Abstract
The human gastrointestinal tract is home to immense and complex populations of microorganisms. Through recent technical innovations, the diversity present in this human body habitat is now being subjected to detailed analyses. This review focuses on the microbial ecology of the gut in inflammatory bowel diseases, and how recent studies provide an impetus for delving into the structure and operations of the gut microbial community, and its interrelationships with the immune system, using carefully designed, comparative metagenomic approaches.
Introduction
Our lives are deeply embedded within a microbe-dominated world. Microbes drive many global geochemical cycles that ultimately provide us with oxygen, food, and water (Committee on Metagenomics, 2007). On a more intimate level, diverse microbial communities assemble, and persist, on the external and internal surfaces of our bodies from the time of our birth until our death (after which time they proceed to consume us!) Our gastrointestinal tract is home to the vast majority of these microorganisms and their viruses. These microbes belong to all three domains of life on Earth -Bacteria, Archaea and Eucarya, and outnumber our own human cells by an order of magnitude (Savage, 1977). This more transcendent perception of ourselves has given rise to the view that we are actually ‘supraorganisms’ whose genome is the sum of genes in our H. sapiens genome and the genomes of ‘our’ microbial partners (microbiome), and whose metabolic features are a synopsis of human and microbial attributes (Gill et al., 2006; Turnbaugh et al., 2007).
Most of the details concerning our gut microbiota remain obscure. The factors that impact its assembly, and that define the spatial distribution of its component members, are largely unknown. In addition, the manner in which the composition and metabolic operations of this microbial ‘organ’ are regulated, and how its functional stability is maintained in the face of varied environmental exposures in a persistently perfused ecosystem are ill-defined. The impact of our modern lifestyles - ranging from our highly synthetic cookery to our use of broad-spectrum antibiotics beginning at early stages of postnatal life - on the gut microbiota are the subjects of active conjecture, but only modest amounts of hard experimental data.
This situation should change rapidly in the coming decade as the recently launched international human microbiome project delves into our gut microbial ecology in health and disease. This project is being propelled by a number of forces (Turnbaugh et al., 2007). They include an evolution in the focus of microbiology from exploring the properties of microbial species in isolation, to characterizing their properties in the context of their natural communities and habitats. In addition, the advent of massively parallel DNA sequencers has dramatically increased the speed of sequencing, markedly reduced its cost, expanded the ability to characterize multiple samples simultaneously (Walker et al., 2008), and helped to democratize (distribute) the process by which hypothesis-directed projects are designed and executed by investigators within their own labs, as well as in partnership with larger genome sequencing centers. One consequence of this change in DNA sequencing capacity has been to spawn a new area of science known as metagenomics. Metagenomics refers to culture-independent studies of the structures and functions of microbial communities, as well as their interactions with the habitats they occupy (Committee on Metagenomics, 2007). It includes sequencing of microbial DNA isolated directly from a community occupying a given environment in order to determine its component microbial lineages and genes (the ‘microbiome’), as well as characterizing the community's expressed RNA and protein products, and its metabolic network.
In this essay, we focus on what culture-independent methods are beginning to teach us about the microbial communities that reside in the intestines of healthy individuals, and those with inflammatory bowel diseases (IBD) - disorders that involve dysregulation of the homeostasis that is forged between our innate and adaptive immune systems and our gut microbiota (Xavier and Podolsky, 2007).
Surveys of the human gut microbiota
Bacteria dominate the gut ecosystem. Much of this world is terra incognita. The fact is that most organisms in complex communities cannot be cultured ex vivo using today's technology. There is hope: new approaches for culturing previously unculturable (gut) microorganisms are being developed (e.g., Duncan et al., 2007), as are methods for amplification and sequencing genomic DNA from minute quantities of starting materials (Marcy et al., 2007). In addition, current culture-independent methods for surveying complex communities are more accessible than ever, thanks to the marked increase in speed and accompanying decrease in cost of DNA sequencing, and the development of computational tools to distill and interpret the data stream.
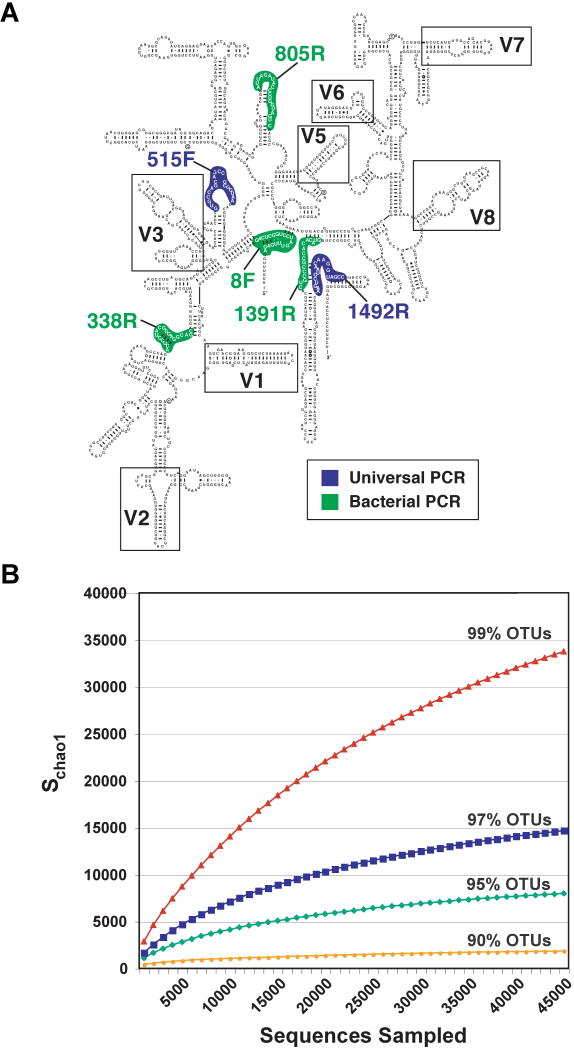
Most of the culture-independent sequencing effort has been directed towards small-subunit rRNA (SSU) genes, which are present in all cellular organisms. 16S rDNA in Bacteria and Archaea can be amplified directly by PCR from DNA isolated from a sample that contains a microbial community - for example, a mucosal biopsy or feces in the case of the gut. PCR reactions use oligonucleotide primers that target highly conserved regions of SSU rDNA (see Figure 1A) and therefore can amplify rDNA from a broad range of organisms. Myriad SSU rDNA primer sets have been devised, thus providing researchers with the ability to target virtually all or selected groups of organisms in a sample. The SSU rRNA gene was chosen for several reasons: it is relatively small (∼1.5 kb); it has a high enough level of sequence conservation between microbial species to permit reliable alignments, and it possesses sufficient variation to infer evolutionary relationships. In addition, it is transmitted vertically within a lineage rather than horizontally (between lineages).
Figure 1. Bacterial SSU rRNA gene-based surveys of the gut microbiota.
(A) Cartoon of the general structure of the bacterial 16S rRNA gene, showing conserved and variable regions (based on http://www.rna.ccbb.utexas.edu/). Targets for PCR primers are shown. The numbers associated with the primers are referenced to the E. coli 16S rDNA gene. The targeted sites for PCR vary depending upon whether the goal is to sequence the entire gene or subdomains containing variable regions. The read length achievable with the current generation of massively parallel sequencers limit the portion of the gene that can be characterized: currently favored regions are V2-V3 (Hamady et al., 2008) and V6 (Huber et al., 2007). (B) Analysis of diversity in the human gut microbial community. Collector's curves of observed and estimated richness are shown. Richness is estimated to be 18,000 (genus-level OTUs with ≥95 %ID) and 36,000 at the species-level (≥99 %ID) using the Chao1 estimator.
The kinds of microbes present in a community specimen can be identified by comparison of their SSU rRNA gene sequences to previously analyzed sequences, including those from phentotypically and genetically well-characterized isolates. Depending on the similarity of the newly isolated sequences to those in databases, microorganisms can be more or less precisely identified. By convention, but somewhat arbitrarily, SSU rRNA sequences with ≥97% identity (%ID) are considered to be from the same ‘species’, while different species of the same genus have ≥95 %ID. More rigorous analyses, exemplified by inference from phylogenetic trees, can provide deeper insights into the evolutionary relationships between newly identified taxa and known relatives.
A cautionary note: although the %ID shared among rRNA sequences is typically used to define a given species, this definition is a matter of some debate, largely because of lateral gene transfer between different organisms. The result is that isolates of the same ‘species’, as defined by rRNA phylogenetics, often differ significantly in their collections of protein-coding genes. This fact has prompted use of the term ‘pan-genome’ to describe all genes associated with all isolates of a ‘species’-level phylogenetic type (phylotype) (Tettelin et al., 2005; Muzzie et al., 2007). This fact has also evoked a provocative series of discussions about how to best define a species (for example, should it be a core set of signaling and/or metabolic pathways conserved between all isolates of that ‘species’?), and conjured up an image of the tree of life as having multiple connections between its branches, giving it a more reticulated appearance (Doolittle and Papke, 2006). Nonetheless, the rRNAs and most other elements of the nucleic acids based genetic machinery have not undergone lateral transfer, and so can be taken to indicate true relationships reliably.
Despite these caveats about the meaning of the term ‘species’, the richness or bio-diversity in a microbial community can be estimated by observing the number of new SSU rRNA sequences, binned into operational taxonomic units (OTUs) with defined %ID cutoff thresholds, that appear with increased depth of sampling (e.g., Figure 1B). Moreover, the degree of relatedness of different communities can be quantified by constructing a common phylogenetic tree composed of all SSU rRNA gene sequences from all communities being analyzed. Pair-wise comparisons of communities are then made based on the degree to which they share branch length on this common tree (the ‘UniFrac metric’; Lozupone and Knight, 2005; Lozupone et al., 2006).
We are unique individuals
Three reports describing the results of SSU rRNA gene sequence-based surveys of the distal small intestinal and colonic microbiota of 139 adult humans living in North America illustrate both the significant inter-personal variation and the similarities that exist among our gut ecosystems (Eckburg et al., 2005; Ley et al., 2006; Frank et al., 2007). These early returns from what will soon be a global human gut survey indicate that at the phylum level only a small fraction of known bacterial diversity is represented in our guts: to date, members of only 10 out of >100 described phyla have been identified. Furthermore, phylum-level diversity is not evenly distributed. The majority of bacteria identified in all human guts surveyed belong to just two phyla: the Firmicutes and the Bacteroidetes. On the other hand, there is great diversity at lower taxonomic levels. Estimates are that at least 1800 genera and 15,000 to 36,000 species of bacteria, (depending on whether species are conservatively or liberally classified) make up the collective human intestinal microbiota (Frank et al., 2007). In addition, the accumulation of new taxa has not reached a plateau (Figure 1B). Moreover, there is considerable interpersonal variation in the bacterial species and strains present in the gut microbiota (Eckburg et al., 2005; Ley et al., 2006; Frank et al., 2007): determination of the degree of variation awaits further sampling of more individuals, as well as longitudinal studies of variation within an individual over time. One challenge for this field is to discern the origins and biological implications of this variation. For example, does it imply a large degree of functional redundancy in the gene lineages and metabolic pathways represented in the gut microbiome? Is it essential for maintaining the robustness, or functional stability, of the gut community: i.e., its ability to adapt to a variety of perturbations ranging from changes in host diet, the state of activation of the innate or adaptive immune system, and/or to invasion by various pathogens?
Legacy and habitat effects shape the gut microbiota
How do we acquire our diverse gut microbiotas? In a pilot study that used DNA fingerprinting, not sequencing, of rDNA amplicons, Zoetendal et al. (2001) noted that the gut microbial communities of monozygotic twins were more similar than between twins and their unrelated marital partners. Moreover, Palmer et al. (2007) utilized bacterial 16S rRNA-based detection methods (DNA microarrays and sequencing) to profile the fecal microbiota of 14 healthy full-term infants during their first year of life. One pair of dizygotic twins was included in their study. They found marked temporal variation in the composition of the microbiota in the first few months of postnatal life. The pattern of community assembly was most similar among the twins, lending strong support to the notion that early environmental exposures (as well as genetic factors) play a major role in determining the distinctive characteristics of each baby's microbiota. These findings, together with the results of reciprocal microbiota transplantation experiments involving transfer of gut microbial communities from conventionally-raised mice or zebrafish to germ-free zebrafish or mouse recipients indicate that the composition of the gut microbiota reflects both legacy effects (a summation of the microbes we are exposed to after birth in our distinctively experienced ecosystems), and habitat effects (largely ill-defined ‘filters’ that operate to select microbes for residency in our different body sites) (Rawls et al., 2006). For example, a role for the immune system and for host genotype was provided by GLC analysis of lipids extracted from the fecal microbiota of isogenic strains of C57Bl/6 mice that differ only at their MHC locus, and in different inbred strains that were congenic for the MHC: in the case of the former, fecal fatty acid profiles cluster according to MHC genotype; in the case of the latter, host genetic background effects are evident (Toivanen et al., 2001).
‘Where’, not just ‘who's there’
The spatial or ‘biogeographic’ features of the gut microbiota are likely to have profound effects on microbe-microbe interactions, including nutrient sharing strategies that underlie syntrophic relationships, and on microbe-host interactions, such as those that affect the way in which microbes are recognized by components of both the innate and adaptive immune systems. Therefore, a great challenge for this field is to develop robust quantitative methods to describe the distribution of phylotypes not only along the length of the gut, but also across its width. By width, we mean from the lumen and its collection of particulate matter (undigested food fragments which can serve as nutrient platforms, exfoliated epithelial cells, and mucus fragments; Sonnenburg et al., 2005), across the mucus slime layer, to the epithelium, which contains multiple cell lineages including resident immune cells.
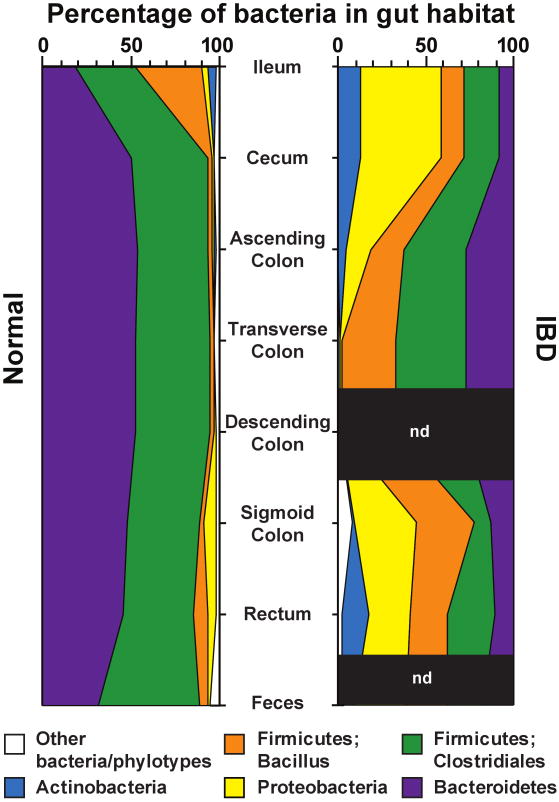
A systematic study of microbial populations sampled from specified regions along the length of the gut has yet to be reported, although data have been obtained from subsets of these regions in a given host - most notably, in the esophagus and stomach (Bik et al., 2006), and in the distal small intestine and colon (plus feces) (Eckburg et al., 2005, Frank et al., 2007). The results reveal that the same bacterial phyla predominate in the stomach, small intestine, and colon, although their relative abundance and details of the component species vary as a function of position along the gut's cephalocaudal axis (e.g., Figure 2).
Figure 2. Bacterial phyla identified in the human gut microbiota.
Distribution of predominant bacterial phylotypes in the human intestinal tract. The graphs depict relative abundance as a function of location along the cephalocaudal axis of the distal gut. DNA sequences were compiled from the studies of Eckburg et al. (2005) and Frank et al. (2007). Phylogenetic classifications were made by parsimony insertion of aligned sequences into an 16S rRNA tree provided by the Greengenes database (DeSantis et al., 2006), using ARB (Ludwig et al, 2004). Abbreviations: IBD, samples from patients with either Crohn's disease or ulcerative colitis (Frank et al., 2007); Normal, data from healthy young adults from Eckburg et al (2005) plus the non-IBD controls reported in Frank et al. (2007); n.d., not done. A total of 18,405 16S rRNA sequences were analyzed (5,405 IBD, 13,000 normal).
Data obtained by Eckburg et al. (2005) indicated that mucosa-associated communities sampled at different positions along the length of the colon from a given individual differ from one another, but not in a systematic manner between individuals. These communities also differ from the fecal microbiota within an individual (Eckburg et al., 2005). UniFrac analysis of these datasets reveals that mucosal and fecal communities from a given individual cluster together: i.e., are more similar to one another compared to samples taken at analogous sites from other hosts. They also show that fecal samples can be used as a readily (and safely) procured proxy for defining interpersonal differences in gut microbial ecology. Nonetheless, the issue of the degree of intra- and interpersonal variation in the biogeography of the human gut microbiota is far from resolved. For example, a recent report focused on distal small intestinal (ileal) and colonic mucosal biopsies obtained from 26 individuals who had not received any bowel preparation or antibiotics prior to sample collection. Using a combination of culture independent methods (denaturing gradient gel electrophoresis, quantitative PCR and sequencing of 16S rRNA genes), Ahmed et al (2007) observed that communities occupying a given location along the length of the distal gut clustered more closely between individuals than all samples from a given host. These findings suggest that the method used for bowel preparation is a very important clinical parameter to note when interpreting such studies. Detailed studies of spatial variation in gut microbial ecology need to be performed in mice where potentially confounding variables such as legacy effects (i.e. kinship relationships), host genotype, diet, and methods used for harvesting and manipulating samples, can be constrained.
Experiments conducted in germ-free mice colonized with members of the human gut microbiota indicate that mucus may be an important factor that influences the pattern of microbial colonization of different regions of the gut, as well as maintenance of microbial community structure and function. The mucosa is a glycan-rich source of nutrients that can be used by saccharolytic members of the gut microbiota (Hooper et al., 1999), especially during periods when the diet is depleted of complex plant polysaccharides (Sonnenburg et al., 2005). [The proteomes of mice and humans contain a very limited arsenal of glycoside hydrolases (Sonnenburg et al., 2006) and therefore they, and we, depend upon glycoside hydrolases encoded in the gut microbiome to process common but otherwise indigestible polysaccharide components of our diets].
Studies of healthy adult humans have shown that at a coarse level of resolution there are regional differences in mucus matrix proteins, as well as differences in the types and amounts of glycans incorporated into mucus (Smithson et al., 1997; Corfield et al, 2001). Unfortunately, mucus- and epithelial surface-associated glycans are notoriously challenging to harvest from different regions of the gut in a precise way, and in sufficient quantities for biochemical analysis. Host glycans are structurally complex and varied, reflecting the combinatorial nature of glycan biosynthesis and modification (including modification by components of the microbiota). Although ‘glycophobia’ (fear of glycan complexity) is common among biologists, new and effective strategies for collecting information about intra- and interpersonal variations in mucus and epithelial glycans are very much needed. Mice with genetically engineered defects in mucus production (An et el., 2006; Heazlewood et al, 2008) could serve as attractive starting points for linking mucus glycan content with gut microbial ecology (see below).
The microbial ecology of the gut in IBD
A variety of mouse models of IBD, including animals with genetically engineered defects in their immune system or mucus glycans, which have been conventionally-raised with a microbiota and then treated with antibiotics, or who have been raised germ-free and then colonized with a gut microbiota from a healthy donor, demonstrate that the presence of a microbiota is instrumental in eliciting pathology (for very recent examples, see Garrett et al., 2007 and Kang et al., 2008; for a review of earlier studies see Sartor, 2008; for a excellent general overview of IBD see Xavier and Podolsky, 2007). It is important to emphasize that although rodent models of colitis are generally dependent on the presence of gut microbes, not all microbes elicit disease (Kim et al, 2007, Stepankova et al., 2007, Dijkstra et al., 2007).
Several studies have documented marked alterations in the gut microbiota of patients with IBD (Giaffer et al., 1991; Gophna et al., 2006; Krook et al., 1981; Lepage et al., 2005; Mangin et al., 2004; Manichanh et al., 2006; Merwe et al., 1988; Prindiville et al., 2004; Scanlan et al., 2006; Seksik et al., 2003). We (Frank et al., 2007) recently described the results of a case-control study of intestinal microbiology ecology in patients with Crohn's disease and ulcerative colitis, and in non-IBD controls (predominantly individuals with cancer of the bowel). Small intestinal and/or colonic biopsy specimens were obtained at surgery, luminal contents were removed and tissue-associated communities were surveyed by 16S rRNA gene sequencing [n=68 Crohn's disease specimens; 61 ulcerative colitis specimens; and 61 non-IBD control samples obtained from 35, 55 and 34 patients respectively (Figure 2)].
The distribution of bacterial species differed only slightly between disease states (i.e. Crohn's disease vs. ulcerative colitis vs. non-IBD ‘controls’) when controlling for anatomic site and pathologic state (p = 0.02 for 99% OTUs, p = 0.11 for 97% OTUs). Each sample was assessed for the presence or absence of all of the OTUs and then analyzed using principal components analysis (PCA) and hierarchical clustering. These clustering methods do not rely on pre-defined categorization of samples, such as into IBD and non-IBD groups: rather, they identify components that explain the greatest amount of the variability within the dataset (although these components may be sensitive to the specific metric chosen for clustering). Based on the principal components analysis, the authors classified the samples into two groups. The major group, which they designated “normal”, contained virtually all of the non-IBD samples (60/61), and the majority of the colitis samples. The second group, which they designated “IBD-specific”, was highly enriched in IBD samples (24 of 68 Crohn's disease samples, 16 of 61 ulcerative colitis samples, and only one of the 61 normal samples). The bacterial lineage distributions in the two groups differed significantly, regardless of taxonomic depth (P<0.001 for OTUs with 99%, 97%, 95%, 90%, and 85%IDs). The IBD-specific group had marked decreases in the representation of two prominent constituents of the gut community: the Bacteroidetes and the Lachnospiraceae (group IV and XIVa Clostridia). Whether diminution of these populations is a cause or consequence of disease, and whether this altered community structure (dysbiosis) persists upon remission, or is a condition that exists in susceptible individuals prior to developing manifest IBD remain to be established. However, there was a significant association between a clinical history of abdominal abscesses and harboring a “IBD specific” type of microbiota in patients with Crohn's disease (Frank et al., 2007). This suggests a testable hypothesis: alterations in the representation of components of the microbiota, such as the Bacteroidetes and Lachnosspiraceae or other phylotypes, may be biomarkers that help predict disease predisposition, activity, severity, and responsiveness to therapy.
Mucosa-associated microbial communities and IBD
The importance of mucosal communities in disease pathogenesis has been highlighted by studies that use fluorescent in situ hybridization (FISH) to examine the distribution of microbes at the interface between the colonic epithelium and its overlying mucus layer in patients with and without IBD. For example, FISH analysis detected bacteria within, or penetrating, the mucus layer more frequently in mucosal biopsies obtained from patients with IBD compared to healthy controls (30% vs 3%, respectively) (Swidsinski et al., 2007). Other studies have demonstrated both thinning of the mucus layer in patients with ulcerative colitis, as well as alterations in mucus glycan content (Pullan et al., 1994 and Smithson et al., 1996). Together, these findings suggest that IBD may be associated with more intimate contact between members of the microbiota and the mucosa. This notion is supported by recent studies in mice: animals unable to manufacture core-3 containing O-glycans are highly susceptible to colitis induced by treatment with dextran sodium sulfate (An et al., 2006), while mice with mutations in the Muc2 gene develop colitis spontaneously (Heazlewood et al., 2008).
Probiotics and IBD
Studies have yet to identify an organism that fulfills Koch's postulates for IBD - a fact consistent with the notion that IBD is generally associated with deranged relationships between a given host and his/her gut microbial community, rather than with a single member of that community. Comprehensive metagenomic analyses of gut microbial communities in mice and humans with known genotypes and well-characterized immune system phenotypes have yet to be reported. Until they are, we are limited to speculation about the cause and consequence of microbial community alterations that have been observed in IBD. Nonetheless, it is intriguing that amelioration of disease has been reported in some controlled clinical studies involving administration of live microbial supplements (probiotics) to patients with IBD. Examples include the use of VSL-3, a mixture of 8 bacterial strains (Bifidobacterium breve, B. longum, B. infantis, Lactobacillus acidophilius, L. plantarum, L. casei, L. bulgaricus and Streptococcus thermophilus) in patients with ileal pouch inflammation (pouchitis) (Gionchetti et al., 2003; Mimura et al., 2004). Various probiotics have demonstrated clinical efficacy in patients with ulcerative colitis but not Crohn's disease (Sartor, 2004).
The impact of these probiotic preparations on gut microbial ecology, and on the functions of the gut microbiota, has not yet been analyzed using metagenomic approaches, either during or after administration. These approaches will have to be applied to patient populations where factors such as antibiotic use, diet, and other potential confounding variables are constrained prior to, during and after probiotic treatment. Proof-of-principle experiments involving administration of probiotics to (gnotobiotic) mouse models of colitis, where genetic and environmental factors can be initially controlled, should help guide clinical studies (see below).
IgA and the regulation of host-microbial homeostasis
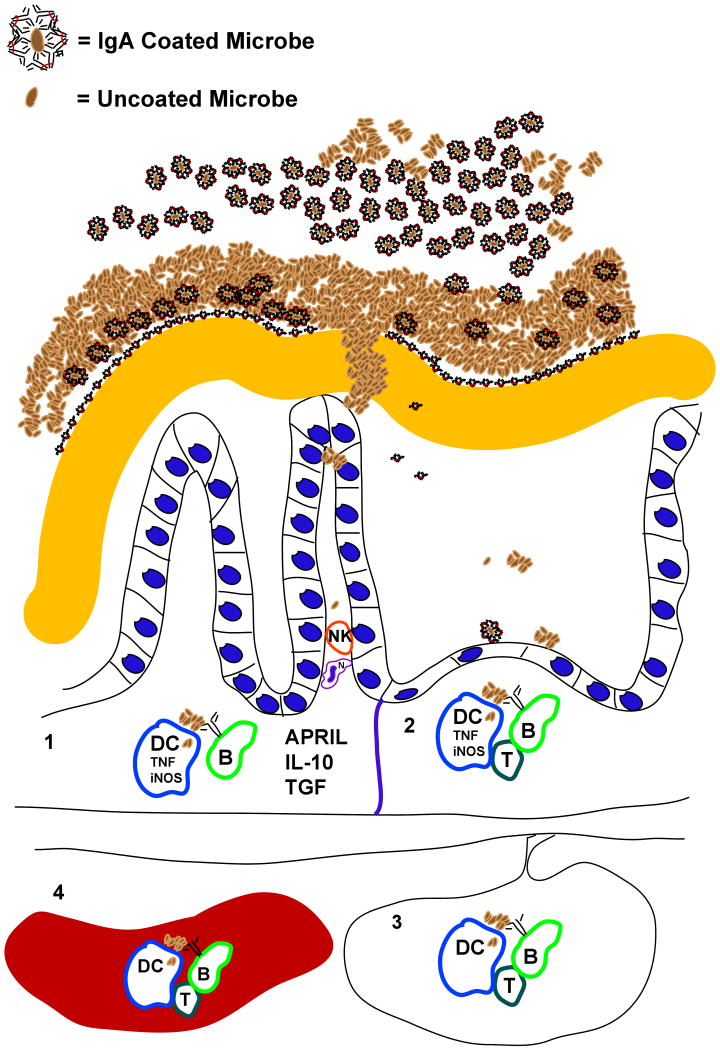
A role for IgA in shaping the gut microbiota has been studied for quite some time (Baklien et al., 1975, van der Waaij et al., 1996). Classical work by John Cebra demonstrated that members of the microbiota elicit production of specific IgAs in the gut, although they vary in their ability to do so (Shroff et al., 1995). Others have shown that innate immune system activation is greater in the absence of adaptive immunity (Cash et al., 2006; Keilbaugh et al. 2005; and Peterson et al., 2007), and that addition of B-cells or immunoglobulin (serum) ameliorates the severity of colitis in several animal models (Mizoguchi et al., 1996; 1997). A number of potential mechanisms in principle could account for this amelioration. For example, it could reflect decreased epithelial interaction with antibody-coated bacteria because they are trapped in the mucus, or a modified host response after bacteria are transported across the M-cell pathway and taken up by dendritic cells, or opsonization and destruction of the bacteria that do translocate across the barrier (Martinolini et al., 2007; Kadaoui et al., 2007) (Figure 3). Decreasing host innate responses to bacteria IgA could also modulate colitis by changing gut microbial composition. IgA is known to produce changes in gut microbial ecology in conventionally-raised mice (Suzuki et al., 2004; Fagarasan et al, 2002). For example, two recent reports have demonstrated that an enigmatic, uncultivated bacterial species known as Segmented Filamentous Bacteria (SFB; a member of the Firmicutes) is important, but not sufficient, to induce colitis in mice (in these studies, colitis was induced by transfer of effector CD4+ T cells in the absence of T regulatory T cells). SFB are consistently enriched in IgA-deficient mice (Suzuki et al., 2004) and are powerful inducers of IgA (Talham et al., 1999). Moreover, colonization of the gut with SFB correlates temporally with maturation of the immune system: development of a mucosal IgA response is associated with decreased levels of SFB colonization while passive transfer of IgA immunity slows colonization (Jiang et al., 2001). It is easy to imagine how a failure to generate an adequate IgA response to SFB could lead to expansion of this phylotype and consequently to the development or aggravation of colitis in genetically predisposed mouse models. Unfortunately, there are no reports describing the results of comprehensive culture-independent surveys of gut microbial ecology in IgA deficient patients with or without colitis.
Figure 3. Host-microbial interactions affected by IgA are modulated as result of stimulation of the immune system at different sites.
Secretory IgA, antimicrobial peptides, and mucus are among the factors that comprise the mucosal barrier. The ability of different microorganisms to develop resistance to these factors, or to degrade mucus, will contribute to greater microbial pattern recognition receptor-mediated signaling in epithelial cells, dendritic cells (DCs), macrophages, and B cells. Where these organisms prime the adaptive immune system will depend on how far they can penetrate the barrier, and where the host traffics microbial cells and their antigens. Numbers in the figure refer to the following processes. (1) The simplest response loop may be the innate immune system's induction of IgA production in the lamina propria: NK, DC, epithelial cell-derived cytokines (APRIL, IL-10, TGF, TNF among others) and membrane-bound receptors (e.g. CD40-CD40 ligand) induce IgA production from recruited B cells. (2) Bacteria, bound or unbound by IgA in the lumen, are actively transported to Peyer's patches and associated lymphoid organs where T cell priming and help are more likely. (3) DCs transport whole bacteria and microbial antigens to mesenteric lymph nodes where priming of T cells and B cells programs them to home to the lamina propria, and where IgA induction is favored. (4) Commensal microbes (and invasive pathogens) are transported and trapped by the innate immune system in the spleen, where a systemic immune response is induced (lymphocytes primed in the spleen are less likely to home to the gut, and more likely to switch to non-IgA subclasses).
A recent study, performed in gnotobiotic mice, provides additional and direct evidence that the IgA antibody response to members of the gut microbiota helps establish a quiescent relationship with the host (Peterson et al., 2007). Hydridoma cells that produce a naturally-primed IgA specific for a carbohydrate epitope present in the capsule of a prominent human gut-associated bacterial symbiont, Bacteroides thetaioataomicron, were implanted into the dorsal subcutaneous tissue of germ-free Rag1-/-mice that lack functional T and B cells. In other words, in this engineered mouse model there was a single antibody repertoire of known specificity, and the gut microbiota was reduced to a single (sequenced) bacterial species. When these animals were colonized with a wild-type strain of B. thetaiotaomicron, levels of epitope expression were inversely correlated with IgA levels (levels reflected the biomass of implanted hybridoma cells). Moreover, when these Rag1-/- hybridoma-bearing (‘backpack’) mice were gavaged with equal numbers of isogenic wild-type epitope-positive, and mutant epitope-negative strains of B. thetaiotaomicron, the presence of the antibody resulted in a marked relative decrease in wild-type bacteria (Rag1-/- animals without implanted hybridoma backpacks served as reference negative controls). Whole genome transcriptional profiling of the bacterium in Rag1-/- mice colonized with just the wild-type strain revealed that in the absence of IgA, B. thetaiotaomicron elicits a more robust oxidative response from the innate immune system in the distal gut, and then adapts to this response by inducing a number of its genes involved in metabolism of the oxidative products of the host response. Whole genome transcriptional profiling of the host response in the distal small intestine disclosed that the presence of IgA reduces intestinal pro-inflammatory signaling (Peterson et al., 2007).
Together, these findings support the following conclusions: (i) IgA responses function to help mediate tolerance to the microbiota; (ii) host-microbial homeostasis in the gut depends on recognition of members of the microbiota by the adaptive immune system; and (iii) the adaptive immune system likely mandates that entrenched residents of the community (autochthonous members) perpetually modify their surface epitopes. This latter type of selective pressure may provide one explanation for the species variation of the gut microbiota, and for why diversification of surface carbohydrates may help promote a quiescent relationship with a microbial community that provides benefits to the host. In this conceptualization of host-microbe homeostasis, tolerance to the microbiota is predicated on immune recognition of its members. The adaptive immune system functions to maintain a ‘connection’ with the gut microbiota by selectively generating responses to bacteria that stimulate the innate system. The result is that the host is able to detect new bacterial phylotypes, ‘ignore’ those that it has already encountered, and support greater diversity in the gut ecosystem without sacrificing the innate immune system's necessary role in maintaining a functional mucosal barrier.
One extrapolation of this concept is that IBD is a manifestation, at least in part, of a failure to generate effective adaptive immune responses to the resident gut microbiota. Such responses may normally prevent presentation of microbial antigens to T cells specific for members of the community (Kadaoui et al., 2007). This hypothesis is supported by the antibody response to gut microbes documented in TCR alpha -/- mice: these responses change from polyclonal to oligoclonal as colitis develops; passive transfer of Ig (serum) ameliorates disease severity (Mizoguchi et al., 1996; 1997).
This concept of host-microbe homeostasis has implications for the immunologist and for the microbial ecologist. If IgA responses to the microbiota are a key component of host-microbial homeostasis in the gut, then understanding the pathway for generating these responses is clearly necessary. Immunologists already know that IgA production can occur without T cells (Macpherson and Uhr, 2004), that class switching to IgA appears to happen locally in the lamina propria (Fagarasan et al., 2001), and that dendritic cells can directly help B-cells class switch to IgA, independent of T cells through TNF, iNOS, Blys and APRIL (Litinsky et al., 2002; He et al., 2007; Tezuka et al., 2007). Thus, the IgA responses to the microbiota are likely to be driven by local phenomena and involve simple feedback loops: dendritic (and epithelial) cells direct IgA production by B cells independent of T cells until IgA levels are sufficient to block microbial stimulation; the systemic immune system is bypassed unless the local feedback loop is not successful in stopping activation (Figure 3).
It is reasonable to suppose that the local feedback loop that results in IgA-mediated homeostasis is impacted by some of the genetic polymorphisms that have been associated with colitis, including those that affect IL23R and ATG16L. For example, ATG16L forms a complex with ATG5: both are involved in autophagy (Fugjta et al., 2008). Studies of genetically engineered mice with cell lineage-specific ATG5 deficiency have shown that they have defects in the development of B1-B cells (Miller et al., 2008), which have a primary role in production of IgA in the gut mucosa (Kroese et al., 1996). IL-23 is an IL-12-like cytokine known to effect immunoinflammatory responses through induction of Th17 CD4+ T cells (McGovern and Powrie, 2007). In the case of IL23R mutations, altered signaling is likely to affect CD4+ T cell differentiation, and could lead to changes in Ig isotype switching, including derangements in the induction of specific IgA responses to gut bacteria.
The implication for the microbial ecologist interested in IBD pathogenesis is that it may not be adequate to simply search for microbial phylotypes that are enriched or depleted in IBD. A key to understanding disease pathogenesis may be to identify defects in the antibody repertoire to microbial epitopes. Comparative metagenomic approaches may be helpful in testing this hypothesis. However, as discussed below, there are significant challenges, including the need to design and stage proof-of-principle tests under carefully controlled circumstances in both mice and humans.
Metagenomics and IBD: a wish list for the future
Future research about the role of the gut microbiota in defining IBD risk, pathogenesis, and treatment strategies will continue to depend upon the intersection of two discovery pipelines - one involving mouse models; the other humans. In the case of conventionally-raised and gnotobiotic mice, an extraordinary opportunity exists to address a number of the unanswered questions alluded to above:
The diversity and biogeography of the gut microbiota needs to be defined at varying scales of resolution, along the length and width of the gut under conditions where diet and host genotype can be constrained
Key questions include: (i) the nature of the interactions of microbial consortia with cellular components of the innate and adaptive immune systems; (ii) the effects of engineered perturbations of the innate and adaptive systems on the acquisition, spatial distribution, diversity, and stability of the gut microbiota (and microbiome); (iii) the impact of antibiotic and/or immunosuppressive therapy on microbial community structure in mice with or without colitis (is the microbiota restored to its original status following these interventions? If so, this opens the door to the possibility of using microbial lineage and gene biomarkers to define disease severity, remission and/or risk of relapse in patients); and (iv) the short and long term effects of probiotics on community ecology in different regions of the gut, and on the activity of components of the immune system in mice with or without genetically engineered perturbations that predispose to development of colitis. These characterizations should be targeted to both SSU rDNA sequences and the microbiome.
Determine whether microbiota transplants from mouse donors with colitis to germ-free recipients (either wild-type or with mutations that sensitize the host to the development of colitis) will produce disease in the recipient
If so, the phenotypic results need to be correlated with comparative metagenomic studies of the harvested donor community and the recipient's community - the latter as a function of time after transplant and in different regions of the recipient's gut with or without disease. Continued development of new algorithms for defining similarities and differences between microbial communities are needed to interpret the results of these experiments. This type of study could lay the groundwork for functional assays of human microbial communities in gnotobiotic mice.
Advance beyond DNA-level characterization of a microbiota to transcriptional, proteomic, and metabolic analyses
These are great challenges. For transcriptional profiling, in the absence of validated DNA microarrays, there is a need to develop reliable methods for reducing the representation of rRNA in a community RNA sample so that expressed tag (EST) sequencing of cDNAs can be conducted using massively parallel sequencers. For shotgun proteomic analyses, technical and bioinformatic lessons need to be gleaned from ongoing studies of environmental communities of varying complexity (e.g., Lo et al., 2007). The formidable task of identifying and classifying known proteins and new protein families will be facilitated by generating more reference genome sequences from representatives of the phylotypes present in a microbiota (one of the goals of the human microbiome project) and from new gut microbiome datasets. Reports of transcriptome-guided or shotgun metabolomic studies of the gut microbiota in various mouse models of the human gut microbiota are just begnnining to appear (Samuel et al., 2007; Li et al., 2008, Martin et al., 2008).
Create a pipeline for characterizing the evolution of microbe-associated carbohydrate epitopes in a model gut ecosystem created in gnotobiotic mice with and without engineered genetic manipulations of their immune systems; expand this analysis to more complex communities
The degree to which adaptive immune responses to surface carbohydrate epitopes play a role in shaping diversity within the microbiota and host-microbial homeostasis needs to be explored further. For example, we know that at least some members of the gut Bacteroidetes dedicate a considerable percentage of their genomes to fashioning surface carbohydrate epitopes (Xu et al., 2007; Krinos et al., 2001; Fletcher et al., 2007), and that certain epitopes have marked effects on the activity of the host immune system (Mazmanian et al., 2005). Capsular polysaccharides, O-antigens, and exopolysaccharides represent a highly diverse group of surface epitopes that can be modified by transcriptional regulation as well as by lateral transfer of glycosyltransferase genes (GT) (Coyne and Comstock, 2008; Comstock et al., 2000; Xu et al., 2007). Unfortunately, surface glycan structures cannot be predicted based only on knowledge of the repertoire of GTs present in a microbial species or microbial community. Therefore, new methods are needed to characterize glycobiomes (genes encoding proteins and enzymes involved in carbohydrate biosynthesis and degradation) and their biochemical products. It is possible that some of these products may be useful for diagnostic and therapeutic purposes.
Characterize the B and T cell repertoire through high-throughput sequencing of the complementary determining regions of their respective antigen receptors
Antigen receptor repertoire diversity, which is based on gene segment combinatorial rearrangement in B and T cells, rivals the complexity of the microbial community. Deep sequencing of these genes should catalyze efforts to associate microbial communities with defined antigen receptor repertoires. This notion is not far-fetched: Hsieh et al. (2004) have already shown that by using a genetically ‘confined’ mouse model and conventional capillary DNA sequencers, T cell receptors associated with Treg cells could be identified. By using the new generation of massively parallel DNA sequencers, it should be possible to associate components of the unrestricted antigen-receptor repertoires in multiple patients with differing genetic backgrounds with their microbial community structures.
The other component of this envisioned intersecting pipeline involves humans with IBD. Among the many directions that this effort can take are:
Characterize monozygotic twins discordant for IBD
Twins who are discordant for pathophysiologic states, such as IBD, provide a very attractive starting point for developing, or testing, hypotheses about the relationships between a gut microbiota/microbiome, host genotype, host phenotypes, and disease pathogenesis. This is generally true of other physiologic and pathophysiologic conditions, where the phenotypically normal co-twin serves as a powerful reference control (e.g., Pietilainen et al., 2008). A key challenge is to develop safe and effective/reliable ways for sampling mucosa-associated communities along the length of the gut, in regions with and without active disease and as a function of disease activity (with activity comprehensively defined by histopathologic criteria and by profiling host gene expression in tissue samples using functional genomic and/or proteomic approaches). A parallel effort is also needed that characterizes the gut microbiota in unrelated individuals, over time and as a function of IBD activity and host genotype. Moreover, do probiotic formulations that exhibit efficacy in well-controlled clinical trials alter the structure and operations of a gut microbiota? If so, how do these effects correlate with disease activity in an individual, or groups of genetically identical or similar individuals?
Evaluate microbial biomarkers of disease status obtained from studies of mouse models
As noted above, these could be DNA, mRNA, protein, immune, metabolic and/or carbohydrate biomarkers. Finding suitable controls for correlating disease activity with these candidate biomarkers will be a challenge: given the interpersonal variations that have been noted in gut microbial ecology, must an individual ultimately serve as his/her own control? A goal for this type of analysis, and for the human microbiome project in general, is to use these biomarkers as new ways for defining health, disease susceptibility, disease status, and responses to therapy.
Acknowledgments
Work in the investigators lab cited in this review was supported by in part by grants from the NIH and from the Crohn's and Colitis Foundation of America. The authors regret that due to space limitations, a number of excellent references relevant to the topics discussed in this essay, could not be cited or discussed. We thank Rob Knight for helpful comments, and Ajit Varki (Univ. of California, San Diego) for allowing us to use the term ‘glycophobia’ which he originally coined.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baklien K, Brandtzaeg P. Comparative mapping of the local distribution of immunoglobulin-containing cells in ulcerative colitis and Crohn's disease of the colon. Clin Exp Immunol. 1975;22:197–209. [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Metagenomics. The new science of metagenomics: Revealing the secrets of our microbial planet. The National Academies Press; 2007. [PubMed] [Google Scholar]
- Comstock LE, Pantosti A, Kasper DL. Genetic diversity of the capsular polysaccharide C biosynthesis region of Bacteroides fragilis. Infect Immun. 2000;68:6182–6188. doi: 10.1128/iai.68.11.6182-6188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield AP, Carroll D, Myerscough N, Probert CS. Mucins in the gastrointestinal tract in health and disease. Front Biosci. 2001;6:D1321–1357. doi: 10.2741/corfield. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Comstock LE. Niche-specific features of the intestinal bacteroidales. J Bacteriol. 2008;190:736–742. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra G, Yuvaraj S, Jiang HQ, Bun JC, Moshage H, Kushnir N, Peppelenbosch MP, Cebra JJ, Bos NA. Early bacterial dependent induction of inducible nitric oxide synthase (iNOS) in epithelial cells upon transfer of CD45RB(high) CD4(+) T cells in a model for experimental colitis. Inflamm Bowel Dis. 2007;13:1467–1474. doi: 10.1002/ibd.20262. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Papke RT. Genomics and the bacterial species problem. Genome Biol. 2006;7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol. 2007;44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci U S A. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Fukuda M, Noda T, Yoshimori T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaffer MH, Holdsworth CD, Duerden BI. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J Med Microbiol. 1991;35:238–241. doi: 10.1099/00222615-35-4-238. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. 2001;69:3611–3617. doi: 10.1128/IAI.69.6.3611-3617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaoui KA, Corthesy B. Secretory IgA mediates bacterial translocation to dendritic cells in mouse Peyer's patches with restriction to mucosal compartment. J Immunol. 2007;179:7751–7757. doi: 10.4049/jimmunol.179.11.7751. [DOI] [PubMed] [Google Scholar]
- Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, Allen PM. An Antibiotic-Responsive Mouse Model of Fulminant Ulcerative Colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilbaugh SA, Shin ME, Banchereau RF, McVay LD, Boyko N, Artis D, Cebra JJ, Wu GD. Activation of RegIIIbeta/gamma and interferon gamma expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut. 2005;54:623–629. doi: 10.1136/gut.2004.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- Kroese FG, de Waard R, Bos NA. B-1 cells and their reactivity with the murine intestinal microflora. Semin Immunol. 1996;8:11–18. doi: 10.1006/smim.1996.0003. [DOI] [PubMed] [Google Scholar]
- Krook A, Lindstrom B, Kjellander J, Jarnerot G, Bodin L. Relation between concentrations of metronidazole and Bacteroides spp in faeces of patients with Crohn's disease and healthy individuals. J Clin Pathol. 1981;34:645–650. doi: 10.1136/jcp.34.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Seksik P, Sutren M, de la Cochetiere MF, Jian R, Marteau P, Dore J. Biodiversity of the Mucosa-associated Microbiota Is Stable Along the Distal Digestive Tract in Healthy Individuals and Patients with IBD. Inflamm Bowel Dis. 2005;11:473–480. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G, Tyson GW, Allen EE, Ram RJ, Detter JC, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, Neut C, Collins MD, Colombel JF, Marteau P, Dore J. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Micro Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333–1336. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwe JPvd, Schroder AM, Wensick F, Hazenberg MP. The obligate anaerobic faecal flora of patients with Crohn's disease and their first-degree relatives. Scand J Gastroenterol. 1988;23:1125–1131. doi: 10.3109/00365528809090179. [DOI] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin HWt. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108–114. doi: 10.1136/gut.53.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi A, Mizoguchi E, Tonoegawa S, Bhan AK. Alteration of a polyclonal to an oligoclonal immune response to cecal aerobic bacteril antigens in TCR alpha mutant mice with inflammatory bowel disease. International Immunology. 1996;8:1387–1394. doi: 10.1093/intimm/8.9.1387. [DOI] [PubMed] [Google Scholar]
- Mizoguchi A, Mizoguchi E, Smith RN, Preffer FI, Bhan AK. Suppressive role of B cells in chronic colitis of T cell receptor alpha mutant mice. J Exp Med. 1997;186:1749–1756. doi: 10.1084/jem.186.10.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzi A, Masignani V, Rappuoli R. The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antimicrobials. Drug Disc Today. 2007;12(11-12):429–39. doi: 10.1016/j.drudis.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Pietilainen KH, Naukkarinen J, Rissanen A, Saharinen J, Ellonen P, Keranen H, Suomalainen A, Gotz A, Suortti T, Yki-Jarvinen H, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn's disease. Inflamm Bowel Dis. 2004;10:824–833. doi: 10.1097/00054725-200411000-00017. [DOI] [PubMed] [Google Scholar]
- Pullan RD, Thomas GA, Rhodes M, Newcombe RG, Williams GT, Allen A, Rhodes J. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;35:353–359. doi: 10.1136/gut.35.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Dore J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–3913. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithson JE, Campbell A, Andrews JM, Milton JD, Pigott R, Jewell DP. Altered expression of mucins throughout the colon in ulcerative colitis. Gut. 1997;40:234–240. doi: 10.1136/gut.40.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, Lochs H, Dorffel Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka H, Abe Y, Iwata M, Takeuchi H, Ishikawa H, Matsushita M, Shiohara T, Akira S, Ohteki T. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- Toivanen P, Vaahtovua J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun. 2001;69:2372–2377. doi: 10.1128/IAI.69.4.2372-2377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]