Abstract
An important event in the development of cervical squamous cell carcinoma (SCC) is deregulated expression of high-risk human papillomavirus (HR-HPV) oncogenes, most commonly related to viral integration into host DNA. Mechanisms of development of the ~15% of SCCs that contain extra-chromosomal (episomal) HR-HPV are poorly understood, due to limited longitudinal data. We therefore employed the W12 model to study mechanisms of cervical carcinogenesis associated with episomal HPV16. In vitro progression of W12 normally occurs through selection of cells containing integrated HPV16. However, in one long-term culture, keratinocytes developed a selective growth advantage and invasive phenotype, while retaining HPV16 episomes at increased copy number in the absence of transcriptionally active integrants. Longitudinal investigations revealed similarities between the episome- and integrant-associated routes of neoplastic progression. Most notable were dynamic changes in viral early gene expression in episome-retaining cells, consistent with continually changing selective pressures. An early increase in viral transcription preceded elevated episome copy number and was followed by a reduction to near baseline after the development of invasiveness. Episomal transcriptional deregulation did not require selection of a specific sequence variant of the HPV16 upstream regulatory region, although increased levels of acetylated histone H4 around the late promoter implicated a role for altered chromatin structure. Interestingly, invasive episome-retaining cells demonstrated high levels of HPV16-E2/E6 proteins (despite decreased transcript levels) and reduced expression of interferon-stimulated genes, adaptations that support viral persistence and cell survival. Our findings suggest a unified working model for events important in cervical neoplastic progression, regardless of HR-HPV physical state.
Keywords: Human papillomavirus, episome, transcription, viral copy number, cervical neoplastic progression
Introduction
Cervical squamous carcinogenesis is characterised by uncoupling of HR-HPV early gene expression from the epithelial differentiation programme, so that the viral oncogenes E6 and E7 are expressed at increased levels in undifferentiated cells (1). Deregulation of viral gene expression in cervical carcinomas is usually associated with integration of HR-HPV into the host DNA (2). However, although transcriptionally active (i.e. ‘selected’) integrants are detected in ~85% of cervical carcinomas, ~15% cases demonstrate only episome-derived early gene expression (3).
Mechanisms of transcriptional deregulation in episome-associated carcinogenesis are relatively poorly understood. DNA sequence alterations in transcription factor binding sites at the viral upstream regulatory region (URR) have been reported in episome-associated SCCs. While some of these alterations are associated with increased viral enhancer activity in reporter assays (4,5), others are not (6,7), suggesting alternative mechanisms of transcriptional deregulation in episome-associated carcinogenesis.
Suitable in vitro systems represent a valuable resource for longitudinal investigations of neoplastic progression. Important insights into viral and host events in cervical carcinogenesis have been obtained previously from the unique W12 cervical keratinocyte model (8-13). W12 is a polyclonal cell population established by explant culture of a cervical low-grade squamous intraepithelial lesion (L-SIL) (14). At early passages the cells retain HPV16 episomes at ~100-200 copies per cell in monolayer culture, are chromosomally stable and reform an L-SIL in organotypic raft culture (12,14). Long-term in vitro cultivation usually leads to spontaneous loss of episomes and selection of cells containing only integrated HPV16. Individual culture ‘series’ retain episomes for different periods and show emergence of cells with different sites of viral integration into the host genome (9,12,13). These events are characterised by development of high-level genomic instability and phenotypic progression through high-grade SIL (H-SIL) to invasiveness (12).
Here, we show that acquisition of in vitro invasiveness in W12 can also be associated with retention of HPV16 episomes in the absence of transcriptionally active HPV16 integrants. Our data indicate that episome-associated neoplastic progression in vitro shows fundamental similarities to the integrant-associated route and emphasise the value of longitudinal investigations in providing new insights into cervical carcinogenesis.
Materials and Methods
Cell culture, growth analysis and immunohistochenistry
Most work was performed on cells grown in monolayer culture, which restricts cell differentiation and maintains the phenotype of the basal epithelial layer, the key site of HR-HPV deregulation in the early stages of cervical squamous carcinogenesis (1,2). The cells used were: W12 [authenticated by the presence of HPV16 episomes and characteristic genomic copy number imbalances (12)], primary cultures of normal ectocervical keratinocytes (Ncx/2, /6, /11; each from a different donor) and the cervical SCC lines CaSki and SiHa (authenticated by short tandem-repeat profiling by the ATCC).
Cells were grown as described (14). Long-term W12 culture series were derived from polyclonal ‘parental’ cells at passage 9 (W12p9) (Figure-1A). In studies of relative viral copy number and viral/host gene expression, base-line levels were obtained using parental cells at W12p9 and also those grown to W12p11 and W12p12 (Figure-1A). The latter maintained the same phenotype as the W12p9 cells and were therefore also referred to as ‘parental’. Colony forming efficiency (CFE) and growth rates were determined using standard techniques (12,15). Organotypic tissue culture was performed as described (12). 9-12 day collagen rafts were formalin-fixed and processed to paraffin for histological analysis. Immunohistochemistry (16), was used to detect cytokeratins 10/13 [KRT10/13; (17)](Dako) and minichromosome maintenance protein 2 [MCM2; (16)] as markers of differentiation and cell cycle entry, respectively.
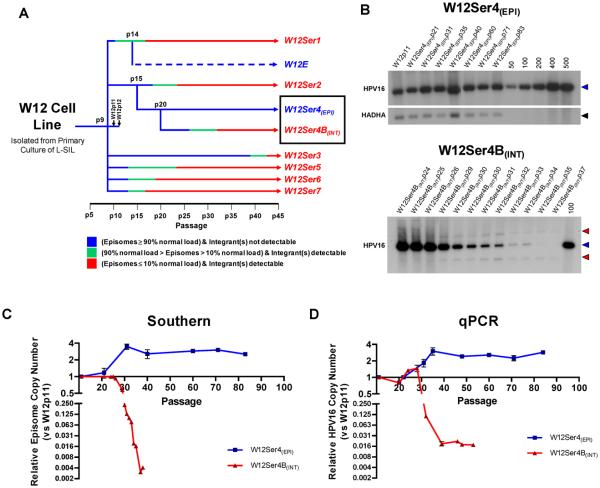
Figure-1. HPV16 physical state and copy number in W12Ser4(EPI) and W12Ser4B(INT).
(A): Summary of HPV16 physical state in long-term cultures generated from early passage parental W12p9. W12 Series1 (W12Ser1), W12Ser2 and W12Ser3 have previously been described (8,12,13). Although W12Ser3 was reported to maintain episomes, they were eventually lost by p42. The W12E cell line, which retains HPV16 episomes at high copy number, was generated from W12Ser1p14 (29). (B): Southern analysis of Series4(EPI) and Series4B(INT) following BamHI restriction. Copy number control lanes represent linearised HPV16 genome copies per diploid cell. Bands generated by linearised unit length HPV16 (7.9 kb) and HADHA loading control (3.9 kb) are indicated by blue and black arrows, respectively. Red arrows indicate virus-host junction fragments. (C and D): Quantification of relative viral copy number by Phosphorimager analysis of Southern blots (C) and qPCR (D). For qPCR, copy numbers were calculated using the relative copy number of either E2 (W12Ser4(EPI)) or E6 (W12Ser4B(INT)). The decreased viral copy number in W12Ser4B(INT) appeared greater by Phosphorimager analysis than qPCR, as episomes (measured by the former) were completely lost while E6 (measured by the latter) was maintained in the viral integrant.
Determination of HPV16 physical state, relative copy number and relative gene expression
Initially, HPV16 physical state and copy number were determined by Southern analysis (8), using HADHA to normalise for DNA loading. Relative viral copy number was also determined by SYBR-Green quantitative PCR (qPCR) (10). Relative levels of the E6 or E2 gene in genomic DNA samples were calculated using the Pfaffl comparative Ct method and normalised against TLR2 at chromosome 4q32, a locus that did not demonstrate copy-number change in the long-term series analysed in this study (unpublished observations). Primers were (5′-3′): TLR2-Fwd, GGCCAGCAAATTACCTGTGTG; TLR2-Rev, AGGCGGACATCCTGAACCT. Gene copy number ratios were calculated as the mean of three technical replicate experiments, with error bars showing the standard error.
HPV16 transcripts were initially detected by amplification of papillomavirus oncogene transcripts (APOT), a 3′ rapid amplification of cDNA ends PCR technique that allows discrimination of HR-HPV mRNAs derived from episomal and integrated viral genomes (3). Nested PCR [DNA-PCR; (9)] was used to confirm and detect the genomic site of viral integration. Relative transcript levels of viral and host genes were determined by SYBR-Green quantitative reverse transcription-PCR (qRT-PCR) (10). Relative expression levels were normalised to four house-keeping genes and ratios expressed as the mean of these four comparisons. Details of the primers for HPV16 E2, E6 and E7, and the host test and house-keeping genes, are given elsewhere (10). Levels of p53 and HPV16 E6, E7 and nuclear E2 proteins were determined by quantitative Western blotting (10). Proteins from total cell and nuclear protein extracts were normalised to levels of beta-actin and TATA-binding protein (TBP), respectively. Anti-HPV16 E6 antibodies were from Euromedex, anti-HPV16 E7 (clone ED17) and p53 (clone DO-1) from Santa Cruz, and anti-HPV16 E2 (clone TVG261) from Cancer Research Technology (18).
Analysis of epigenetic changes at the HPV16 URR
DNA methylation at the HPV16 URR and surrounding region was determined by sequence analysis of sodium bisulphite-modified gDNA, prepared as described (19). Primers were designed using MethPrimer (20), and used to amplify and sequence between nucleotides 6801-559 (Supplementary Table-S1).
To test for differences in histone acetylation around the URR, cells were initially screened for differential responses to the histone deacetylase (HDAC) inhibitor trichostatin-A (TSA), at 200nM or 400nM. Expression of HPV16 early genes in treated cells (versus dimethyl sulphoxide vehicle-only) controls was analysed by qRT-PCR at an early time point (2 hours) following treatment, to eliminate confounding effects of changes in viral copy number and/or host gene expression (21,22). Expression ratios were normalised to RPL13A, which is unresponsive to TSA in many cell types (23). All treatments were performed in duplicate and mean relative expression levels calculated, with error bars indicating standard error.
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP-IT Express Enzymatic kit (Active Motif). Briefly, cells were cross-linked in culture medium containing 1% formaldehyde, lysed and then incubated with an enzymatic shearing cocktail that cuts DNA randomly (Active Motif) at 37°C for 10 minutes. ChIP was performed by incubating 10μg chromatin per reaction with Protein G magnetic beads and ChIP-validated antibody for 4 hours with agitation at 4°C. We used antibodies against acetyl-histone H3 and acetyl-histone H4 (Active Motif), with pre-immune rabbit serum (Dako) as a negative control for background signal. Beads were pelleted and washed to reduce non-specific binding. Chromatin was eluted and cross-links reversed, followed by incubation with proteinase K for 1 hour at 37°C. Enrichment of HPV16 sequences at the enhancer, early promoter (P97) and late promoter (P670) was analysed by qPCR of purified DNA (10,24), with all reactions performed in triplicate. Primers were (5′-3′): 7555-F, CCAAATCCCTGTTTTCCTGA; 7681-R, CGTTGGCGCATAGTGATTTA; 7854-F, GCAAACCGTTTTGGGTTACA; 65R, ACTAACCGGTTTCGGTTCAA; 649F, GACAGCTCAGAGGAGGAGGA; 765R, GCACAACCGAAGCGTAGAGT. For each sample, fold enrichment (FE) of target sequence in ChIP samples versus negative control was calculated using the equation: FE = EInput Ct – ChIP Ct/EInput Ct – Negative Control Ct, where E = primer efficiency. All ChIP reactions were performed in duplicate and mean levels of fold enrichment calculated, with error bars indicating standard error.
Results
Generation of W12 Series 4 and W12 Series 4B
To date, we have generated eight long-term culture series under normal in vitro conditions (i.e. in the absence of specific selection pressures) from the same starting population of polyclonal parental W12p9 (Figure-1A). The majority of these series demonstrated spontaneous selection of integrated HPV16, via a ‘mixed’ episome and integrant state (8,12,13). Here, we describe a detailed analysis of two series that originated from W12 Series2 at passage 15 (W12Ser2p15), when only episomal HPV16 was detectable by Southern blotting (13). The cells were grown to p20 and then split into two independent series, W12 Series4 (W12Ser4) and W12 Series4B (W12Ser4B), each of which was cultured for >60 further passages (>300 population doublings).
W12Ser4 maintains episomal HPV16 at increased copy number during long-term culture
The physical state of HPV16 in W12Ser4 and W12Ser4B was determined by Southern analysis. For W12Ser4, restriction with the HPV16 single cutter BamHI generated only a single band of 7.9kb at all passages analysed (Figure-1B; top), while restriction with the HPV16 non-cutter HindIII generated a banding pattern reflecting the presence of supercoiled, nicked and open circular forms of episomal HPV16, with no evidence of concatamerised integrants (data not shown). These data indicate that episomal HPV16 was retained throughout W12Ser4 (hereafter referred to as W12Ser4(EPI)). Quantification by Phosphorimager analysis and relative qPCR (Figures-1C and 1D) demonstrated an increase in episome copy number between W12Ser4(EPI)p20 and W12Ser4(EPI)p35 (to ~500-600 copies per cell, ~3.5-fold higher than in parental W12), which was subsequently maintained.
In contrast, BamHI analysis of W12Ser4B demonstrated a marked decrease in episome number by p30, concomitant with the emergence of two virus-host junction fragments representing integrated HPV16 (Figure-1B; bottom). Integrated virus was subsequently retained at ~2 copies per cell, as suggested by relative qPCR (Figure-1D). W12Ser4B (hereafter referred to as W12Ser4B(INT)) therefore progressed via an integrant-associated route, similar to previous long-term W12 series (8,13).
W12Ser4(EPI) cells acquire a strong selective growth advantage and an invasive phenotype
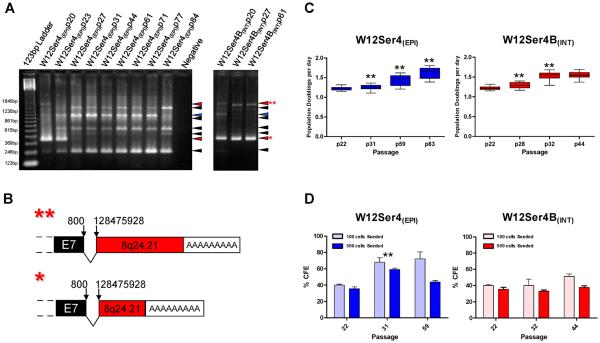
APOT demonstrated that only episome-derived oncogene transcripts were expressed in established W12Ser4(EPI) cells (Figure-2A). Thus, if any integrants were present below the detection limits of Southern blotting, they were transcriptionally inactive and made no apparent contribution to cell phenotype. In contrast, established W12Ser4B(INT) cells expressed two transcripts derived from HPV16 integrated at 8q24.21, with differences in transcript length attributable to use of different host poly(A) sites (Figures-2A and 2B). DNA-PCR demonstrated that the integrant was identical to that selected spontaneously in W12Ser2 following episome clearance (9) (Supplementary Figure-S1). It was therefore interesting that at the point of divergence of W12Ser4(EPI) and W12Ser4B(INT) (i.e. p20) both episome- and 8q24.21-integrant-derived transcripts were present. This suggested that W12Ser4(EPI) cells acquired a strong selective advantage, which enabled them to outcompete cells expressing selectable integrant-derived transcripts. Consistent with this, W12Ser4(EPI) cells at p27 and beyond demonstrated increased growth rate and CFE compared to cells at p20, whereas W12Ser4B(INT) cells only showed an increase in growth rate (Figures-2C and 2D).
Figure-2. Retention of HPV16 episome-derived transcripts and acquisition of a growth advantage in W12Ser4(EPI).
(A): APOT analysis of W12Ser4(EPI) (left panel) and W12Ser4B(INT) (right panel). Blue arrows indicate the expected size of episome-derived transcripts (1050 bp), black arrows indicate non-specific products and red arrows indicate integrant-derived transcripts. The far left and right lanes of the left panel show DNA molecular weight markers and a no-template negative control, respectively. As the panels represent two independent APOT assays, there are differences in band intensities and background signal in the two samples at the point of divergence of W12Ser4(EPI) and W12Ser4(INT) (passage 20; lane 2 in left panel; lane 1 in right panel) (B): Schematics of the low molecular weight (*) and high molecular weight (**) integrant-derived transcripts detected throughout long-term culture in W12Ser4B(INT) and at early passages in W12Ser4(EPI). Viral and host sequences are shaded black and red, respectively. Numbers above each schematic indicate the genomic positions of the viral and host sequence at the splice junction. (C): Growth rate of W12Ser4(EPI) and W12Ser4B(INT), expressed as population doublings per day. Boxes show the median growth rate and interquartile range, and whiskers represent the maximum and minimum values. (D): CFE of W12Ser4(EPI) and W12Ser4B(INT), calculated after seeding either 100 or 500 cells. Error bars indicate the standard error of three independent platings. In both (C) and (D), asterisks (**) indicate a significantly different growth rate/CFE compared to the previous passage analysed (two-sample t-test, p<0.05).
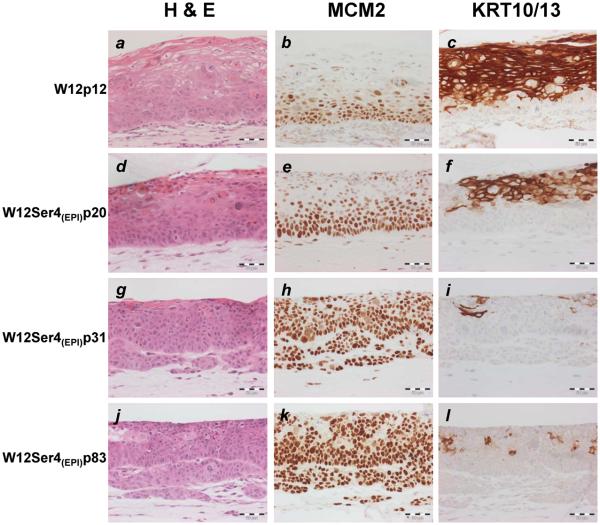
In organotypic raft culture, which represents a validated system for analysing stratified cell phenotype in vitro (25,26), parental W12p12 recapitulated an L-SIL, with cycling (MCM-positive) atypical cells being confined to the lower third of the epithelium, and extensive terminal differentiation (KRT10/13-positive cells) in the upper layers (Figure-3). In contrast, W12Ser4(EPI) showed striking phenotypic progression from p31 onwards. While the reformed epithelium resembled H-SIL, nests of cells also extended irregularly into the underlying collagen, indicating acquisition of in vitro invasiveness. W12Ser4B(INT) showed broadly similar features, although invasiveness was not observed until p85 (Supplementary Figure-S2).
Figure-3. Phenotype of W12Ser4(EPI) cells in organotypic culture.
The images show histological appearances following haematoxylin and eosin (H & E) staining, together with expression of minichromosome maintenance protein 2 (MCM2) and cytokeratins 10/13 (KRT10/13), in organotypic cultures reformed by parental W12p12 (a-c), and W12Ser4(EPI) cells at p20 (d-f), p31 (g-i) and p83 (j-l). Scale bars represent 50μM.
Neoplastic progression in W12Ser4(EPI) is associated with dynamic changes in HPV16 early gene transcription
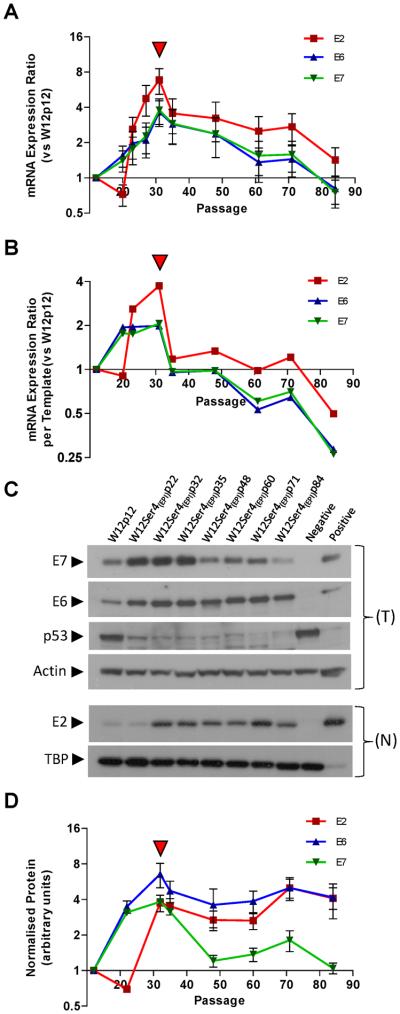
We next quantified viral transcription during the full course of phenotypic progression in W12Ser4(EPI). We measured the oncogenes E6 and E7, and also E2 (deleted in the W12Ser4B(INT) integrant) as a specific marker of episome-derived transcription (Figure-4A). At the point of divergence of W12Ser4(EPI) and W12Ser4B(INT) (p20), E2 levels were reduced relative to parental W12p12 cells, while E6 and E7 levels were higher, consistent with the presence of the transcriptionally active 8q24.21 integrant at this point (see Figure-2A). During selection of episome-only cells in W12Ser4(EPI) (between p20 and p31), levels of all transcripts increased, with E2 consistently higher than E6 and E7 thereafter. Interestingly, after levels of viral transcription peaked at p31 (when cells had acquired in vitro invasiveness), they subsequently declined despite the high episome copy number, with E6/E7 transcripts decreasing to just below base-line at p84. Maximal levels of elevation were 6.9-fold for E2 and 3.7/3.8-fold for E6/E7, the latter elevations being similar to those that occurred in W12Ser4B(INT) following episome loss (Supplementary Figure-S3A).
Figure-4. Levels of HPV16 early gene mRNA and protein in W12Ser4(EPI).
(A and B): Relative HPV16 early gene mRNA expression in W12Ser4(EPI), showing levels overall (A) and per viral copy, calculated using the formula [mean relative mRNA expression ratio ÷ mean relative HPV16 copy number] (B). (C): Western analysis of p53 and the HPV16 E6, E7, and E2 proteins. Beta-actin and TBP represent loading controls for total (T) and nuclear (N) protein extracts, respectively. For detection of E6/E7 protein, controls were total protein from Ncx/2 (negative) and W12Ser2p25 (positive). Since the p53 blot was generated by re-probing the E6 blot, the negative and positive lanes are reversed. For detection of E2 protein, controls were HeLa nuclear protein (negative) and full-length E2 purified from Sf21 insect cells (positive; a kind gift from Norman Maitland). (D): Viral protein quantification (arbitrary units) by densitometry. Error bars represent the standard error of technical replicate experiments. In (A), (B) and (D), the red arrowheads indicate the point at which invasiveness was acquired.
To determine if levels of viral transcription in W12Ser4(EPI) changed in proportion to viral copy number or instead represented true deregulation of viral transcription, we next measured relative mRNA expression per episome copy (using the relative PCR copy number data shown in Figure-1D). Compared to parental W12p12, mRNA expression per template increased until the development of invasiveness (i.e. between p20 and p31), indicating that transcriptional deregulation was an early event in W12Ser4(EPI) (Figure-4B). This was followed by a sharp reduction to base-line levels and then to below base-line from p61. Despite this, overall transcript levels remained above base-line or only just below throughout W12Ser4(EPI) (Figure-4A), due to stable maintenance of increased viral copy number. Together, these data demonstrated that dynamic changes in levels of transcription per template were key determinants of the overall fluctuations in viral early gene expression levels. Maximal levels of transcriptional deregulation per episome (3.7-fold for E2 and 2.0/2.1-fold for E6 and E7, respectively) were much less than for the de-repressed integrant in W12Ser4B(INT), where >150-fold increases in relative viral oncogene expression per template copy were seen following episome loss (Supplementary Figure-S3B).
Retention of high episome copy number in W12Ser4(EPI) associates with high levels of E6 and nuclear E2 proteins and decreased transcription of interferon-stimulated genes (ISGs)
The observed decreases in transcription of all early genes in W12Ser4(EPI) (including the replication factor E2) were surprising given the maintenance of high episome copy number at later passages. We therefore quantified viral proteins in W12Ser4(EPI). Normalised levels of E7 protein peaked at p31 and then returned to baseline (Figures-4C and 4D), correlating with E7 transcript levels (p=0.0179, Pearson's correlation), but not with viral copy number (Supplementary Figure-S4). In contrast, normalised levels of E6 and nuclear E2 protein did not return to base-line after p31, remaining high at later passages (Figures-4C and 4D). Consistent with E6 function (27), levels of p53 protein were reciprocal to those of E6 (Figure-4C). There was no correlation between protein and transcript levels for E2 or E6, although E2 protein did correlate with viral copy number (p=0.0421, Pearson's correlation) (Supplementary Figure-S4).
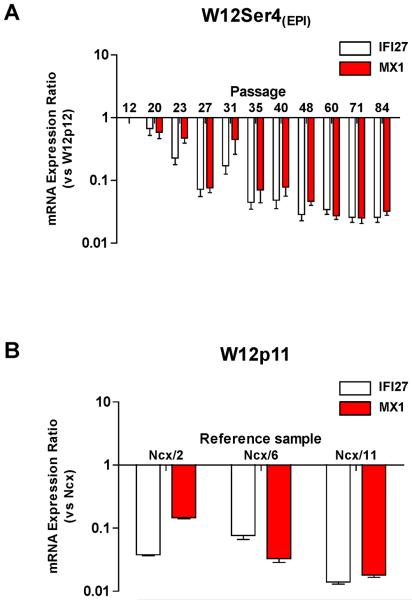
We also quantified relative expression of IFI27 and MX1, as representative ISGs [which can mediate episomal HPV16 clearance (10)]. Levels of both transcripts were greatly reduced in W12Ser4(EPI) cells compared to parental W12p12 (Figure-5A), while levels in parental W12p11 were themselves lower than in normal cervical keratinocytes (Figure-5B).
Figure-5. Expression of ISGs in W12Ser4(EPI).
(A and B): qPCR analysis of IFI27 and MX1 mRNA expression in W12Ser4(EPI) relative to parental W12p12 (A) and in parental W12p11 relative to three normal ectocervical keratinocyte (Ncx) primary cultures (B).
Deregulated episomal early gene transcription is associated with epigenetic changes at the URR
We studied the mechanisms underlying the deregulated episomal expression in W12Ser4(EPI), addressing genetic and epigenetic changes, both of which can influence viral transcript regulation (28). For W12Ser4(EPI)p20, p31 and p84, genetic analysis of the URR and flanking regions (including E6 and E7) was performed by TA cloning and sequencing of PCR products spanning nucleotides 6864-1358. Sequences were compared to the reference HPV16 sequence determined from W12E (29). Selection of a specific mutation was not observed and there were no significant differences between the three populations (data not shown). In addition, we found no mutation of the E2 gene (nucleotides 2756-3853)(data not shown).
Epigenetic changes in the URR were first analysed by bisulfite sequencing to determine methylation of CpG sites (Supplementary Table-S2). There was no evidence of methylation in W12Ser4(EPI) in any passage tested. We observed partial methylation at the proximal promoter of the integrated URR in W12Ser4B(INT)p39 (later passages were not examined) and methylation patterns in CaSki and SiHa that were consistent with previous reports (30,31).
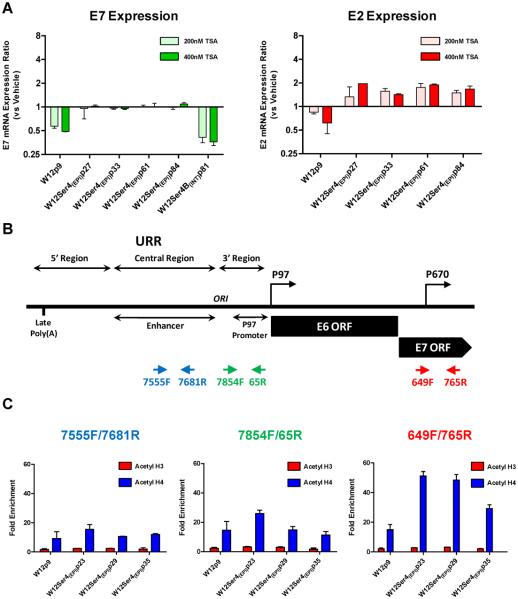
Next, we investigated differences in chromatin structure at the URR. In preliminary screening using the histone deacetylase inhibitor TSA, parental W12p9 cells showed a significant reduction in expression of E7 at 2 hours post-treatment, whereas W12Ser4(EPI) cells showed minimal changes (Figure-6A; left panel). W12Ser4B(INT)p81 showed reduced levels of E7 after TSA treatment, consistent with previous data (32, 33). In addition, while levels of E2 reduced in W12p9 in response to TSA, albeit by less than E7, they increased in W12Ser4(EPI) cells (Figure-6A; right panel). In view of the differential responses to TSA in W12p9 and W12Ser4(EPI), we performed ChIP analysis of acetylated histones around the URR, and quantified pull-down of sequences spanning the HPV16 enhancer, early promoter and late promoter (the last being within the E7 open reading frame) (Figure-6B). There were no differences in levels of acetylated histone H3, which were low in both W12p9 and W12Ser4(EPI). However, in comparison to W12p9 there was a significant increase in acetylated histone H4 in W12Ser4(EPI), particularly around the late promoter (two-sample t-test for W12p9 versus W12Ser4(EPI)p23 and W12Ser4(EPI)p29, p<0.01), when early gene transcription per template was maximal (Figure-6C). However, there was no significant difference between W12p9 and W12Ser4(EPI)p35, when transcription levels had reduced. Together these data imply that transcriptional deregulation in W12Ser4(EPI) was due, at least in part, to alterations in chromatin structure downstream of the URR.
Figure-6. Analysis of changes in histone acetylation state around the HPV16 URR in W12Ser4(EPI).
(A): qPCR analysis of E7 and E2 mRNA expression (versus vehicle-only controls) following treatment of cells with TSA at either 200nM or 400nM for 2 hours. (B): Schematic of the HPV16 URR, early promoter (P97) and late promoter (P670) regions. Arrowheads in blue, green and red indicate viral sequences quantified by qPCR following ChIP. (C): qPCR analysis of ChIP assays for binding of acetylated histone H3 and H4 around the HPV16 URR. For each cell population analysed, graphs show fold enrichment (versus pre-immune serum negative control samples) for sequences at the enhancer (7555F/7681R), early promoter (7854F/65R) or late promoter (649F/765R), following ChIP with antibodies against either acetylated histone H3 or acetylated histone H4.
Discussion
While in vitro progression of keratinocytes containing integrated HR-HPV has been described previously (for example, see (25)), we describe here the first model of HPV16 episome-associated cervical carcinogenesis. We observed a number of parallels with integrant-associated progression (2), although the underlying mechanisms differ.
First, we found that deregulation of viral early gene mRNA expression, leading to elevated transcript levels, was an early event in episome-associated progression. Deregulation preceded the increase in viral copy number, which was likely to have resulted from greater availability of viral replication factors. The level of deregulation per template was relatively modest in W12Series4(EPI) compared to W12Ser4B(INT), perhaps attributable to the different transcriptional ‘contexts’ of episomal versus integrated viral DNA (32), as well as deletion of the transcriptional inhibitor E2 in the latter (9). Nevertheless, the overall increases in early gene expression per cell – the more important determinant of phenotype – were similar in both culture series, due to markedly higher viral copy numbers in W12Ser4(EPI). Selection of a specific URR sequence alteration was not required for transcriptional deregulation. Instead, the kinetics of viral transcriptional responses to TSA and increased acetylation, especially of the late promoter region, in W12Ser4(EPI) indicated that altered chromatin structure may have been responsible, at least in part, for the transcriptional deregulation in episome-associated neoplastic progression [as well as having a role in the productive viral life cycle (21,24,34)]. Interestingly, other studies have shown that acetylation around the HPV31 late promoter is associated with early gene transcription in undifferentiated keratinocytes (24).
Second, the episome-containing cells acquired a growth advantage in vitro, similar to integrant containing cells (29). Interestingly, at the point of divergence of W12Ser4(EPI) and W12Ser4B(INT), the cell population contained a transcriptionally active integrant that came to dominate in W12Ser4B(INT) and was also independently selected in W12Ser2 (9). This integrant was therefore present at an early point in the history of W12 and had a considerable growth advantage in a mixed cell population. The fact that W12Ser4(EPI) cells outcompeted the transcriptionally active integrant-containing cells indicates that they acquired an even stronger selective advantage, as supported by the increased clonogenicity of W12Ser4(EPI) cells compared to the integrant-containing cells. Given that the overall increases in E6 and E7 expression during the period of selection were similar in both W12Ser4(EPI) and W12Ser4B(INT), it is unlikely that increased clonogenicity was a direct effect of viral oncogene levels. Indeed, increased cell proliferation rate, a known consequence of higher viral oncogene expression (29,35), was similar in both culture series.
Third, reduced transcription of the viral early genes, leading to reduced levels of the E7 oncoprotein (but not E2 or E6), were observed in W12Ser4(EPI) after acquisition of invasiveness. This mirrors the late stages of integrant-associated carcinogenesis, where reduction in viral oncogene expression levels is attributable to URR methylation (31). In W12Ser4(EPI), we found no evidence of URR methylation, although there was some reduction of histone H4 acetylation, particularly at the late promoter, suggesting that epigenetic mechanisms may also have contributed to the reduction in viral oncogene expression. Interestingly, reduced E7 expression had no effect on cell proliferation in W12Ser4(EPI), suggesting alternative contributions to growth during the late stages of progression, from host genes or possibly the viral E5 protein (36).
Dynamic changes in viral expression in episome- and integrant-associated cervical carcinogenesis are likely to reflect responses to the continually changing selection pressures encountered during neoplastic progression. For example, in established malignancy continued high-level E7 expression and consequent chromosomal instability may be unnecessary for progression and even deleterious, so that cells with reduced oncogene expression would have a survival advantage (31). The selective downregulation of E7 protein (relative to E2 and E6) that we observed in W12Ser4(EPI) is consistent with this hypothesis. The dynamic changes we observed contrast with a recent cross-sectional clinical study, which suggested that HR-HPV oncogene expression levels are similar across the spectrum of cervical disease, regardless of viral physical state (37). We believe that the previous conclusions may have been confounded by the nature of the samples analysed. The key issue in these studies is how gene expression changes in relation to normally regulated episomes in basal-like cells (which can be addressed in our in vitro system). The previous study grouped expression data from H-SILs and L-SILs, preventing identification of transcriptional changes that we predict would occur in basal-like cells in the progression from L-SIL to H-SIL (by both episome- and integrant-associated routes). Furthermore, the L-SILs analysed were not subjected to micro-dissection and therefore measurements of episome-derived oncogene expression in basal cells may have been masked by high-level expression in the differentiated layers during the late stages of the viral life cycle. Thus, it would be interesting to restrict analysis of clinical material to the proliferating layer and consider H-SILs and L-SILs separately, to determine if consistencies with our in vitro data emerge. Longitudinal clinical studies of individual patients would also help to address this issue, although such studies are difficult to undertake.
Fourth, W12Ser4(EPI) showed adaptations for persistence of viral episomes and early gene expression, allowing continued cell survival. This again mirrors integrant-associated carcinogenesis, where persistence of E6/E7 is required to prevent activation of senescence pathways (38,39) and is achieved by insertion into (and replication in concert with) the host genome. This renders integrants insensitive to viral clearance mechanisms such as ISGs (10,40). The requirements for persistence were more complex in W12Ser4(EPI), since episomal genomes remain vulnerable to clearance mechanisms and (unlike integrants) require viral early gene expression for replication. The emergent population was therefore required to achieve an appropriate balance of early gene expression, to maintain those proteins required for replication, cell survival and inhibition of viral clearance mechanisms, yet reduce those that can exert deleterious effects (see above). Our data suggest that this was achieved through a striking dissociation between protein and transcript levels of both E2 and E6. Consistent with its function as a replication protein, increased levels of E2 protein correlated with viral copy number, suggesting an important role in episomal persistence. Retention of high E6 would be likely to mediate continued cell survival through inhibition of p53 and other pro-apoptotic factors (41). As E6 can also inhibit interferon regulatory factor 3 (42), it may also have contributed to the decreased levels of ISGs, further supporting viral persistence. Nevertheless, despite stable E6 protein levels, we saw further decreases in ISG levels at later passages, suggesting that additional mechanisms may also have contributed to ISG reduction. It will be important in future work to determine how HR-HPV proteins inhibit endogenous and exogenous ISG induction pathways and functionally test their significance in regulating episome numbers.
The increase in E2/E6 proteins was not due to mutations of the respective genes in W12Ser4(EPI). Dissociation between protein and mRNA levels may therefore be due to changes in protein stability, altered splicing of early transcripts or, in the case of E2, use of a different transcription start site downstream of the viral oncogenes (43). The latter could account for our observation of higher relative levels of E2 transcripts compared to E6 and E7 in W12Ser4(EPI). Expression of E2 from heterologous promoters can have deleterious effects in some experimental systems, including repression of E6/E7 transcription and apoptosis (38,44). However, overall levels of E2 expressed from the homologous viral promoter in W12Ser4(EPI) remained low, with enrichment of nuclear protein being required for Western blot detection. Nevertheless, it may be that the increased levels of E2 protein seen after acquisition of invasiveness in W12Ser4(EPI) may have contributed to the associated reduction in early gene transcription.
It will be interesting to determine if similar observations can be made using other cell lines containing episomes of HPV16 (45-47) or other HR-HPV types (48, 49). W12E retains HPV16 episomes but may be unsuitable for investigations of early events in episome-associated progression since it has a high copy number [~1000 per cell, (29)] and reforms an H-SIL in organotypic culture (6). Given that episome-derived oncogene transcripts are detected at a much higher frequency in HPV31/33-positive cervical carcinomas (50), it is possible that features of episome-associated carcinogenesis vary according to HR-HPV type.
Overall, our results demonstrate that HPV16 episome-associated (as well as integrant-associated) cervical neoplastic progression in vitro are characterised by fluctuations in viral and host gene expression, consistent with adaptation to continually evolving selection pressures in order to acquire or retain a competitive advantage. Given that monolayer cultures cannot support the late stages of the HR-HPV life cycle, events selected spontaneously during W12 evolution are of no direct benefit to the virus. In view of the relative simplicity of our model system, it is striking that long-term cultures of W12 can lead to selection of viral and host events of known importance in cervical carcinogenesis in vivo (8,11-13). While long-term culture cannot recapitulate all the selection pressures that occur in tumours (e.g. effects of stromal factors and the immune system), our data indicate that many of the selection pressures that apply in vitro mirror those encountered in vivo. Accordingly, exploitation of models such as W12 is an attractive initial strategy to obtain new insights into the natural history of cervical neoplasia, particularly the temporal associations between key events. Our present data suggest that despite the complex and varied ways in which HPV16-positive cervical keratinocytes can progress to the invasive phenotype, there are fundamental commonalities.
Supplementary Material
Acknowledgements
We thank Stephen Inglis and Norman Maitland for reagents, Nicolas Wentzensen for advice regarding APOT and Pamela Stacey for assistance with immunohistochemistry.
1. Financial support: Medical Research Council, Cancer Research UK and Gates Cambridge Trust.
References
- 1.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992;23(2):117–28. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- 2.Pett M, Coleman N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J Pathol. 2007;212(4):356–67. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 3.Klaes R, Woerner SM, Ridder R, et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59(24):6132–6. [PubMed] [Google Scholar]
- 4.Dong XP, Stubenrauch F, Beyer-Finkler E, Pfister H. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int J Cancer. 1994;58(6):803–8. doi: 10.1002/ijc.2910580609. [DOI] [PubMed] [Google Scholar]
- 5.May M, Dong XP, Beyer-Finkler E, Stubenrauch F, Fuchs PG, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. Embo J. 1994;13(6):1460–6. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lace MJ, Isacson C, Anson JR, et al. Upstream regulatory region alterations found in human papillomavirus type 16 (HPV-16) isolates from cervical carcinomas increase transcription, ori function, and HPV immortalization capacity in culture. J Virol. 2009;83(15):7457–66. doi: 10.1128/JVI.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veress G, Szarka K, Dong XP, Gergely L, Pfister H. Functional significance of sequence variation in the E2 gene and the long control region of human papillomavirus type 16. J Gen Virol. 1999;80(Pt 4):1035–43. doi: 10.1099/0022-1317-80-4-1035. [DOI] [PubMed] [Google Scholar]
- 8.Alazawi W, Pett M, Arch B, et al. Changes in cervical keratinocyte gene expression associated with integration of human papillomavirus 16. Cancer Res. 2002;62(23):6959–65. [PubMed] [Google Scholar]
- 9.Dall KL, Scarpini CG, Roberts I, et al. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68(20):8249–59. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 10.Herdman MT, Pett MR, Roberts I, et al. Interferon-beta treatment of cervical keratinocytes naturally infected with human papillomavirus 16 episomes promotes rapid reduction in episome numbers and emergence of latent integrants. Carcinogenesis. 2006;27(11):2341–53. doi: 10.1093/carcin/bgl172. [DOI] [PubMed] [Google Scholar]
- 11.Muralidhar B, Goldstein LD, Ng G, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212(4):368–77. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 12.Pett MR, Alazawi WO, Roberts I, et al. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res. 2004;64(4):1359–68. doi: 10.1158/0008-5472.can-03-3214. [DOI] [PubMed] [Google Scholar]
- 13.Pett MR, Herdman MT, Palmer RD, et al. Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proc Natl Acad Sci U S A. 2006;103(10):3822–7. doi: 10.1073/pnas.0600078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley MA, Browne HM, Appleby M, Minson AC. Properties of a non-tumorigenic human cervical keratinocyte cell line. International Journal of Cancer. 1989;43:672–6. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 15.Freshney RI. Culture of Animal Cells: A Manual of Basic Technique. 4th ed Wiley-Liss; New York: 2000. [Google Scholar]
- 16.Gonzalez MA, Pinder SE, Callagy G, et al. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J Clin Oncol. 2003;21(23):4306–13. doi: 10.1200/JCO.2003.04.121. [DOI] [PubMed] [Google Scholar]
- 17.Smedts F, Ramaekers FC, Vooijs PG. The dynamics of keratin expression in malignant transformation of cervical epithelium: a review. Obstet Gynecol. 1993;82(3):465. [PubMed] [Google Scholar]
- 18.Hibma MH, Raj K, Ely SJ, Stanley M, Crawford L. The interaction between human papillomavirus type 16 E1 and E2 proteins is blocked by an antibody to the N-terminal region of E2. Eur J Biochem. 1995;229(2):517–25. doi: 10.1111/j.1432-1033.1995.0517k.x. [DOI] [PubMed] [Google Scholar]
- 19.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89(5):1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 21.del Mar Pena LM, Laimins LA. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. J Virol. 2001;75(20):10005–13. doi: 10.1128/JVI.75.20.10005-10013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 23.Mogal A, Abdulkadir SA. Effects of Histone Deacetylase Inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Mol Cell Probes. 2006;20(2):81–6. doi: 10.1016/j.mcp.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Wooldridge TR, Laimins LA. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology. 2008;374(2):371–80. doi: 10.1016/j.virol.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurlin PJ, Kaur P, Smith PP, Perez-Reyes N, Blanton RA, McDougall JK. Progression of human papillomavirus type 18-immortalized human keratinocytes to a malignant phenotype. Proc Natl Acad Sci U S A. 1991;88(2):570–4. doi: 10.1073/pnas.88.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rader JS, Golub TR, Hudson JB, Patel D, Bedell MA, Laimins LA. In vitro differentiation of epithelial cells from cervical neoplasias resembles in vivo lesions. Oncogene. 1990;5(4):571–6. [PubMed] [Google Scholar]
- 27.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 28.Kalantari M, Bernard HU. Gene expression of papillomaviruses. In: Campo MS, editor. Papillomavirus Research: From natural history to vaccines and beyond. Caister Academic Press; 2006. pp. 11–8. [Google Scholar]
- 29.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69(5):2989–97. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badal V, Chuang LS, Tan EH, et al. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77(11):6227–34. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Tine BA, Kappes JC, Banerjee NS, et al. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol. 2004;78(20):11172–86. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bechtold V, Beard P, Raj K. Human papillomavirus type 16 E2 protein has no effect on transcription from episomal viral DNA. J Virol. 2003;77(3):2021–8. doi: 10.1128/JVI.77.3.2021-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finzer P, Ventz R, Kuntzen C, Seibert N, Soto U, Rosl F. Growth arrest of HPV-positive cells after histone deacetylase inhibition is independent of E6/E7 oncogene expression. Virology. 2002;304(2):265–73. doi: 10.1006/viro.2002.1667. [DOI] [PubMed] [Google Scholar]
- 34.Zhao W, Noya F, Chen WY, Townes TM, Chow LT, Broker TR. Trichostatin A up-regulates human papillomavirus type 11 upstream regulatory region-E6 promoter activity in undifferentiated primary human keratinocytes. J Virol. 1999;73(6):5026–33. doi: 10.1128/jvi.73.6.5026-5033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Knebel Doeberitz M, Oltersdorf T, Schwarz E, Gissmann L. Correlation of modified human papilloma virus early gene expression with altered growth properties in C4-1 cervical carcinoma cells. Cancer Res. 1988;48(13):3780–6. [PubMed] [Google Scholar]
- 36.Bouvard V, Matlashewski G, Gu ZM, Storey A, Banks L. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology. 1994;203(1):73–80. doi: 10.1006/viro.1994.1456. [DOI] [PubMed] [Google Scholar]
- 37.Hafner N, Driesch C, Gajda M, et al. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. 2008;27(11):1610–7. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed] [Google Scholar]
- 38.Goodwin EC, Yang E, Lee CJ, Lee HW, DiMaio D, Hwang ES. Rapid induction of senescence in human cervical carcinoma cells. Proc Natl Acad Sci U S A. 2000;97(20):10978–83. doi: 10.1073/pnas.97.20.10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall AH, Alexander KA. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol. 2003;77(10):6066–9. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang YE, Pena L, Sen GC, Park JK, Laimins LA. Long-term effect of interferon on keratinocytes that maintain human papillomavirus type 31. J Virol. 2002;76(17):8864–74. doi: 10.1128/JVI.76.17.8864-8874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78(21):11451–60. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12(13):2061–72. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lace MJ, Anson JR, Thomas GS, Turek LP, Haugen TH. The E8--E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J Virol. 2008;82(21):10841–53. doi: 10.1128/JVI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webster K, Parish J, Pandya M, Stern PL, Clarke AR, Gaston K. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J Biol Chem. 2000;275(1):87–94. doi: 10.1074/jbc.275.1.87. [DOI] [PubMed] [Google Scholar]
- 45.Flores ER, Allen-Hoffmann BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262(2):344–54. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, Yi SM, Anderson ME, et al. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc Natl Acad Sci U S A. 2004;101(7):2094–9. doi: 10.1073/pnas.0308615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprague DL, Phillips SL, Mitchell CJ, et al. Telomerase activation in cervical keratinocytes containing stably replicating human papillomavirus type 16 episomes. Virology. 2002;301(2):247–54. doi: 10.1006/viro.2002.1542. [DOI] [PubMed] [Google Scholar]
- 48.Bedell MA, Hudson JB, Golub TR, et al. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65(5):2254–60. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frattini MG, Lim HB, Laimins LA. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci U S A. 1996;93(7):3062–7. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vinokurova S, Wentzensen N, Kraus I, et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68(1):307–13. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.