Abstract
Nonsense-mediated mRNA decay (NMD) is of universal biological significance1-3. It has emerged as an important global RNA, DNA and translation regulatory pathway4. By systematically sequencing 737 genes (annotated in the Vertebrate Genome Annotation database) on the human X chromosome in 250 families with X-linked mental retardation, we identified mutations in the UPF3 regulator of nonsense transcripts homolog B (yeast) (UPF3B) leading to protein truncations in three families: two with the Lujan-Fryns phenotype5,6 and one with the FG phenotype7. We also identified a missense mutation in another family with nonsyndromic mental retardation. Three mutations lead to the introduction of a premature termination codon and subsequent NMD of mutant UPF3B mRNA. Protein blot analysis using lymphoblastoid cell lines from affected individuals showed an absence of the UPF3B protein in two families. The UPF3B protein is an important component of the NMD surveillance machinery8,9. Our results directly implicate abnormalities of NMD in human disease and suggest at least partial redundancy of NMD pathways.
Mental retardation is a common debilitating condition of the human brain present in 1%–2% of the population. Major advances have been made in unraveling the molecular basis of this disease in cases in which the causes are primarily genetic. X-linked mental retardation (XLMR) is defined simply as the presence of mental retardation (defined as an IQ <70) where the causative gene is on the X chromosome. Classical linkage analysis together with positional cloning has led to the identification of a number of genes10,11, but many remain elusive. In order to identify the remaining genes, we embarked on a systematic and comprehensive approach of resequencing of 737 genes, annotated in the Vertebrate Genome Annotation (VEGA) database, in probands from 250 families with mental retardation, which are compatible with X linkage and which did not have mutations in the currently known XLMR-linked genes12-14.
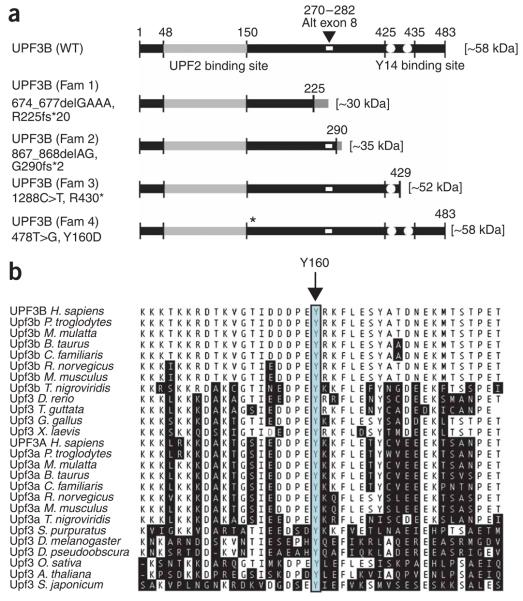
As part of this effort, we identified mutations in the UPF3 regulator of nonsense transcripts homolog B (yeast) (UPF3B) gene. The UPF3B gene is composed of 11 exons and is located on Xq24 (at ~118.8 Mb, build 36). We identified three different protein-truncating mutations in three families. One family had a clinical diagnosis of FG syndrome (OMIM 305450; family 407/K8890). Two others had a clinical diagnosis of Lujan-Fryns syndrome (LFS; OMIM 309520). In family 1 (which had a diagnosis of FG syndrome), we identified a deletion of four nucleotides, 674_677delGAAA, in exon 7 of UPF3B, leading to a translational frameshift and subsequent protein truncation, R225fs*20 (Fig. 1a). Family 2 (LFS) had a two-nucleotide deletion, 867_868delAG, in exon 9. This mutation causes a frameshift and UPF3B protein truncation G290fs*2 (Fig. 1b). In family 3 (with LFS), we identified a nonsense mutation, 1288C>T, in exon 10, leading to a premature termination codon (PTC) and subsequent protein truncation, R430* (Fig. 1c). A subsequent analysis of 118 additional probands from a cohort of new families with putative XLMR (collected by the Greenwood Genetic Center) identified a single-nucleotide substitution, 478T>G, in exon 5 in a family K9170 (family 4, Fig. 1d) with nonsyndromic XLMR. This nucleotide change is predicted to cause a missense amino acid change, Y160D (Fig. 2a). The tyrosine residue at this position is invariant in UPF3B and UPF3A orthologs from animals and plants, suggesting that it is of crucial importance for UPF3B and UPF3A protein function (Fig. 2b). The UPF3B mutation in each family was found only in affected individuals. We did not find any other sequence variants in the 250-case cohort. Moreover, complete sequencing of the coding sequences and splice site junctions of UPF3B in 730 control X chromosomes did not show these or any further variants (data not shown). We did not observe the Y160D alteration in an additional 422 normal males. Taken together, these results strongly suggest that the three protein-truncating mutations and the mutation resulting in the missense Y160D change are the disease-causing mutations in these families.
Figure 1.
UPF3B mutations identified in this study. cDNA (NM_080632) and protein (NP_542199) annotation of individual mutations is shown for each family. (a) Family 1 (previously reported)29. A four-nucleotide deletion, 674_677delGAAA, has been identified in this family; it was present in the two affected brothers and their mother (II-2; 100% X-chromosome inactivation skewing) but was absent in the grandmother (I-2; random, 61:39 skewing), indicating a probable de novo mutation event in the mother (II-2). (b) Family 2 (ascertained in Australia). The mutation was present in both affected boys and the mother. (c) Family 3, from the UK. Three generations with affected males have been recorded. This mutation, which causes a PTC, was identified in four affected males (no sample was available from II-3) and three obligate female carrier mothers (II-1, III-1 and III-3) and was absent from three unaffected males (II-5, III-4 and IV-2). (d) Family 4 (two generations of affected males; from the USA). The X chromosome inactivation assay showed moderately to highly skewed inactivation in all carrier females in this family, whereas noncarriers showed random skewing (data not shown; see also Methods). Open symbols represent normal individuals, and filled squares represent affected males. Probands in each family are indicated with arrows. Individual generations are numbered with Roman numerals (I, II, III). WT, wild-type allele; MUT, mutant allele; NT, not tested. DNA sequence chromatograms of individual mutations and wild-type alleles are given in Supplementary Figure 1 online.
Figure 2.
Schematic of wild-type UPF3B and UPF3B from affected individuals. (a) There are two recognized domains within the UPF3B protein: one at residues 48–150 (light gray), which is involved in binding to UPF2 (ref. 30), and the other, spanning residues 425–435 (white dots), through which UPF3B interacts with the components of the exon junction complex and Y14 in particular18. The three protein-truncating mutations (in families 1, 2 and 3) and one missense mutation (in family 4) of UPF3B, and the resulting proteins, are also shown. The position of the alternatively spliced exon 8 (residues 270–282) is indicated as a small white rectangle. (b) ClustalW multiple protein alignment of partial UPF3A and UPF3B orthologs. The amino acid residues that differ from the sequence of the human UPF3B are shaded. The highly conserved tyrosine (Y) at position 160 is indicated with an arrow and outlined with a box.
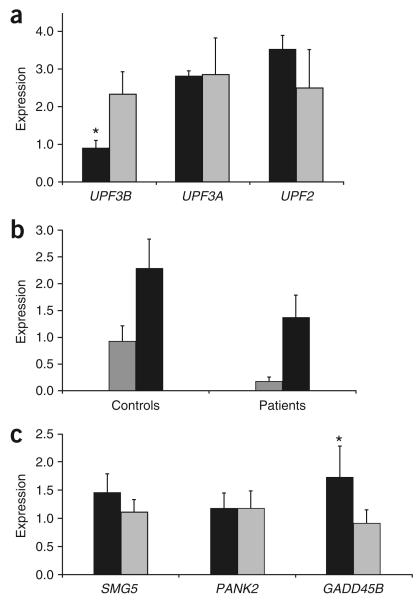
Three of the four mutations identified introduce a PTC into the UPF3B mRNA ORF (Fig. 2a). Such PTCs often trigger NMD of mRNA1. Given that the UPF3B protein itself is an important component of NMD surveillance protein complexes8,9,15, we investigated the consequence of the probable lack of UPF3B function on NMD of its own mutant UPF3B PTC-containing mRNA. For this purpose, we used RNA isolated from the Epstein-Barr virus–transformed lymphoblastoid cell lines (LCLs) of affected individual III-1 from family 1 and individuals III-4 and III-5 from family 2. Real-time quantitative RT-PCR (qRT-PCR) analysis showed significantly reduced levels of the UPF3B PTC-containing mRNAs from all three individuals (Fig. 3a,b). This result suggested that the NMD-mediated mRNA degradation of UPF3B PTC-containing mRNA was not substantially compromised by the lack of UPF3B protein function. Blocking protein translation in LCLs with 100 μg ml−1 cycloheximide restored UPF3B PTC-containing mRNA expression (Fig. 3b). To investigate further the functionality of NMD in the cells of these individuals, we selected (based on their expression in LCLs) three known NMD targets: SMG5, PANK2 and GADD45B (ref. 16). Expression of GADD45B mRNA (part of the classical NMD pathway target) was significantly higher in individuals with UPF3B PTCs compared with controls (P < 0.05), indicating that the classical NMD pathway is compromised in these individuals. By contrast, mRNA expression of SMG5 and PANK2 (alternative NMD pathway targets) did not significantly differ between individuals with UPF3B PTCs and controls, suggesting that the alternative NMD pathway is intact (Fig. 3c). Thus, these results demonstrate that NMD is only partially compromised in individuals with UPF3B PTCs and that this is primarily due to dysfunction of the classical NMD pathway. Moreover, the data suggest that although UPF3B seems to be more important in the classical NMD pathway, the UPF3B transcript itself may be at least partially regulated by the alternative pathway.
Figure 3.
Analysis of RNA expression of the UPF3B and other genes in controls and affected individuals. (a) Mean expression (± s.d.) of UPF3B, UPF3A and UPF2 in affected individuals (n = 3, black box) and controls (n = 4, gray box), as determined by real-time PCR. Expression was measured in three independent real-time PCRs and was normalized against expression of the ACTB gene in the same sample using the relative standard curve method. * P = 0.01, Student's t test. (b) UPF3B mRNA expression is significantly downregulated in affected individuals (n = 3) with UPF3B PTCs compared with unaffected controls (n = 3), as shown by real-time qRT-PCR. Inhibition of translation by treatment of LCLs with 100 mg ml−1 cycloheximide for 6 h released the UPF3B mRNA from NMD. Although the expression of UPF3B also increased in controls (n = 3) as a consequence of cycloheximide treatment, this increase was not as marked as in affected individuals. The differential increase of UPF3B expression in affected individuals versus controls as a consequence of cycloheximide treatment was statistically significant (P = 0.008) at the 5% significance level (analysis of variance (ANOVA)). Measurements of expression were normalized against the expression of the ACTB gene in the same sample. Gray boxes indicate untreated LCLs; black boxes indicate cycloheximide-treated LCLs. All samples were run in triplicate. Bars indicate s.d. (c) Mean expression (± s.d.) of SMG5, PANK2 and GADD45B mRNA in affected individuals (n = 3, black box) and controls (n = 4, gray box) by real-time PCR. Expression was measured in two independent real-time PCRs and was normalized against ACTB expression using the relative standard curve method. * P < 0.01 (Student's t test).
We also investigated the effect of these mutations on UPF3B protein expression. Protein blot analysis using two peptide-based antibodies targeted to the N terminus (hUPF3B_901 and hUPF3B_913) did not detect wild-type or truncated UPF3B proteins in lysates from LCLs of four affected individuals (Fig. 4) (a truncated protein of ~30 kDa was theoretically possible in the affected males from family 1, and a protein of ~35 kDa was possible in affected males from family 2; Fig. 2a). Irrespective of NMD-mediated degradation of UPF3B PTC-containing mRNAs, which could be tissue specific17 and thus different in the brain, the resulting UPF3B proteins of affected individuals from each of the three families would lack either the entire C-terminal Y14 protein-interacting domain (in families 1 and 2) or the major part of it (in family 3) (Fig. 2a). This domain is essential for NMD through proper assembly of the exon junction complex18,19. As such, the three different protein-truncating mutations are probably complete loss-of-function mutations of UPF3B.
Figure 4.
Protein blot analysis of UPF3B. Protein lysates from two control LCLs and LCLs from four affected individuals are shown. Cellular proteins were separated on a 8% acrylamide gel and transferred onto nitrocellulose membrane. The membrane was probed with the UPF3B_901 (peptide 1) primary polyclonal sheep antibody and subsequently probed with horseradish peroxidase (HRP)-conjugated secondary donkey anti-sheep. The signal was detected using the enhanced chemiluminescence (ECL) method (upper panel). The position of the full-length, ~58-kDa UPF3B protein is indicated by an arrow. The UPF3B protein was detected in protein lysates from controls but not from any of the four affected individuals tested. The additional, larger protein species detected by the hUPF3B_901 antibody in lysates LCLs from controls and affected individuals represents nonspecific binding of the donkey anti-sheep and/or immunoglobulin cross-reactivity. A control protein blot using the monoclonal mouse antibody SC-32233 (Santa Cruz Biotechnology) directed to GAPDH is also shown (lower panel). The same result was also achieved with the hUPF3B_913 antibody (specific to peptide 2; data not shown).
Previously published analyses9 and our RT-PCR (Fig. 5) and EST (data not shown) analyses show that UPF3B is expressed in a variety of tissues and developmental stages. The only characterized alternative splicing involves exon 8. Inclusion of exon 8 seems to be tissue specific (Fig. 5). The larger ORF of 1,452 bp (including exon 8) is translated to a 483-residue protein (~58 kDa, Figs. 2a and 4).
Figure 5.
Tissue expression profile of human UPF3B and UPF3A mRNA isoforms. Selected RNAs from the Total Human RNA Master Panel II (Clontech) were subjected to reverse transcription. The efficiency of the reaction was tested by PCR using primers specific to the ubiquitously expressed ESD gene (bottom panel). The expression of the two isoforms of either UPF3B or UPF3A were assessed by semiquantitative RT-PCR across the alternatively spliced exon 8 of UPF3B with primers UPF3B Ex5F and UPF3B Ex9R (upper panel), and across exon 4 in UPF3A with primers UPF3A Ex1F and UPF3A Ex6n7R (middle panel). The alternative isoforms, labeled isoform 1 and isoform 2, and the resulting two heteroduplexes, migrating as one product above isoforms 1 and 2, are indicated with arrowheads.
We have shown that the UPF3B PTC-containing mRNA from the affected individuals we studied is subject to NMD, even in the absence of the UPF3B protein itself. Thus, UPF3B is an important but probably nonessential component of the NMD protein machinery (see also ref. 15). Redundancy of the NMD pathways has recently been uncovered15,20. UPF3B shares high similarity and comparable tissue specificity, but not necessarily the same level of expression (Fig. 5 and ref. 9), with its autosomal paralog UPF3A (partial alignment shown in Fig. 2b). Therefore, we tested whether UPF3A expression was altered to compensate for the reduction of UPF3B expression in LCLs. We did not find any significant differences in UPF3A mRNA expression (or in UPF2 mRNA expression) by qRT-PCR (Fig. 3a).
Eleven of the thirteen affected individuals from the four families were available for examination. The clinical features of these affected individuals are summarized in Table 1. Almost all of the clinical features frequently observed in the affected males with truncating UPF3B mutations were absent in the three affected males with the Y160D missense change (Table 1 and Fig. 6). With respect to the eight males with the truncating mutations, the features present at >50% frequency were slender build with poor musculature (87%), long, thin face (75%), high arched palate (75%), high nasal bridge (75%) and pectus (62%). Notably, we observed autistic features (4/8) and behavioral problems (3/8) as well, although these are common coincident findings in mental retardation. Overall, the clinical phenotype is variable, although many of the clinical features are suggestive of LFS and FG phenotypes. Apart from family 1 and perhaps family 3, most individuals are not overtly dysmorphic. All carrier females examined had normal intelligence and physical examinations. For family 4 with the Y160D missense change, the males had rather normal physical examinations, consistent with the designation of nonsyndromic XLMR.
Table 1. Summary of clinical features in affected males from families with UPF3B mutations.
| Family 1 (n = 2) |
Family 2 (n = 2) |
Family 3 (n = 4) |
Total, families 1–3 (n = 8) |
Family 4 (n = 3) |
Total, all families (n = 11) |
|
|---|---|---|---|---|---|---|
| Height >97th centile | 0 | 2 | 0 | 2/8 | 0 | 2/11 |
| Slender build | 2 | 2 | 3 | 7/8 | 0 | 7/11 |
| Head circumference >97th centile | 2 | 1 | 0 | 3/8 | 1 | 4/11 |
| Poor musculature | 2 | 2 | 2 | 6/8 | 0 | 6/11 |
| Long, thin face | 1 | 2 | 3 | 6/8 | 0 | 6/11 |
| Maxillary hypoplasia | 2 | 0 | 2 | 4/8 | 0 | 4/11 |
| High arched palate | 0 | 2 | 4 | 6/8 | 0 | 6/11 |
| High nasal bridge | 0 | 2 | 4 | 6/8 | 0 | 6/11 |
| Prominent jaw | 0 | 0 | 4 | 4/8 | 0 | 4/11 |
| Hypernasal voice | 0 | 0 | 4 | 4/8 | 0 | 4/11 |
| Pectus | 2 | 2 | 1 | 5/8 | 0 | 5/11 |
| Hypotonia | 1 | 1 | 1 | 3/8 | 0 | 3/11 |
| Constipation | 2 | 0 | 0 | 2/8 | 0 | 2/11 |
| Corpus callosum abnormality | 2a | Unknown | 0/2 | 2/4 | Unknown | 2/4 |
| Autistic features | 1 | 0 | 3 | 4/8 | 1 | 5/11 |
| Behavioral problems | 0 | 1 | 2 | 3/8 | 0 | 3/11 |
| No MR | 1 | 1/11 | ||||
| Mild MR | 1 | 1 | 1 | 3/8 | 2 | 5/11 |
| Moderate MR | 1 | 1/8 | 1/11 | |||
| Severe MR | 1 | 1 | 2 | 4/8 | 4/11 | |
MR, mental retardation.
See Supplementary Figure 2 online for futher details.
Figure 6.
Facial features of the probands from families 1, 2, 3 and 4. (a) Individual III-1 from family 1, showing a long, thin face, broad forehead and maxillary hypoplasia. (b) Individual III-4 from family 2, showing a long, thin face. (c) Individual IV-1 from family 3, showing a long, thin face, prominent forehead, facial asymmetry, high nasal bridge and a prominent chin. (d) Individual II-5 from family 4, demonstrating elongated but otherwise normal facial appearance. We obtained informed consent to publish photos of affected individuals from all four families.
LFS (OMIM 309520), or XLMR with marfanoid habitus syndrome, is a syndromal form of XLMR predominantly affecting males. The main features include mild to moderate mental retardation, distinct facial dysmorphism (prominent forehead, maxillary hypoplasia, small mandible, long nose with short and deep philtrum, thin upper lip and low-set normally shaped ears), hypernasal voice, marfanoid-like features (tall stature after puberty and long, thin hyper-extensible fingers and toes) and hypotonia5,6. Behavioral problems and psychiatric disorders are frequently present21,22. FG syndrome was first described by Opitz and Kaveggia in 1974 in a family with five affected males that presented with mental retardation, macrocephaly, imperforate anus and hypotonia7. Facial anomalies include a high broad forehead with frontal cowlick, hypertelorism and a prominent lower lip23. Clinical diagnosis of both conditions remains challenging. None of the physical features in either syndrome is a prerequisite, and many of the features overlap and are commonly seen in individuals with mental retardation.
Until recently the genetic basis for both FG and LFS syndromes was not known, although genetic heterogeneity was expected22,23. Others24 have identified a recurrent mutation, 2881C>T/R961W, in the MED12 gene in the original Opitz-Kaveggia (FG) syndrome family7 and five other FG families. Notably, another mutation in the same gene has been found25 in the original family described in ref. 5, suggesting that LFS and FG syndrome are allelic. Mutations in ZDHHC9 have also been reported in XLMR with marfanoid habitus and in one family where FG was suggested as a clinical diagnosis14. Our findings of UPF3B mutations in families ascertained as having LFS (two families) or FG (one family) provide further evidence for the overlap of these two clinical phenotypes.
The UPF3A and UPF3B proteins are crucial but are probably alternative components of the exon junction complex, which assembles about 20–24 bp upstream of each exon-exon junction on processed mRNA8. A recent study15 demonstrated that although the UPF3A and UPF3B proteins show considerable similarity, they differ functionally. They showed that the UPF3B protein is much more active in NMD and translation regulation than UPF3A15. UPF3B gene expression is also considerably higher than that of UPF3A in most human tissues, including adult and fetal brain (Fig. 5 and ref. 9). More recent work16 suggests the existence of an alternative NMD pathway, which is very likely to be independent of UPF3B and UPF3A. Our results on naturally occurring mutations in UPF3B strongly support the existence of such alternative NMD pathway(s). The molecular mechanism by which loss-of-function mutations of UPF3B lead to mental retardation and associated features in the individuals we studied is not known. It is conceivable that the absence of UPF3B leads to a partial, although apparently not complete, loss of NMD, especially in tissues where UPF3A expression levels are low. This may result in subtle deregulation of mRNA surveillance of multiple genes or their isoforms, which are otherwise physiologically controlled by NMD. There may also be an effect on the translation of certain mRNAs, nuclear export, RNA localization or a general increase in RNA noise (for example, expression of pseudogenes26 and viral sequences27). It is also possible, however, that the redundancy of UPF3B in NMD is complete in some tissues and that the phenotype associated with UPF3B mutations is due to a loss of functions other than those of NMD. The identification of naturally occurring loss-of-function mutations of the human UPF3B gene may therefore offer an opportunity to identify bona fide targets of UPF3B-activated NMD and translation control and may provide a glimpse into the complexities of human NMD pathways.
METHODS
Subjects and families
A detailed description of affected individuals and families is provided in Supplementary Methods online. This study was approved by the relevant institutional research ethics committees (Women's and Children's Hospital, Cambridge Medical Research Institute and Self Regional Hospital), and signed written consent was obtained from all participants.
UPF3B antibody design and application
Three UPF3B specific peptides were selected. Peptide 1 (amino acid residues 31–50: GDSSKGEDKQDRNKEKKEAL) was selected from the N-terminal end of the UPF3B protein from the region before the UPF2 binding site; peptide 2 (residues 209–230: RMREEKREERRRREIERKRQRE) was selected from the middle portion of UPF3B without known domains; and peptide 3 (residues 363–380: RERLKRQEEERRRQKERY) was selected from the C-terminal end of UPF3B just before the Y14 binding domain. We compared the sequences of all three peptides with publicly available human sequences in order to avoid possible cross-reactivity with UPF3A and other proteins. The sequence of peptide 1 diverged between human and mouse UPF3B proteins, but the sequence of peptide 2 is 100% conserved and the sequence of peptide 3 is highly similar (2/19 differences). The UPF3B synthetic peptides (Mimotopes) were conjugated to diphtheria toxoid linked to the peptide via maleimidocaproyl-N-hydoxysuccinimide. Cross-bred Merino sheep were immunized in four to six sites at three weekly intervals with 2 mg protein per sheep in a 400 μl bolus administered subcutaneously. A priming injection of peptide in Freund's complete adjuvant was followed by three additional boosts of peptide in Freund's incomplete adjuvant. Two weeks after the last boost, the sheep were exsanguinated and the blood clotted to obtain serum. Polyclonal antibodies were purified by protein G affinity chromatography.
The polyclonal UPF3B-901 antibody (peptide 1) was used at a final concentration of 75 ng ml−1, and the polyclonal UPF3B-913 (peptide 2) antibody was used at a final concentration of 195 ng ml−1. The secondary horseradish peroxidase (HRP)-conjugated donkey antibody to sheep immunoglobulin G (IgG) (heavy plus light chains) were purchased from Chemicon and used at a dilution of 1:6,000.
Protein blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitors) on ice for 30 min, and debris was removed by high-speed centrifugation for 10 min. Cleared lysates were separated on an 8% acrylamide gel and transferred onto nitrocellulose membranes by semidry electroblotting using a Trans-Blot SD Semi-Dry Transfer Cell (Biorad). Membranes were blocked with 5% skim milk and 5% horse serum and then incubated with appropriate primary antibody and subsequently with secondary HRP-conjugated antibody. The blots were developed using the enhanced chemiluminescence (ECL) method (Pierce).
RT-PCR analyses
Total RNA was extracted from LCL with the RNeasy mini kit (Qiagen) and treated with DNase I (Qiagen). We primed 2 μg of RNA with 1 μg of random hexanucleotides and then subjected it to reverse transcription for 90 min at 42 °C using Superscript II (Invitrogen). The efficiency of the reaction was tested by PCR using primers specific to the ubiquitously expressed ESD gene. cDNAs were amplified with Taq DNA polymerase (Roche) and specific single-stranded DNA primers (35 cycles of denaturation, 94 °C for 30 s; annealing for 30 s (for specific Tm for each pair of primers, see Supplementary Table 1 online); extension, 72 °C for 30 s). PCR products were separated on agarose gel stained with 1% ethidium bromide.
Tissue culture and LCLs
The Epstein-Barr virus–immortalized B cell lines (B-LCLs) used in this study were established from peripheral blood lymphocytes of individuals III-1 and III-3 (family 1) and III-4 and III-5 (family 2), as described previously28. Once established, the LCL cell lines were cultured in RPMI 1640 (GIBCO/BRL) supplemented with 10% FCS, 2 mM l-glutamine, 0.017 mg ml−1 benzylpenicillin (CLS) and grown at 37 °C with 5% CO2.
Cycloheximide treatment of LCL lines
Approximately 3 × 106 cultured LCL cells in RPMI with 10% FCS were incubated with 100 μg ml−1 cycloheximide (Sigma) or medium alone for 6 h. LCL cells were harvested by centrifugation at 1,500g and then washed once in PBS before total RNA extraction and processing to cDNA as described above.
Quantitative real-time PCR
The relative standard curve technique was used to analyze the expression of the gene of interest normalized to ACTB expression in the same sample. The standard curves were prepared from equally pooled control cDNAs at 5, 10, 102, 103, 104, 105 and 106 dilution factors for each primer pair. Each reaction well contained 2 μl of template cDNA at appropriate concentrations for linear amplification based on the standard curve, 50 pmol of each primer and 1× SYBR green PCR Master Mix (Applied Biosystems) to a final volume of 25 μl. Reactions were carried out using a 7300 Real-time PCR System for 40 cycles (92 °C for 30 s and 60 °C for 30 s). Absorbance was measured at 494 nm at the end of each cycle. The purity of the PCR products was determined by melting curve analysis. Data were analyzed using System 7300 Software v.1.2.2 (Applied Biosystems).
GenBank accession numbers
Human UPF3B mRNA isoform 1, NM_080632; human UPF3B mRNA isoform 2, NM_023010; human UPF3B protein isoform 1, NP_542199; human UPF3B protein isoform 2, NP_075386; human UPF3A mRNA isoform 1, NM_023011; human UPF3A mRNA isoform 2, NM_080687; human UPF3A protein isoform 1, NP_075387; human UPF3A protein isoform 2, NP_542418.
ACKNOWLEDGMENTS
We thank the families studied for their participation and K. Dowling of the Faculty of Health Sciences, University of Adelaide for assistance with statistical analysis of the data. This work was supported by grants from the Australian National Health and Medical Research Council project grant 453457, the State of New South Wales (NSW) Health Department through their support of the NSW GOLD Service, a US National Institute of Child Health and Human Development grant (HD26202) to C.E.S., a grant from the South Carolina Department of Disabilities and Special Needs and the Wellcome Trust. Dedicated to the memory of E.F. Schwartz (1996–1998).
Footnotes
Requests for materials: jozef.gecz@adelaide.edu.au
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
URLs. NCBI: http://www.ncbi.nlm.nih.gov/; Ensembl: http://www.ensembl.org/
References
- 1.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 2.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Medghalchi SM, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 4.Weischenfeldt J, Lykke-Andersen J, Porse B. Messenger RNA surveillance: neutralising natural nonsense. Curr. Biol. 2005;15:R559–R562. doi: 10.1016/j.cub.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Lujan JE, Carlin ME, Lubs HA. A form of X-linked mental retardation with Marfanoid habitus. Am. J. Med. Genet. 1984;17:311–322. doi: 10.1002/ajmg.1320170124. [DOI] [PubMed] [Google Scholar]
- 6.Fryns JP, Buttiens M. X-linked mental retardation with Marfanoid habitus. Am. J. Med. Genet. 1987;28:267–274. doi: 10.1002/ajmg.1320280202. [DOI] [PubMed] [Google Scholar]
- 7.Opitz JM, Kaveggia EG. Studies of malformation syndromes of man 33: the FG syndrome. An X-linked recessive syndrome of multiple congenital anomalies and mental retardation. Z. Kinderheilkd. 1974;117:1–18. doi: 10.1007/BF00439020. [DOI] [PubMed] [Google Scholar]
- 8.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 9.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol. Cell. Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gecz J, Mulley JC. Genes for cognitive function: developments on the X. Genome Res. 2000;10:157–163. doi: 10.1101/gr.10.2.157. [DOI] [PubMed] [Google Scholar]
- 11.Raymond FL, Tarpey P. The genetics of mental retardation. Hum. Mol. Genet. 2006;15:R110–R116. doi: 10.1093/hmg/ddl189. [DOI] [PubMed] [Google Scholar]
- 12.Tarpey PS, et al. Mutations in the gene encoding the sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X–linked mental retardation. Am. J. Hum. Genet. 2006;79:1119–1124. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarpey PS, et al. Mutations in CUL4B, which encodes a ubiquitin e3 ligase subunit, cause an x-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 2007;80:345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond FL, et al. Mutations in ZDHHC9, a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am. J. Hum. Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUPF3a and hUPF3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan WK, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum. Mol. Genet. 2003;12:217–225. doi: 10.1093/hmg/ddg054. [DOI] [PubMed] [Google Scholar]
- 18.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 19.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 20.Gehring NH, et al. Exon-junction complex components specify distinct routes of nonsense mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 21.De Hert M, Steemans D, Theys P, Fryns JP, Peuskens J. Lujan-Fryns syndrome in the differential diagnosis of schizophrenia. Am. J. Med. Genet. 1996;67:212–214. doi: 10.1002/(SICI)1096-8628(19960409)67:2<212::AID-AJMG13>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.VanBuggenhout G, Fryns JP. Lujan-Fryns syndrome (mental retardation, X-linked, marfanoid habitus) Orphanet J. Rare Dis. 2006;1:26. doi: 10.1186/1750-1172-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battaglia A, Chines C, Carey JC. The FG syndrome: report of a large Italian series. Am. J. Med. Genet. A. 2006;140:2075–2079. doi: 10.1002/ajmg.a.31302. [DOI] [PubMed] [Google Scholar]
- 24.Risheg H, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz C, et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J. Med. Genet. 2007;44:472–477. doi: 10.1136/jmg.2006.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 27.LeBlanc JJ, Beemon KL. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 2004;78:5139–5146. doi: 10.1128/JVI.78.10.5139-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- 29.Graham JM, Jr., et al. FG syndrome: report of three new families with linkage to Xq12-q22.1. Am. J. Med. Genet. 1998;80:145–156. [PubMed] [Google Scholar]
- 30.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]