 2001;37 (Suppl 4):S16–S22. doi: 10.1016/s0959-8049(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 19.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–59. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 20.Chow NH, Liu HS, Yang HB, Chan SH, Su IJ. Expression patterns of erbB receptor family in normal urothelium and transitional cell carcinoma. An immunohistochemical study. Virchows Arch. 1997;430:461–6. doi: 10.1007/s004280050056. [DOI] [PubMed] [Google Scholar]
- 21.Mellon JK, Cook S, Chambers P, Neal DE. Transforming growth factor α and epidermal growth factor levels in bladder cancer and their relationship to epidermal growth factor receptor. Br J Cancer. 1996;73:654–8. doi: 10.1038/bjc.1996.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black PC, Dinney CP. Bladder cancer angiogenesis and metastasis--translation from murine model to clinical trial. Cancer Metastasis Rev. 2007;26:623–34. doi: 10.1007/s10555-007-9084-9. [DOI] [PubMed] [Google Scholar]
- 23.Black PC, Agarwal PK, Dinney CP. Targeted therapies in bladder cancer--an update. Urol Oncol. 2007;25:433–8. doi: 10.1016/j.urolonc.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Nishi H, Nishi KH, Johnson AC. Early Growth Response-1 gene mediates up-regulation of epidermal growth factor receptor expression during hypoxia. Cancer Res. 2002;62:827–34. [PubMed] [Google Scholar]
- 25.Johnson AC, Ishii S, Jinno Y, Pastan I, Merlino GT. Epidermal growth factor receptor gene promoter. Deletion analysis and identification of nuclear protein binding sites. J Biol Chem. 1988;263:5693–9. [PubMed] [Google Scholar]
- 26.Xu J, Thompson KL, Shephard LB, Hudson LG, Gill GN. T3 receptor suppression of Sp1-dependent transcription from the epidermal growth factor receptor promoter via overlapping DNA-binding sites. J Biol Chem. 1993;268:16065–73. [PubMed] [Google Scholar]
- 27.Shrader M, Pino MS, Brown G, et al. Molecular correlates of gefitinib responsiveness in human bladder cancer cells. Mol Cancer Ther. 2007;6:277–85. doi: 10.1158/1535-7163.MCT-06-0513. [DOI] [PubMed] [Google Scholar]
- 28.Weihua Z, Tsan R, Huang WC, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–93. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paglin S, Hollister T, Delohery T, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–44. [PubMed] [Google Scholar]
- 32.Ranson M, Hammond LA, Ferry D, et al. ZD1839, a selective oral epidermal growth factor receptor-tyrosine kinase inhibitor, is well tolerated and active in patients with solid, malignant tumors: results of a phase I trial. J Clin Oncol. 2002;20:2240–50. doi: 10.1200/JCO.2002.10.112. [DOI] [PubMed] [Google Scholar]
- 33.Cappellen D, De OC, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 34.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–58. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 35.Kelly K, Chansky K, Gaspar LE, et al. Phase III trial of maintenance gefitinib or placebo after concurrent chemoradiotherapy and docetaxel consolidation in inoperable stage III non-small-cell lung cancer: SWOG S0023. J Clin Oncol. 2008;26:2450–6. doi: 10.1200/JCO.2007.14.4824. [DOI] [PubMed] [Google Scholar]
- 36.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 37.Chou TY, Chiu CH, Li LH, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:3750–7. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 38.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 39.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 40.Blehm KN, Spiess PE, Bondaruk JE, et al. Mutations within the kinase domain and truncations of the epidermal growth factor receptor are rare events in bladder cancer: implications for therapy. Clin Cancer Res. 2006;12:4671–7. doi: 10.1158/1078-0432.CCR-06-0407. [DOI] [PubMed] [Google Scholar]
- 41.Somers-Edgar TJ, Scandlyn MJ, Stuart EC, Le Nedelec MJ, Valentine SP, Rosengren RJ. The combination of epigallocatechin gallate and curcumin suppresses ER α-breast cancer cell growth in vitro and in vivo. Int J Cancer. 2008;122:1966–71. doi: 10.1002/ijc.23328. [DOI] [PubMed] [Google Scholar]
- 42.Patel BB, Sengupta R, Qazi S, et al. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122:267–73. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 43.Kim JH, Xu C, Keum YS, Reddy B, Conney A, Kong AN. Inhibition of EGFR signaling in human prostate cancer PC-3 cells by combination treatment with β-phenylethyl isothiocyanate and curcumin. Carcinogenesis. 2006;27:475–82. doi: 10.1093/carcin/bgi272. [DOI] [PubMed] [Google Scholar]
- 44.Qiu L, Wang Q, Di W, et al. Transient activation of EGFR/AKT cell survival pathway and expression of survivin contribute to reduced sensitivity of human melanoma cells to betulinic acid. Int J Oncol. 2005;27:823–30. [PubMed] [Google Scholar]