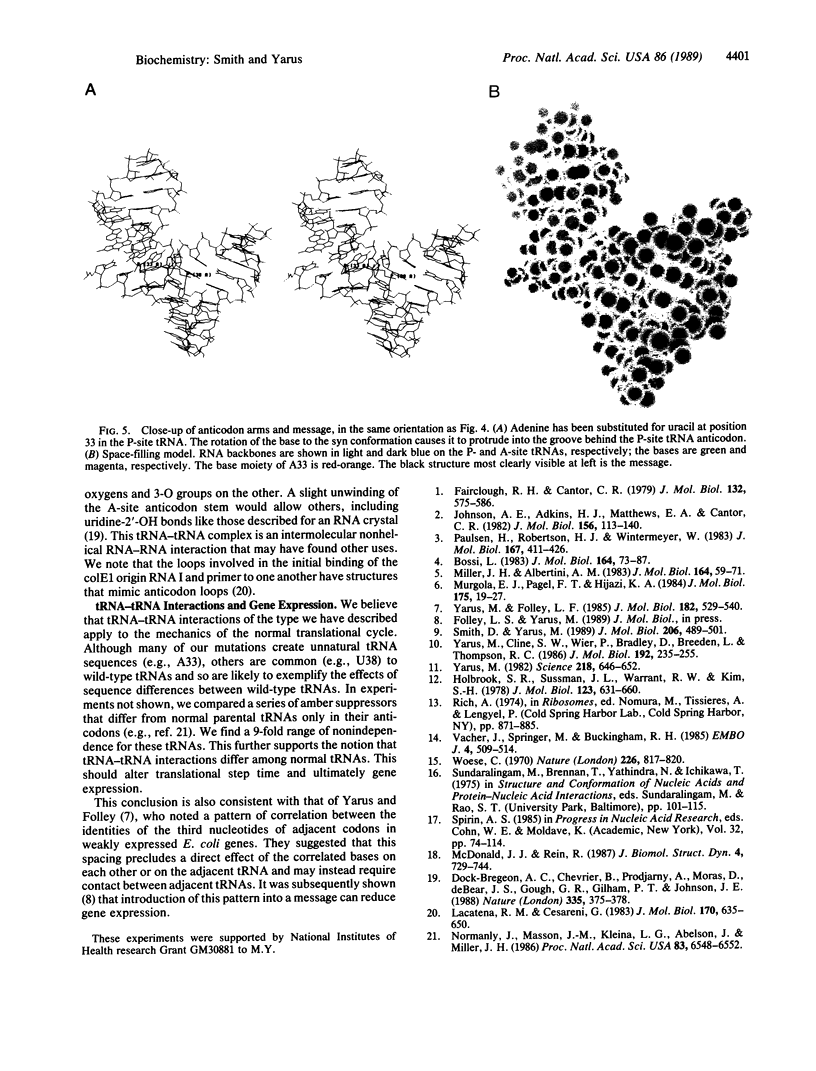
Abstract
We describe an assay that converts the effects of tRNA-tRNA contacts at two particular codons into a quantitative effect on beta-galactosidase level. The assay measures the separate and combined efficiency of suppression at adjacent nonsense codons in vivo using a set of specially created homologous messages. In a survey of distal anticodon arm substitutions, we find that particular mutant tRNAs occupying the P-site reduce the apparent efficiency of the suppressor tRNA reading the A-site codon by factors of 2-170. By using measured tRNA-tRNA distances and the crystallographic tRNA structure, we propose a model of the tRNA-tRNA-mRNA complex. In the model, the anticodon loops of the P-site and A-site tRNAs contact one another in a way that is consistent with our combined tRNA efficiency data. These results suggest that tRNA-tRNA interactions that modulate tRNA action are an inevitable feature of translation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983 Feb 15;164(1):73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Chevrier B., Podjarny A., Moras D., deBear J. S., Gough G. R., Gilham P. T., Johnson J. E. High resolution structure of the RNA duplex [U(U-A)6A]2. Nature. 1988 Sep 22;335(6188):375–378. doi: 10.1038/335375a0. [DOI] [PubMed] [Google Scholar]
- Fairclough R. H., Cantor C. R. The distance between the anticodon loops of two tRNAs bound to the 70 S Escherichia coli ribosome. J Mol Biol. 1979 Aug 25;132(4):575–586. doi: 10.1016/0022-2836(79)90375-9. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Johnson A. E., Adkins H. J., Matthews E. A., Cantor C. R. Distance moved by transfer RNA during translocation from the A site to the P site on the ribosome. J Mol Biol. 1982 Mar 25;156(1):113–140. doi: 10.1016/0022-2836(82)90462-4. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Interaction between RNA1 and the primer precursor in the regulation of Co1E1 replication. J Mol Biol. 1983 Nov 5;170(3):635–650. doi: 10.1016/s0022-2836(83)80125-9. [DOI] [PubMed] [Google Scholar]
- McDonald J. J., Rein R. A stereochemical model of the transpeptidation complex. J Biomol Struct Dyn. 1987 Apr;4(5):729–744. doi: 10.1080/07391102.1987.10507675. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T., Hijazi K. A. Codon context effects in missense suppression. J Mol Biol. 1984 May 5;175(1):19–27. doi: 10.1016/0022-2836(84)90442-x. [DOI] [PubMed] [Google Scholar]
- Normanly J., Masson J. M., Kleina L. G., Abelson J., Miller J. H. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H., Robertson J. M., Wintermeyer W. Topological arrangement of two transfer RNAs on the ribosome. Fluorescence energy transfer measurements between A and P site-bound tRNAphe. J Mol Biol. 1983 Jun 25;167(2):411–426. doi: 10.1016/s0022-2836(83)80342-8. [DOI] [PubMed] [Google Scholar]
- Smith D., Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J Mol Biol. 1989 Apr 5;206(3):489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- Vacher J., Springer M., Buckingham R. H. Functional mutants of phenylalanine transfer RNA from Escherichia coli. EMBO J. 1985 Feb;4(2):509–513. doi: 10.1002/j.1460-2075.1985.tb03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature. 1970 May 30;226(5248):817–820. doi: 10.1038/226817a0. [DOI] [PubMed] [Google Scholar]
- Yarus M., Cline S. W., Wier P., Breeden L., Thompson R. C. Actions of the anticodon arm in translation on the phenotypes of RNA mutants. J Mol Biol. 1986 Nov 20;192(2):235–255. doi: 10.1016/0022-2836(86)90362-1. [DOI] [PubMed] [Google Scholar]
- Yarus M., Folley L. S. Sense codons are found in specific contexts. J Mol Biol. 1985 Apr 20;182(4):529–540. doi: 10.1016/0022-2836(85)90239-6. [DOI] [PubMed] [Google Scholar]
- Yarus M. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science. 1982 Nov 12;218(4573):646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]



