Parkin catalyzes mitochondrial ubiquitination, recruiting autophagic components that clear damaged mitochondria. Defects in this pathway are implicated in Parkinson's disease.
Abstract
Mutations in parkin, a ubiquitin ligase, cause early-onset familial Parkinson's disease (AR-JP). How parkin suppresses Parkinsonism remains unknown. Parkin was recently shown to promote the clearance of impaired mitochondria by autophagy, termed mitophagy. Here, we show that parkin promotes mitophagy by catalyzing mitochondrial ubiquitination, which in turn recruits ubiquitin-binding autophagic components, HDAC6 and p62, leading to mitochondrial clearance. During the process, juxtanuclear mitochondrial aggregates resembling a protein aggregate-induced aggresome are formed. The formation of these “mito-aggresome” structures requires microtubule motor-dependent transport and is essential for efficient mitophagy. Importantly, we show that AR-JP–causing parkin mutations are defective in supporting mitophagy due to distinct defects at recognition, transportation, or ubiquitination of impaired mitochondria, thereby implicating mitophagy defects in the development of Parkinsonism. Our results show that impaired mitochondria and protein aggregates are processed by common ubiquitin-selective autophagy machinery connected to the aggresomal pathway, thus identifying a mechanistic basis for the prevalence of these toxic entities in Parkinson's disease.
Introduction
Parkinson's disease (PD) is the second most common progressive neurodegenerative disorder. The neurological lesions are frequently accompanied by cytoplasmic inclusion bodies, termed Lewy bodies, which contain ubiquitin-positive protein aggregates (McNaught et al., 2002). The prevalence of Lewy bodies has led to a central proposal that aberrant accumulation of protein aggregate is a key contributing factor to the development of Parkinsonism. In addition to protein aggregates, mitochondrial dysfunction has emerged as another prominent pathological feature associated with PD. It has been long recognized that inhibitors of mitochondrial complex I can elicit Parkinsonian syndrome in human and rodent models. Indeed, complex I deficiency has been observed in PD patients (for review see Abou-Sleiman et al., 2006). The commonality of mitochondrial defects and protein aggregates in PD suggest that the accumulation of these toxic entities might be mechanistically linked. Uncovering this potential link could provide an important clue to the fundamental defects underlying PD.
Mutations in parkin (PARK2) account for almost 50% of familial autosomal recessive juvenile PD (AR-JP; Lücking et al., 2000). The prevalence of parkin mutations in AR-JP suggests a dominant role for parkin in suppressing Parkinsonism (Abou-Sleiman et al., 2006). Parkin encodes for a RING domain–containing ubiquitin E3 ligase (Shimura et al., 2000; Zhang et al., 2000). Several disease-associated parkin mutants are deficient in E3 ligase activity, suggesting that parkin suppresses Parkinsonism by promoting ubiquitination. In the cell-based model, parkin can poly-ubiquitinate mutant DJ-1 and α-synuclein–associated synphilin, which are both involved in PD (Chung et al., 2001; Lim et al., 2005; Olzmann et al., 2007). Interestingly, parkin-mediated ubiquitination of synphilin or DJ-1 does not lead to rapid degradation by the proteasome (Lim et al., 2005). For DJ-1, ubiquitination recruits a ubiquitin-binding protein deacetylase, HDAC6, which facilitates the transport of DJ-1 to the microtubule-organizing center, forming the inclusion body, aggresome (Olzmann et al., 2007). Evidence indicates that HDAC6-dependent aggresomal pathway concentrates toxic protein aggregates for subsequent clearance by autophagy (Kawaguchi et al., 2003; Iwata et al., 2005; Pandey et al., 2007; Lee et al., 2010). Aggresomes share many features with Lewy bodies, suggesting that active concentration of protein aggregate might be a neuroprotective mechanism in PD patients (McNaught et al., 2002). Accordingly, parkin might protect neurons by promoting the clearance of toxic protein aggregates, although there is not yet definitive experimental support to this proposition.
Characterization of parkin mutant animal models has revealed prominent mitochondrial defects (for review see Abou-Sleiman et al., 2006). How parkin affects mitochondrial functionality remains poorly understood. Recently, parkin was found to associate with functionally impaired and depolarized mitochondria. Remarkably, parkin-marked mitochondria were subsequently cleared by autophagy, which sequesters and delivers damaged mitochondria via autophagosomes to lysosomes for elimination (Narendra et al., 2008). In principle, this mitophagy activity could protect neurons by eliminating dysfunctional mitochondria, which could produce toxic reactive oxygen species that damage neurons. How parkin promotes the clearance of impaired mitochondria and whether mitophagy is important in suppressing Parkinsonism are not known.
Historically, autophagy has been principally characterized as a nonselective degradative pathway activated by starvation. However, independent of nutrient status, autophagy has emerged as main quality control machinery that selectively disposes protein aggregates and damaged organelles (for review see Mizushima et al., 2008). This so-called quality control (QC) autophagy is fundamentally different from starvation-induced autophagy in its unique involvement of ubiquitinated substrates and the ubiquitin-binding HDAC6 and p62 (Lee et al., 2010). In QC autophagy, it was proposed that HDAC6 recruits a cortactin-dependent actin-remodeling machinery to ubiquitinated protein aggregates, where the assembly of F-actin facilitates autophagosome–lysosome fusion and clearance of autophagic substrates (Lee et al., 2010). Whether mitophagy operates by a similar molecular mechanism is a fundamental question that remains to be answered.
In this report, we provide evidence that parkin induces mitophagy by promoting the ubiquitination of dysfunctional mitochondria. The parkin-mediated ubiquitination serves to recruit ubiquitin-binding deacetylase HDAC6 and p62, which assemble autophagy machinery that clears impaired mitochondria. We also found that parkin-dependent mitophagy requires the formation of juxtanuclear mitochondrial inclusion bodies that resemble the aggresome. Importantly, we showed that several disease-causing parkin mutants are defective in mitophagy, thereby supporting a critical role for mitophagy in suppressing Parkinsonism.
Results and discussion
Disease-associated parkin mutants are deficient in promoting mitophagy
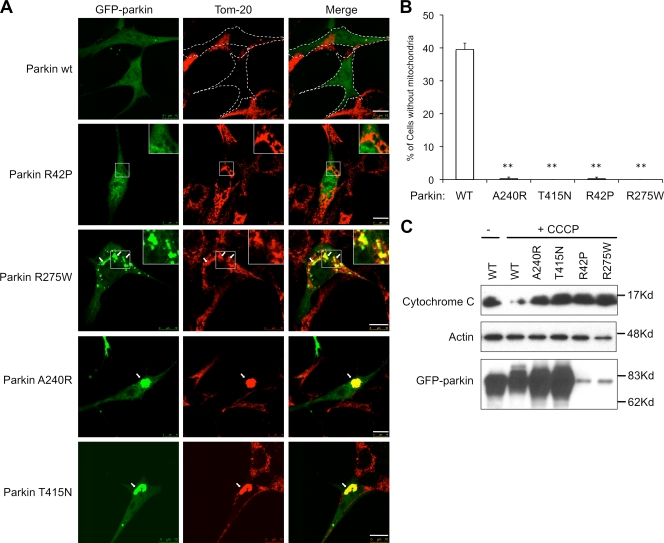
Overexpressed parkin was recently reported to promote the clearance of depolarized mitochondria by mitophagy (Narendra et al., 2008). We found that SH-SYY5 neuroblastoma cells, which express high levels of endogenous parkin, indeed lost mitochondria in response to the mitochondrial uncoupler, carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. S1). Thus, endogenous parkin can support mitophagy. To investigate whether the mitophagy activity is linked to development of PD, we determined if parkin mutations associated with familial AR-JP are defective in supporting mitophagy. We chose to study representative parkin mutants R42P, R275W, A240R, and T415N that affect three conserved domains in parkin: ubiquitin-like (UBL) domain, RING-1, and RING-2 domains. We expressed wild-type or disease-associated parkin mutants in mouse embryo fibroblasts (MEFs), which have undetectable levels of parkin protein, and assessed their ability to clear impaired mitochondria induced by CCCP. As expected, in the absence of CCCP, wild-type and parkin mutants were largely cytosolic, did not colocalize with mitochondria, and had little effect on mitochondrial abundance or morphology (Fig. S2). Upon CCCP treatment, a prominent loss of mitochondria was observed in a significant percentage of cells expressing wild-type parkin (Fig. 1, A and B). In stark contrast, mitochondria were retained in cells expressing disease-associated parkin mutants after CCCP treatment (Fig. 1, A and B). The defect in mitophagy was further confirmed by immunoblotting analysis for the loss of mitochondrial markers (Fig. 1 C). Unlike wild-type parkin, whose expression resulted in significant loss of mitochondrial cytochrome c upon CCCP treatment, none of the disease-associated parkin mutants showed such an activity. We conclude that the disease-associated parkin mutations are defective in supporting mitophagy.
Figure 1.
Disease-associated Parkin mutations are defective in mitophagy. (A) MEFs were transfected with GFP-tagged wild-type (WT) or mutant Parkin expression plasmid followed by an 18-h treatment of CCCP. Cells are immunostained with a Tom20 antibody to visualize mitochondria (red). GFP-parkin–transfected cells are marked by dotted lines. Arrows indicate parkin-positive mitochondria or mitochondrial aggregates. Bar, 10 µm. (B) The average percentages of mitochondria-free cells from three independent experiments from A are presented with standard deviation as error bar. **, P < 0.01 (C) MEFs were transfected and treated with CCCP as described in A, followed by an immunoblotting analysis with antibodies for cytochrome c, actin, and parkin. Note that levels of parkin R275W and R42P mutant were lower, as previously reported (Wang et al., 2005).
Disease-causing parkin mutants arrest mitophagy at distinct steps
Although every parkin mutant tested in our assay was defective in clearing impaired mitochondria, their phenotypes were distinct. Although wild-type parkin prominently colocalized with mitochondria upon CCCP treatment (see below), UBL domain mutant parkin-R42P did not, suggesting that the UBL domain is involved in recruitment of parkin to damaged mitochondria (Fig. 1 A). We found that A240R and T415N mutants, which are deficient in the ubiquitin E3 ligase activity (Zhang et al., 2000; Sriram et al., 2005), prominently associated with depolarized mitochondria and, remarkably, induced the formation of large mitochondrial aggregates in the perinuclear region (Fig. 1 A). In contrast, parkin R275W mutant, while apparently localized to mitochondria, did not induce the formation of perinuclear mitochondrial aggregates (Fig. 1 A). These results show that all PD-associated parkin mutants tested are defective in supporting mitophagy but with distinct phenotypes.
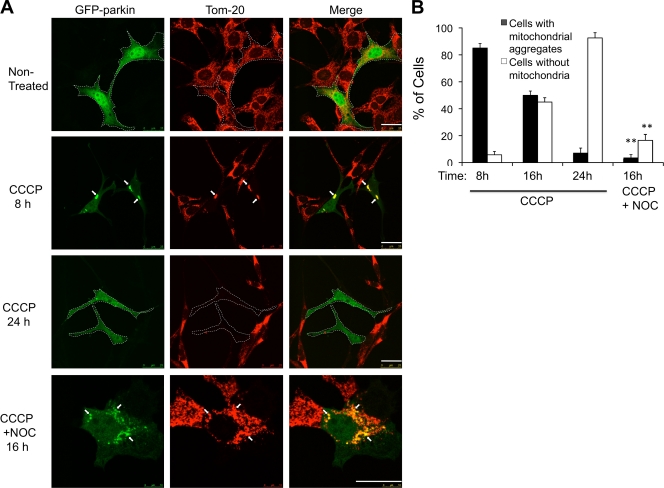
The distinct mitochondrial phenotypes caused by different parkin mutations suggest that the clearance of impaired mitochondria involves multiple discrete steps. We further investigated the formation of perinuclear mitochondrial aggregates, as they show some features of aggresomes. We first determined if mitochondrial aggregate formation is part of mitophagy induced by wild-type parkin and CCCP. To this end, we assessed mitochondrial status at different time points after CCCP treatment. As shown in Fig. 2 A, although mitochondria were largely cleared after 24 h treatment, prominent perinuclear-localized mitochondrial aggregates were observed in the majority of parkin-expressing cells (80%) at 8 h (Fig. 2). This result suggests that formation of perinuclear mitochondrial aggregates is an intermediate step for parkin-CCCP–induced mitophagy (Fig. 2).
Figure 2.
CCCP-induced parkin-mitochondrial aggregate formation is an intermediate step for mitophagy. (A) MEFs expressing WT GFP-parkin were treated with CCCP or CCCP and nocodazole (NOC, 10 µM) for 8, 16, and 24 h as indicated. Cells were immunostained with anti-Tom20 to visualize mitochondria. GFP-parkin–positive cells are marked by dotted lines. Arrows indicate parkin-positive mitochondria or mitochondrial aggregates. Bar, 25 µm. (B) The average percentages of cells with mitochondrial aggregates or without mitochondria from three independent experiments from A are presented with standard deviation as error bar. **, P < 0.01.
Microtubule dynein motors are required for parkin to induce aggregation and clearance of impaired mitochondria
The perinuclear mitochondrial aggregates are reminiscent of the aggresome, an inclusion body where protein aggregates are concentrated by the microtubule dynein motor (Johnston et al., 2002). To determine if impaired mitochondria are similarly concentrated to the perinuclear region by dynein-dependent transport, parkin-expressing MEFs were treated with a microtubule-destabilizing reagent, nocodazole. As shown in Fig. 2 A (bottom) and 2 B, nocodazole significantly inhibited the formation of juxtanuclear mitochondrial aggregates but did not affect parkin localization to mitochondria. Further, overexpression of dynamitin, which inhibits dynein motor activity, also suppressed mitochondrial aggregate formation (Fig. S3 A). Importantly, nocodazole treatment significantly inhibited parkin-CCCP–induced mitochondrial clearance (Fig. 2 B), indicating that dynein motor–dependent aggregate formation is required for efficient clearance of impaired mitochondria.
Parkin induces ubiquitination of impaired mitochondria
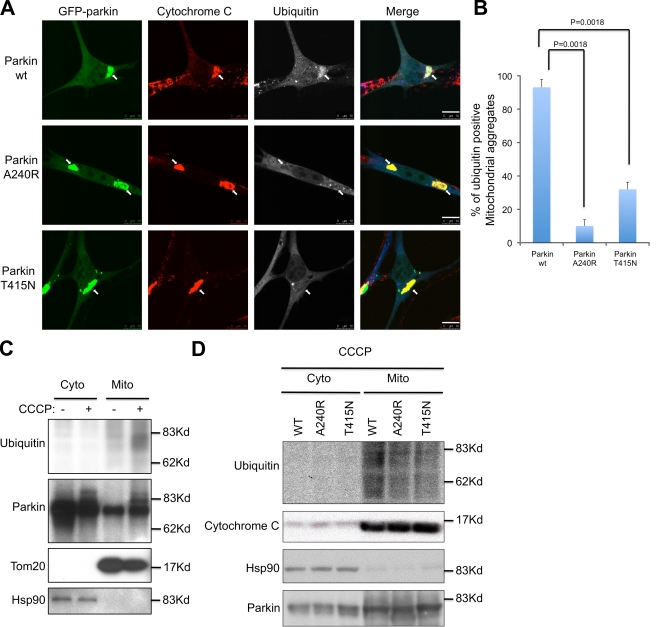
We next determined how parkin promotes the clearance of mitochondria by autophagy. We noticed that although parkin A240R and T415N mutants induced perinuclear mitochondrial aggregate formation, these mitochondria are not cleared (Fig. 1). As A240R and T415N parkin mutants share a common biochemical defect in E3 ubiquitin ligase activity, we asked if an ubiquitination step is required for the final clearance of impaired mitochondria. To test this, we immunostained parkin-expressing cells with an antibody for polyubiquitin. The ubiquitin antibody reacted strongly with the majority of perinuclear mitochondrial aggregates, indicating that mitochondria are ubiquitinated (Fig. 3, A and B). Indeed, ubiquitinated protein species were detected in purified mitochondria after CCCP treatment (Fig. 3 C). In stark contrast, the majority of mitochondrial aggregates in parkin E3 ligase-deficient A240R- and T415N-expressing cells lacked ubiquitin signals (Fig. 3, A and B). Immunoblotting of purified mitochondria from parkin-A240R and T415N-expressing cells confirmed a reduction in ubiquitination (Fig. 3 D). These results indicate that parkin induces ubiquitination in impaired mitochondria and this ubiquitination is required for efficient clearance of impaired mitochondria.
Figure 3.
Parkin promotes mitochondrial ubiquitination. (A) MEFs were transfected with GFP wild-type (WT), A240R, and T415N mutant parkin followed by CCCP treatment for 8 h. Cells were immunostained with antibodies for cytochrome c (red) and ubiquitin (blue). Arrows indicate mitochondrial aggregates. Bar, 10 µm. (B) The average percentages of ubiquitin-positive mitochondrial aggregates from three independent experiments from A are presented with standard deviation as error bar. (C and D) MEFs were transfected and treated with CCCP as described in A. Cells were subjected to the fractionation to isolate crude mitochondria. Cytoplasmic and mitochondrial fractions were subjected to immunoblotting analysis using antibodies for ubiquitin, parkin, cytochrome c, and cytosolic Hsp90.
Mitochondrial ubiquitination recruits ubiquitin-binding p62 and HDAC6
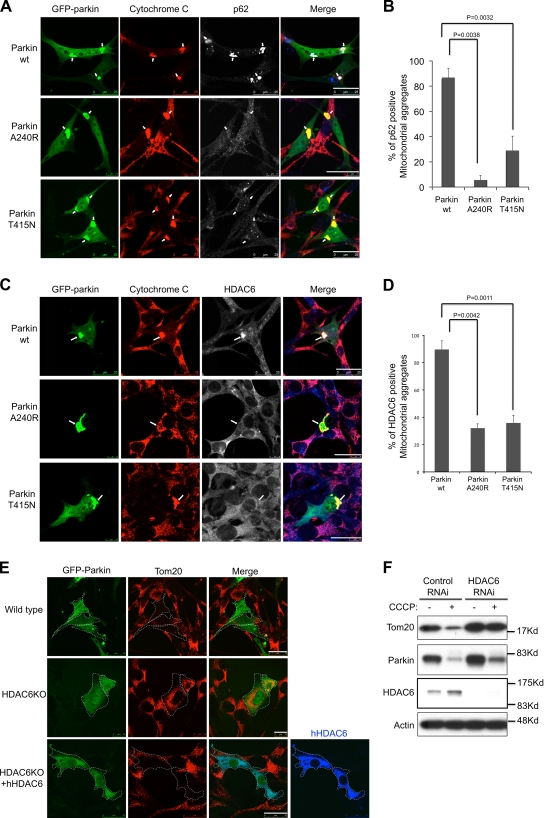
We have recently shown that ubiquitin modification in protein aggregates serves to recruit two key regulatory components of autophagy machinery, p62 and HDAC6, both with intrinsic ubiquitin-binding activity. p62 binds the key autophagosome component LC3, whereas HDAC6 activates an actin-remodeling machinery that promotes autophagosome–lysosome fusion, thereby enhancing autophagy activity (Pankiv et al., 2007; Lee et al., 2010). As E3 ligase–deficient parkin A240R and T415N mutants failed to complete mitophagy (Fig. 1), we asked if mitochondrial ubiquitination is involved in recruiting autophagy machineries. As shown in Fig. 4, both p62 and HDAC6 are prominently localized to ubiquitinated mitochondrial aggregates in wild-type parkin-expressing cells. In contrast, the mitochondrial localization of p62 and HDAC6 is significantly less abundant in parkin A240R and T415N mutant-expressing cells. This result indicates that parkin-CCCP–dependent mitochondrial ubiquitination recruits p62 and HDAC6 to damaged mitochondria.
Figure 4.
Parkin-mediated ubiquitination recruits p62 and HDAC6. MEFs were transfected with WT, A240R, and T415N mutant GFP-parkin followed by CCCP treatment for 8 h. Cells were double immunostained with cytochrome c (red) and p62 antibody (blue) in A, and cytochrome c (red) and HDAC6 antibody in C. Arrows indicate mitochondrial aggregates. (B and D) The average percentages of p62- or HDAC6-positive mitochondrial aggregates from three independent experiments are presented with standard deviation as error bar. (E) Wild-type and HDAC6 knockout (KO) MEFs were transfected with parkin-GFP or cotransfected with a Flag-tagged HDAC6 followed by CCCP treatment for 16 h as indicated. Cells are immunostained with Tom20 (red) and Flag (blue) antibodies. Bar, 25 µm. (F) Wild-type MEFs were transfected with control or HDAC6 siRNA and parkin-GFP, and treated with or without CCCP for 16 h. Cell lysates were subjected to immunoblotting analysis using antibodies for Tom20, parkin, HDAC6, and actin.
HDAC6, cortactin, and p62 are required for the clearance of impaired mitochondria
We next determined if HDAC6 and p62 are required for the clearance of dysfunctional mitochondria. Parkin failed to clear CCCP-treated mitochondria in HDAC6 knockout (KO) MEFs (Fig. 4 E) or p62 knockdown U2OS cells (Fig. S3). It is notable that large perinuclear mitochondrial aggregates did not form in HDAC6 KO MEFs, suggesting that HDAC6 is also required for the transport and concentration of damaged mitochondria. The parkin-CCCP–induced mitophagy can be effectively restored upon the reintroduction of wild-type HDAC6 (Fig. 4 E, bottom). To confirm the role of HDAC6 in mitophagy, we knocked down HDAC6 by specific siRNA in parkin-expressing cells and determined mitochondrial Tom20 levels by immunoblotting after CCCP treatment. As shown in Fig. 4 G, Tom20 levels were greatly reduced in control but not in HDAC6 knockdown cells, demonstrating an important role for HDAC6 in parkin-mediated mitophagy.
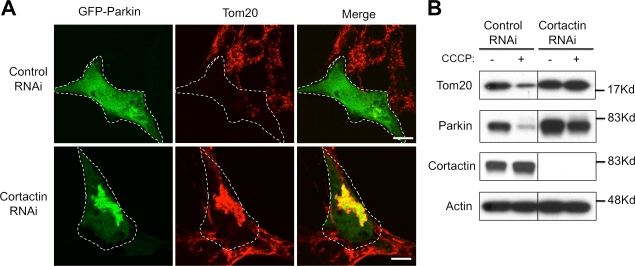
We have shown that HDAC6 facilitates the clearance of protein aggregates by recruiting cortactin-dependent actin remodeling machinery, which promotes the fusion of autophagosome and lysosome. To determine if cortactin is also required for impaired mitochondrion clearance, we knocked down cortactin by siRNA and assessed parkin-dependent mitophagy. As shown in Fig. 5 A, inactivation of cortactin led to prominent accumulation of parkin-positive mitochondrial aggregates at the perinuclear region, indicative of a mitophagy failure. Indeed, immunoblotting analysis showed that cortactin knockdown, similar to HDAC6 inactivation, prevents the loss of mitochondrial markers induced by CCCP (Fig. 5 B). We conclude that, similar to protein aggregate processing, HDAC6, cortactin, and p62 are required for the clearance of damaged mitochondria.
Figure 5.
Cortactin is required for parkin-CCCP–induced mitochondrial clearance. MEFs were transfected with control or cortactin RNAi and parkin-GFP, and incubated with or without CCCP for 16 h as indicated. (A) Cells were subjected to immunostaining with antibodies for Tom20 and parkin. Bar, 10 µm. (B) Cell lysates were subjected to immunoblotting with antibodies for Tom20, parkin, cortactin, and actin.
In this study, we found that several disease-associated mutations located in different functional domains all abrogate the ability of parkin to clear impaired mitochondria, demonstrating a tight link between parkin-dependent mitophagy and the suppression of Parkinsonism. Importantly, analyses of parkin mutants revealed distinct mitochondrial phenotypes, indicating a complex activity of parkin in mitophagy (Fig. 1). The phenotype of the R42P mutant indicates that the UBL domain is required for parkin recruitment to mitochondria (Fig. 1). The ubiquitin E3 ligase–deficient A240R and T415N mutants are capable of promoting mitochondrial aggregate formation, termed mito-aggresomes, but cannot promote clearance of mitochondria (Figs. 1 and 3). In contrast, the R275W mutant can associate with depolarized mitochondria but is deficient in promoting mito-aggresome formation. Thus, these disease-causing parkin mutants arrest mitophagy at distinct stages and demonstrate that association and clearance of impaired mitochondria represent different activities of parkin. These findings allow the reconstruction of parkin-dependent mitophagy into temporal events consisting of marking the damaged mitochondria by parkin association, transporting the “marked” mitochondria to form mito-aggresomes, and finally the clearance of concentrated mitochondria by ubiquitin-mediated autophagy. Failure in any of these steps would abrogate mitophagy, resulting in toxicity emanating from damaged mitochondria.
The physiological function of parkin E3 ligase activity has been elusive. Our results indicate that impaired mitochondria are substrates of parkin. Interestingly, parkin-mediated mitochondrial ubiquitination apparently is not obligatory for mito-aggresome formation (Fig. 3 A); however, ubiquitin-negative mitochondria are not further processed and become prominently accumulated (Figs. 1 and 3). Therefore, parkin-dependent ubiquitination appears to provide the signal for the final clearance of impaired mitochondria concentrated at the perinuclear region. Our results indicate that mitochondrial ubiquitination acts to recruit HDAC6 and p62, two ubiquitin-binding proteins required for efficient autophagy that targets protein aggregates (Lee et al., 2010). Indeed, HDAC6- and p62-deficient cells are defective in parkin-dependent mitophagy (Fig. 4 and Fig. S3 B). Altogether, these results provide strong support that ubiquitin modification is a critical basis for the specific and effective clearance of damaged mitochondria by QC autophagy.
The parkin-mediated formation of CCCP-depolarized mitochondrial aggregate is intriguing, as this structure resembles the aggresome. Similar to the aggresome, the formation of mitochondrial aggregate (mito-aggresome) depends on microtubule dynein motors and HDAC6 (Figs. 2 A, 4 E, and S3 A). Interestingly, a recent study found that PINK1 overexpression induced parkin association with mitochondria and also led to mitochondrial aggregate formation, although the significance of mitochondrial aggregates was not addressed (Vives-Bauza et al., 2010). We found that mito-aggresome formation is required for efficient mitochondrial clearance (Fig. 2). We propose that impaired mitochondria under pathological conditions are concentrated by parkin- and HDAC6-dependent aggresomal pathway to the juxtanuclear region where they are cleared by QC autophagy. Indeed, paraquat, a mitochondrial complex I inhibitor linked to PD, could induce mito-aggresome formation (Fig. S1 C). Our study revealed that protein aggregates and impaired mitochondria are processed by a common pathway involving HDAC6- and parkin-dependent ubiquitin-selective autophagy and aggresomal machinery, thus providing a unifying model toward understanding the two most common pathological features in the pathogenesis of PD.
Materials and methods
Cell lines and plasmids
Immortalized wild-type and HDAC6 KO MEFs were obtained from E14.5 wild-type and HDAC6 KO littermate embryos using 3T3 protocol as described previously (Lee et al., 2010) and maintained in DME with 10% FBS. U2OS and SH-SYY5 cells were purchased from the American Type Culture Collection and maintained in DME (U2OS) and Ham's F12/DME (SH-SYY5) with 10% FBS. All cell lines were cultured in 37°C with 5% CO2. GFP-tagged wild-type and mutant parkin expression plasmids were provided by Dr. Michael Ehlers (Duke University, Durham, NC).
Antibodies and reagents
Anti–mouse HDAC6 antibody was generated against aa 991–1149. The following antibodies/reagents were also used: anti-HDAC6 (H-300; Santa Cruz Biotechnology, Inc.), anti-ubiquitin (Enzo Life Sciences, Inc. and EMD), anti-p62 (Santa Cruz Biotechnology, Inc.), anti-mitochondrial Hsp70 (Thermo Fisher Scientific), anti-COX IV (Invitrogen), anti-cortactin (Millipore), anti-cytochrome c (BD), and nocodazole (Sigma-Aldrich).
Immunofluorescence microscopy
Immunostaining was performed as described previously (Hubbert et al., 2002; Lee et al., 2004). Cells were cultured on a glass cover slide for 1 d and transfected with wild-type or mutant Parkin expression plasmid. Cells were treated with the mitochondrial uncoupler CCCP at 25 µM for required amounts of time (8, 16, and 24 h). Cells were fixed in 4% paraformaldehyde made in phosphate-buffered saline (PBS) for 15 min at room temperature. Cells were stained with the following primary antibodies: mouse monoclonal cytochrome c (BD), rabbit polyclonal Tom20 (Santa Cruz Biotechnology, Inc.), rabbit polyclonal ubiquitin (Enzo Life Sciences, Inc.), rabbit polyclonal p62 (Santa Cruz Biotechnology, Inc.), rabbit polyclonal HDAC6 (generated against aa 991–1149 of HDAC6); and with the following secondary antibodies: mouse and/or rabbit Alexa 488 (Invitrogen), rhodamine (Jackson ImmunoResearch Laboratories, Inc.), and Cy5 (Jackson ImmunoResearch Laboratories, Inc.). Samples are mounted with Anti-Fade Fluoromount G media (SouthernBiotech) for imaging. Images were acquired by a spinning-disk confocal microscope (DMI6000C; Leica) at room temperature equipped with a charge-coupled device camera (ORCA ER; Hamamatsu Photonics), using 100x/1.4–0.70 NA oil (Plan Apochromat; Leica) or 40x/1.25–0.75 oil NA (Plan NeoFluar; Leica) objectives. Images were acquired and processed with the Leica LAS AF program version 1.8.2.
Mitochondrial fractionation
Mitochondria purification was performed with a modified method from previous methods (Schwer et al., 2002). In brief, cells were homogenized in ice-cold buffer H (210 mM mannitol, 70 mM sucrose, 0.1 mM EGTA, and 2 mM Hepes-KOH, pH 7.5). Cell homogenates were incubated with anti-Tom20 immobilized magnetic beads for 2 h. Mitochondria were isolated with Tom20 immobilized magnetic beads using magnetic field.
Western blotting
MEFs stably expressing GFP-Parkin were harvested. Samples were run on SDS-PAGE and immunoblotted with the following antibodies: polyclonal rabbit anti-Parkin (Invitrogen), rabbit anti-ubiquitin (Enzo Life Sciences, Inc.), rabbit anti-HDAC6 (generated against aa 991–1149 of HDAC6), rabbit anti-Tom20 (Santa Cruz Biotechnology, Inc.), and mouse anti-cytochrome c (BD).
Statistical analysis
A two-tailed Student's t test was conducted for statistic analysis of quantitative data.
Online supplemental material
Fig. S1. Endogenous parkin triggers mitophagy in SH-SYY5 neuroblastoma cells under CCCP treatment and paraquat treatment induces mito-aggregate. Fig. S2. Wild-type and mutant parkins are mainly localized in cytosol without CCCP treatment. Fig. S3. The formation of mitochondrial aggregate is inhibited by overexpressed dynamitin and requires p62. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001039/DC1.
Acknowledgments
We thank Dr. M. Ehlers for reagents and A. Wagner for comments.
This work is supported by a National Institutes of Health grant (NS053825) to T.-P. Yao.
Footnotes
Abbreviations used in this paper:
- AR-JP
- autosomal recessive juvenile PD
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- KO
- knockout
- MEF
- mouse embryo fibroblast
- PD
- Parkinson's disease
- QC
- quality control
- UBL
- ubiquitin-like
References
- Abou-Sleiman P.M., Muqit M.M., Wood N.W. 2006. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat. Rev. Neurosci. 7:207–219 10.1038/nrn1868 [DOI] [PubMed] [Google Scholar]
- Chung K.K., Zhang Y., Lim K.L., Tanaka Y., Huang H., Gao J., Ross C.A., Dawson V.L., Dawson T.M. 2001. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 7:1144–1150 10.1038/nm1001-1144 [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P. 2002. HDAC6 is a microtubule-associated deacetylase. Nature. 417:455–458 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Iwata A., Riley B.E., Johnston J.A., Kopito R.R. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280:40282–40292 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Illing M.E., Kopito R.R. 2002. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskeleton. 53:26–38 10.1002/cm.10057 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Kim H., Ryu C.H., Kim J.Y., Choi B.H., Lim Y., Huh P.W., Kim Y.H., Lee K.H., Jun T.Y., et al. 2004. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J. Biol. Chem. 279:30265–30273 10.1074/jbc.M312083200 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.S., Pandey U.B., Kaushik S., Tresse E., Lu J., et al. 2010. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29:969–980 10.1038/emboj.2009.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.L., Chew K.C., Tan J.M., Wang C., Chung K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A., et al. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25:2002–2009 10.1523/JNEUROSCI.4474-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking C.B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B.S., Meco G., Denèfle P., Wood N.W., et al. ; French Parkinson's Disease Genetics Study Group; European Consortium on Genetic Susceptibility in Parkinson's Disease. 2000. Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl. J. Med. 342:1560–1567 10.1056/NEJM200005253422103 [DOI] [PubMed] [Google Scholar]
- McNaught K.S., Shashidharan P., Perl D.P., Jenner P., Olanow C.W. 2002. Aggresome-related biogenesis of Lewy bodies. Eur. J. Neurosci. 16:2136–2148 10.1046/j.1460-9568.2002.02301.x [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., Li L., Chudaev M.V., Chen J., Perez F.A., Palmiter R.D., Chin L.S. 2007. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178:1025–1038 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey U.B., Batlevi Y., Baehrecke E.H., Taylor J.P. 2007. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 3:643–645 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Schwer B., North B.J., Frye R.A., Ott M., Verdin E. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647–657 10.1083/jcb.200205057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25:302–305 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- Sriram S.R., Li X., Ko H.S., Chung K.K., Wong E., Lim K.L., Dawson V.L., Dawson T.M. 2005. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 14:2571–2586 10.1093/hmg/ddi292 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tan J.M., Ho M.W., Zaiden N., Wong S.H., Chew C.L., Eng P.W., Lim T.M., Dawson T.M., Lim K.L. 2005. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson's disease-linked point mutations. J. Neurochem. 93:422–431 10.1111/j.1471-4159.2005.03023.x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao J., Chung K.K., Huang H., Dawson V.L., Dawson T.M. 2000. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA. 97:13354–13359 10.1073/pnas.240347797 [DOI] [PMC free article] [PubMed] [Google Scholar]