Abstract
Transcriptional regulation is dependent on layers of interactions between transcription factors and coactivators, controlling the specificity, temporal regulation, and extent to which transcriptional programs are executed. A key issue in the field of transcriptional regulation is to identify structural mechanisms by which transcription factors and coactivators build hierarchical protein assemblies. The bHLH-PAS (basic helix-loop-helix Per-ARNT-Sim domain) family of transcriptional regulators is comprised of both transcription factors and coactivators, which have different functions despite conserved domain architecture. Within this family, the tandem PAS domains typically mediate dimerization of the transcription factors, while C-terminal transactivation domains facilitate the dynamic interplay between transcription factors and coactivators. However, recent studies have shown that the modular PAS domains play an important role in regulating coactivator recruitment and oligomerization status. Here we provide a brief overview of the structural and functional studies that have identified a novel protein interaction interface on PAS domains utilized by both transcription factors and coactivators within the bHLH-PAS family.
Keywords: PAS domain, Transcription factor, Coactivator, Coiled coil
Work over the last two decades has developed a molecular understanding of the diverse transcriptional responses to environmental stress coordinated by members of the bHLH-PAS (basic helix-loop-helix Per-ARNT-Sim domain) family. These proteins are united by sharing the common architecture of a bHLH DNA-binding domain, tandem PAS domains, and member-specific regulatory and transcriptional control elements (Letunic et al., 2006). Consistent with this organization, bHLH-PAS proteins act as either DNA-bound transcription factors or transcription factor-interacting coactivators to regulate gene expression networks underlying many essential physiological and developmental processes. Our understanding of transcriptional regulation by these proteins has been largely defined by protein interaction and functional studies that identified the basic composition of active bHLH-PAS containing complexes in cells (Kewley et al., 2004; Xu and Li, 2003). While these data provide a sound conceptual foundation for bHLH-PAS function, they cannot explain either the dynamic interplay of coactivator/transcription factor interactions that exist within living cells or the functional differentiation that has occurred among these homologous proteins. However, recent studies have identified additional regulatory mechanisms that exploit conserved PAS domains within these proteins to mediate additional coactivator interactions, building more complexity and potential crosstalk among signaling pathways. Here we will address a key aspect of these new findings: the emerging structural description of PAS domain interactions with coiled coil or helical motifs found in coactivators that play critical roles in transcriptional activation by bHLH-PAS family proteins.
PAS domains: A versatile fold for regulation of protein interactions
PAS domains are a widespread class of small, modular domains presently identified in thousands of proteins in all three kingdoms of life (Letunic et al., 2006). These domains exhibit a remarkable array of functions depending on their context, acting as dimerization modules, photosensors, and/or ligand-binding domains to convey external environmental cues to the organism (Möglich et al., 2009). Much of this functional diversity arises from a subset of PAS domains that bind small molecule cofactors or ligands such as heme, FMN, or citrate. However, the unifying function of PAS domains is to modulate protein function by participating in protein-protein interactions, whether regulated by protein abundance or localization, light, or reversible ligand binding.
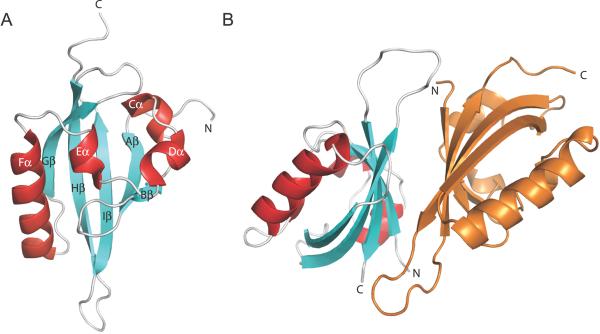
A structural basis for this functional diversity has been provided by X-ray crystallographic and NMR studies of over 45 PAS domains within the last decade. Collectively, these studies have demonstrated that PAS domains adopt strikingly similar α/βfolds despite less than 20% average sequence identity among individual domains (Möglich et al., 2009). The core PAS domain fold is defined by an antiparallel β-sheet with five strands flanked on one side by a series of α-helices (Fig. 1A). The β-sheet structure is critical for PAS function as it is involved in essentially all known intra- and interprotein interactions made by these domains. In contrast, the α-helical region is poorly structurally conserved between different classes of PAS domains and may represent an important element of structural diversity within these otherwise structurally similar proteins. The characterized role of this α-helical surface has predominantly been limited to interactions with internally-bound cofactors or routes used by these cofactors to bind within the domains (Amezcua et al., 2002; Key et al., 2009). Some PAS proteins may self-associate by using long loops that bind to these helical surfaces, as observed in the crystal structures of Drosophila PERIOD PAS-AB (Yildiz et al., 2005) or the KinA PAS-A domain from B.subtilis (Lee et al., 2008), but the functional relevance of these interactions are unclear in general. This situation appears to be changing due to recent studies suggesting that this helical surface may play an integral role in mediating transcriptional regulation by proteins throughout the bHLH-PAS family.
Figure 1. PAS domain structure and prototypical β-sheet-mediated dimerization.
A, PAS domain architecture is demonstrated by the PAS-B domain of the aryl hydrocarbon nuclear translocator, ARNT (1×0O.pdb). B, Heterodimerization of the HIF-1 transcription factor is mediated in part by the antiparallel association of ARNT (cyan/red) and HIF-2α (orange) PAS-B domain β-sheets (3F1P.pdb).
bHLH-PAS family: Multiple roles in transcriptional regulation
The largest class of PAS-containing proteins in eukaryotes is the bHLH-PAS family of transcriptional regulatory factors, which have a basic helix-loop-helix DNA binding domain followed by two PAS domains (noted as PAS-A and PAS-B) and a C-terminal activation domain (Letunic et al., 2006). Most members of this family are DNA-binding transcription factors that dimerize via their bHLH and PAS domains in response to environmental stimuli, which allows binding to cognate DNA sequences for transcriptional regulation. The PAS domains contribute to selectivity of dimer formation, with PAS β-sheet interfaces defining the formation of specific heterodimeric configurations of transcription factor complexes (Fig. 1B) (Card et al., 2005; Erbel et al., 2003; Scheuermann et al., 2009; Yang et al., 2005). However, some members of this family lack the ability to dimerize via their bHLH and PAS domains. Incapable of binding to DNA themselves, they act instead as transcriptional coactivators, interacting with DNA-bound transcription factors and nuclear receptors to facilitate gene transactivation by recruiting additional histone-modifying coactivators. Notably, functionally important roles for PAS domain α-helical surfaces have been identified for both subclasses and will be described below.
bHLH-PAS Transcriptional Coactivators
The bHLH-PAS coactivator family of Nuclear Coactivators (NCoA; also known as p160/Steroid Receptor Coactivators, SRC) contains three related members: NCoA-1, NCoA-2 (also known as GRIP1), and NCoA-3 (also known as p/CIP, RAC3, ACTR, AIB1 or TRAM-1). Although the highest degree of sequence conservation exists within their bHLH and PAS domains, the coactivator function of the NCoA family has been ascribed to their C-terminal activation domains. These are employed to interact with a variety of nuclear receptors, other transcription factors, and different coactivators through conserved Nuclear Receptor boxes (NR boxes) defined as LXXLL motifs (where L is a conserved leucine and X represents any amino acid) (Heery et al., 1997; Xu and Li, 2003). The redundant interaction of NCoA members with an array of DNA-bound factors serves to efficiently recruit general coactivators such as CBP/p300 and CARM1/PRMT1 that possess potent histone modification activity needed for transcriptional activation.
Dimerization of NCoA family members plays an important role in the temporal regulation of gene transactivation by estrogen and androgen receptors. These nuclear receptors exhibit a bimodal pattern of gene regulation with rapid activation of a subset of genes that lack a classical hormone responsive element (HRE), and delayed activation of HRE-containing genes that require prolonged ligand-dependent stimulation (Perissi and Rosenfeld, 2005). NCoA proteins are recruited as monomers to early gene, non-HRE-containing promoters, while dimeric complexes of NCoA-1/NCoA-3 and NCoA-2/NCoA-3 are preferentially recruited to late gene promoters containing HREs (Zhang et al., 2004). This suggests that the mode in which the nuclear receptor is recruited to DNA, either indirectly through interaction with a previously bound transcription factor or through direct binding to the HRE, creates different requirements for NCoA coactivator interactions.
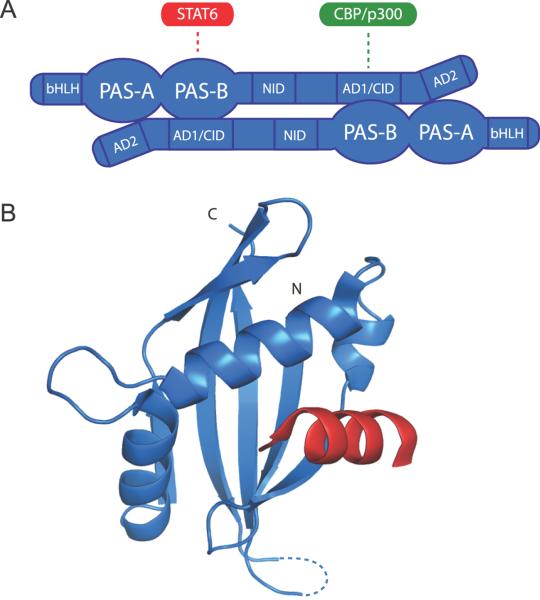
Recruitment of dimeric NCoA complexes to HRE-bound nuclear receptors requires NCoA PAS-B domains (Zhang et al., 2004); however, unlike their bHLH-PAS transcription factor counterparts, the NCoA PAS-B domains do not interact with one another (Lodrini et al., 2008). Instead, NCoA heterodimerization is mediated by the intermolecular association of the PAS-B domain with conserved C-terminal NR boxes within activation domain 1 (AD1, also known as the CBP-interaction domain, CID) (Fig. 2A) (Lodrini et al., 2008). The PAS-B domain of NCoA-1 also directly interacts with a NR box from the STAT6 (signal transducer and activator of transcription 6) transcription factor (Litterst and Pfitzner, 2001; Litterst and Pfitzner, 2002). Importantly, NCoA heterodimerization via the PAS-B/NR box intermolecular interaction effectively competes for interactions with both CBP/p300 and STAT6, demonstrating that the oligomeric status of NCoA proteins is likely capable of regulating the affinity for subsequent transcription factor and/or coactivator recruitment (Lodrini et al., 2008).
Figure 2. Regulation of NCoA coactivators by PAS domain/helical interactions.
A, Model for PAS-B-mediated NCoA dimerization via C-terminal LXXLL motifs in the activation domain 1/CBP-interaction domain (AD1/CID). NCoA dimerization competes with STAT6 and CBP/p300 interactions. Other abbreviations: NID, nuclear receptor interaction domain; AD2, activation domain 2. B, Interaction of NCoA-1 PAS-B domain (blue) with the STAT6 LXXLL motif (red) utilizes the PAS α-helical surface (1OJ5.pdb).
The molecular basis for the NCoA PAS-B/NR box interaction has been elucidated with the co-crystal structure of the NCoA-1 PAS-B domain complexed with the STAT6 LXXLL peptide (Razeto et al., 2004). Surprisingly, the interaction is mediated by the α-helical surface of the PAS-B domain, with the LXXLL motif bound within a hydrophobic cleft formed by a rearrangement of two short helices into a long, colinear helix (Fig. 2B). The colinear arrangement of the Dα and Eα helices is unique to this system among all the PAS structures solved to date (Möglich et al., 2009), suggesting that binding of the NR box may induce an allosteric conformational change within the canonical PAS domain fold. LXXLL motifs fold into amphipathic helices upon interaction (Shiau et al., 1998), with the conserved leucines oriented on the same face of the helix to interact in a knobs-and-holes configuration with helices within the target protein, as seen in typical coiled coil interactions. Residues that flank the conserved LXXLL motifs also make energetically important contacts with the PAS-B domain (Seitz et al., 2008) and thus are likely important for defining specificity within NCoA heterodimer pairs. As hetero- and homodimeric NCoA complexes may have differing capacities to recruit additional coactivators through their AD1/CID domains, it is likely that the interaction of NCoA PAS-B domains with LXXLL motifs has introduced a new level of complexity within the combinatorial code of transcriptional regulation by bHLH-PAS coactivators.
bHLH-PAS Transcription Factors
bHLH-PAS transcription factors function as obligate dimers (chiefly heterodimeric), binding DNA at specified promoter elements and recruiting coactivators via their unstructured C-terminal transcriptional activation domains to regulate transcriptional responses to diverse stimuli. Within this functional subclass, two proteins are of central importance: ARNT and BMAL1 (brain and muscle ARNT-like 1). These proteins act as general heterodimeric partners for bHLH-PAS proteins that define the specificity of target gene activation in response to stimuli such as hypoxia, xenobiotics, or timing of circadian rhythmicity (Kewley et al., 2004). Dimerization of bHLH-PAS transcription factors is mediated in large part by the association of PAS domains in vivo, since point mutations on a single PAS domain β-sheet can reduce or eliminate transcriptional activation by disrupting dimer formation (Erbel et al., 2003; Yang et al., 2005). Domain-swapping experiments in Drosophila bHLH-PAS proteins have shown that the PAS domains also contribute to the specificity of target gene induction (Zelzer et al., 1997), suggesting that they also participate in downstream signaling events on DNA, such as coactivator recruitment.
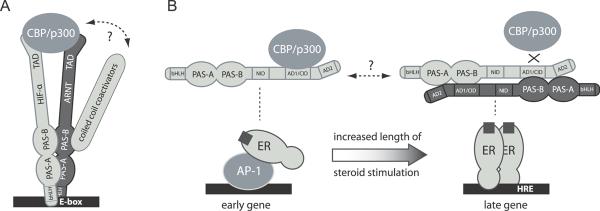
The recent discovery of coiled-coil coactivator proteins that directly target PAS domains, as opposed to the C-terminal transactivation domains, demonstrates that multiple structural motifs are used by bHLH-PAS transcription factors to recruit coactivators. CoCoA (coiled-coil coactivator) (Kim and Stallcup, 2004) and TRIP230 (thyroid hormone receptor interacting protein 230) (Beischlag et al., 2004) are both recruited to endogenous promoters after hypoxia or xenobiotic stress and are required for ARNT heterodimer function in vivo. Coiled coil fragments from both coactivators directly interact with ARNT PAS-B; more specifically, TRIP230 utilizes an LXXLL-like motif to interact with ARNT, with residues flanking this motif making additional energetic contributions (Partch et al., 2009). Structural mapping of the interaction using solution NMR spectroscopy indicates that use of the ARNT PAS-B α-helical surface is conserved with the NCoA family (Partch et al., 2009). Moreover, mutation of ARNT PAS-B residues that undergo perturbation by coactivator binding selectively disrupts the coactivator interaction while not affecting the β-sheet-mediated interaction with HIF-2α PAS-B, suggesting a way in which ARNT PAS-B can simultaneously engage both coactivators and HIF-α proteins (Fig 3A) (Partch et al., 2009). These data demonstrate a remarkable conservation of specificity in PAS-B/helical peptide interactions from functionally diverse members of the bHLH-PAS family.
Figure 3. PAS domain α-helical surface interactions with helical peptides modulate transcriptional responses by the bHLH-PAS family.
A, ARNT PAS-B utilizes β-sheet and α-helical surfaces to assemble complexes required for the transcriptional response to xenobiotics and hypoxia. Secondary interactions of coactivators recruited by transcriptional activation domains (TADs) and PAS domains may influence the specificity and/or timing of transcriptional regulation. B, PAS-B-mediated dimerization of NCoA family members plays a role in the temporal gating of ligand-dependent nuclear receptor signaling. Early gene induction by recruitment of the estrogen receptor (ER) to AP-1 (activator protein 1) promoters favors binding of monomeric NCoA proteins, while DNA-bound ER dimers preferentially recruit dimeric NCoA complexes. Competition for NR boxes within the NCoA AD1/CID may affect recruitment of subsequent coactivators such as CBP/p300.
What about other bHLH-PAS transcription factors? We believe that the PAS-B α-helical surface of other members of this family may interact specifically with helical motifs. At least one of the ARNT PAS-B-targeting coiled coil coactivat CoCoA, has been shown to interact directly with the aryl hydrocarbon receptor (AhR) in a region containing the PAS domains (Kim and Stallcup, 2004). Furthermore, the N-terminal bHLH-PAS region of AhR is also reported to interact with other coactivators such as GAC63 and the activation domain of BRCA1 (Chen et al., 2006; Kang et al., 2008). While numerous coactivators have now been identified that target AhR and ARNT, none of the coiled coil coactivators tested thus far interact directly with HIF-α proteins, suggesting a possible mode of pathway-specific regulation. Furthermore, it is unknown whether BMAL1, a key bHLH-PAS transcription factor within the mammalian circadian clock, will utilize its PAS-B domain to recruit coiled coil coactivators or other helical proteins, as might be anticipated from the fact that it shares the highest degree of homology with ARNT among all bHLH-PAS proteins.
Towards an understanding of PAS domain interactions in building transcription factor/coactivator complexes
PAS domains within the bHLH-PAS family act as scaffolds to build complex assemblies of transcription factors and coactivators, often through the simultaneous utilization of multiple interfaces on these small, modular domains. This raises the possibility that recruitment of multiple coactivators by the activation and PAS domains of bHLH-PAS heterodimers may play a role in the crosstalk that is observed in bHLH-PAS signaling. Crosstalk is functionally important, as it allows cells to fine-tune transcriptional responses by integrating multiple stimuli. Nearly half of the genes regulated by AhR or HIF-α are significantly affected by stimulus from the other pathway (i.e. a reduction of AhR-mediated gene induction under hypoxia) (Lee et al., 2006). Although ARNT is required for the activity of both AhR and HIF-α, it is not limiting under physiological conditions (Pollenz et al., 1999), demonstrating that crosstalk between the two pathways is not likely due to competition for ARNT.
Crosstalk may result from recruitment of different combinations of coactivators or corepressors, perhaps by competition or synergy with other bHLH-PAS proteins. In addition to acting a primary coactivator for ARNT and AhR, CoCoA also interacts with the N-terminal bHLH-PAS region of NCoA-1 and NCoA-2, as well as possessing the capability to independently recruit CBP/p300 (Kim et al., 2003; Yang et al., 2006). Importantly, the same transcription factor can demonstrate preference for different coactivators depending on the context of the core promoter (Marr et al., 2006). This promoter-dependent selection of coactivator recruitment is also observed with the PAS-mediated dimerization of NCoA coactivators (Fig. 3B), which helps to impart temporal regulation and/or specificity of target gene induction on the transcriptional response to steroid hormones (Zhang et al., 2004). Further studies that examine cellular mechanisms regulating NCoA dimerization are needed to understand the role of NCoA oligomeric status in transcriptional regulation.
Beyond the bHLH-PAS family of transcriptional regulators, these structural studies have also provided valuable information on the versatility of the PAS domain fold in mediating protein-protein interactions, and build on the idea that PAS domains are ideal scaffolds for creating supramolecular protein assemblies.
Acknowledgements
The authors wish to apologize to those colleagues whose work could not be cited due to space constraints.
Contract grant sponsor: NIH
Contract grant number: GM081875 (K.H.G.) and CA130441 (C.L.P.)
Literature Cited
- Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10(10):1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee WH, Rosenfeld MG, Hankinson O. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J Biol Chem. 2004;279(52):54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353(3):664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Chen YH, Beischlag TV, Kim JH, Perdew GH, Stallcup MR. Role of GAC63 in transcriptional activation mediated by the aryl hydrocarbon receptor. J Biol Chem. 2006;281(18):12242–12247. doi: 10.1074/jbc.M512537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2003;100(26):15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim HJ, Cho CH, Hu Y, Li R, Bae I. BRCA1 transcriptional activity is enhanced by interactions between its AD1 domain and AhR. Cancer Chemother Pharmacol. 2008;62(6):965–975. doi: 10.1007/s00280-008-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc. 2009;131(48):17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12(6):1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. J Biol Chem. 2004;279(48):49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- Lee J, Tomchick DR, Brautigam CA, Machius M, Kort R, Hellingwerf KJ, Gardner KH. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47(13):4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- Lee K, Burgoon LD, Lamb L, Dere E, Zacharewski TR, Hogenesch JB, LaPres JJ. Identification and characterization of genes susceptible to transcriptional cross-talk between the hypoxia and dioxin signaling cascades. Chem Res Toxicol. 2006;19(10):1284–1293. doi: 10.1021/tx060068d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34(Database issue):D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litterst CM, Pfitzner E. Transcriptional activation by STAT6 requires the direct interaction with NCoA-1. J Biol Chem. 2001;276(49):45713–45721. doi: 10.1074/jbc.M108132200. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Pfitzner E. An LXXLL motif in the transactivation domain of STAT6 mediates recruitment of NCoA-1/SRC-1. J Biol Chem. 2002;277(39):36052–36060. doi: 10.1074/jbc.M203556200. [DOI] [PubMed] [Google Scholar]
- Lodrini M, Munz T, Coudevylle N, Griesinger C, Becker S, Pfitzner E. P160/SRC/NCoA coactivators form complexes via specific interaction of their PAS-B domain with the CID/AD1 domain. Nucleic Acids Res. 2008;36(6):1847–1860. doi: 10.1093/nar/gkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20(11):1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17(10):1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Card PB, Amezcua CA, Gardner KH. Molecular basis of coiled coil coactivator recruitment by the aryl hydrocarbon receptor nuclear translocator (ARNT) J Biol Chem. 2009;284(22):15184–15192. doi: 10.1074/jbc.M808479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6(7):542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Pollenz RS, Davarinos NA, Shearer TP. Analysis of aryl hydrocarbon receptor-mediated signaling during physiological hypoxia reveals lack of competition for the aryl hydrocarbon nuclear translocator transcription factor. Mol Pharmacol. 1999;56(6):1127–1137. doi: 10.1124/mol.56.6.1127. [DOI] [PubMed] [Google Scholar]
- Razeto A, Ramakrishnan V, Litterst CM, Giller K, Griesinger C, Carlomagno T, Lakomek N, Heimburg T, Lodrini M, Pfitzner E, Becker S. Structure of the NCoA-1/SRC-1 PAS-B domain bound to the LXXLL motif of the STAT6 transactivation domain. J Mol Biol. 2004;336(2):319–329. doi: 10.1016/j.jmb.2003.12.057. [DOI] [PubMed] [Google Scholar]
- Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106(2):450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz M, Maillard LT, Obrecht D, Robinson JA. Molecular characterization of the NCoA-1-STAT 6 interaction. Chembiochem. 2008;9(8):1318–1322. doi: 10.1002/cbic.200700773. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17(9):1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Yang CK, Kim JH, Stallcup MR. Role of the N-terminal activation domain of the coiled-coil coactivator in mediating transcriptional activation by beta-catenin. Mol Endocrinol. 2006;20(12):3251–3262. doi: 10.1210/me.2006-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang L, Erbel PJ, Gardner KH, Ding K, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J Biol Chem. 2005;280(43):36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- Yildiz O, Doi M, Yujnovsky I, Cardone L, Berndt A, Hennig S, Schulze S, Urbanke C, Sassone-Corsi P, Wolf E. Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol Cell. 2005;17(1):69–82. doi: 10.1016/j.molcel.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Wappner P, Shilo BZ. The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev. 1997;11(16):2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yi X, Sun X, Yin N, Shi B, Wu H, Wang D, Wu G, Shang Y. Differential gene regulation by the SRC family of coactivators. Genes Dev. 2004;18(14):1753–1765. doi: 10.1101/gad.1194704. [DOI] [PMC free article] [PubMed] [Google Scholar]