The myosin Myo4 is nonprocessive by itself, but when several motor subunits team up with She2 and She3, it forms an active RNP transport unit that moves ASH1 mRNA to the bud tip in dividing yeast.
Abstract
In Saccharomyces cerevisiae, ASH1 mRNA is transported to the bud tip by the class V myosin Myo4. In vivo, Myo4 moves RNA in a rapid and continuous fashion, but in vitro Myo4 is a nonprocessive, monomeric motor that forms a complex with She3. To understand how nonprocessive motors generate continuous transport, we used a novel purification method to show that Myo4, She3, and the RNA-binding protein She2 are the sole major components of an active ribonucleoprotein transport unit. We demonstrate that a single localization element contains multiple copies of Myo4 and a tetramer of She2, which suggests that She2 may recruit multiple motors to an RNA. Furthermore, we show that increasing the number of Myo4–She3 molecules bound to ASH1 RNA in the absence of She2 increases the efficiency of RNA transport to the bud. Our data suggest that multiple, nonprocessive Myo4 motors can generate continuous transport of mRNA to the bud tip.
Introduction
mRNA localization generates asymmetric distributions of proteins that are essential for cell motility, cell fate determination, and synaptic plasticity (Du et al., 2007). Localized mRNAs contain cis-acting sequences, called localization elements (LE) or zipcodes, that recruit proteins, which mediate the transport of mRNA within the cytosol (Jambhekar and Derisi, 2007; Martin and Ephrussi, 2009). Motor proteins, including myosins, kinesins, and dynein, have been shown to transport localized mRNAs in vivo, and live-cell imaging revealed that transport of most localized mRNAs is continuous over several micrometers (St Johnston, 2005; Bullock, 2007; Müller et al., 2007). Although several cis-acting elements and proteins involved in mRNA transport have been identified, it remains unclear how these components assemble to generate sustained transport of mRNA in vivo.
In the budding yeast Saccharomyces cerevisiae, ASH1 mRNA is localized to the bud tip by the class V myosin motor Myo4 (Long et al., 1997; Takizawa et al., 1997). Localization of ASH1 mRNA confines Ash1 to the daughter cell nucleus, where it represses expression of HO endonuclease, preventing mating-type switching specifically in the daughter cell. ASH1 mRNA contains four localization elements; three are located within the coding region and the fourth, U3 (E3), starts seven nucleotides upstream of the stop codon and extends into the 3′ untranslated region (UTR). These localization elements, as well as those from several other bud-localized transcripts, form stem–loop structures, and each element is sufficient to localize a reporter RNA to the bud (Chartrand et al., 1999; Gonzalez et al., 1999; Jambhekar et al., 2005; Olivier et al., 2005). Genetic and biochemical data suggest that the RNA-binding protein She2 binds these elements and recruits a complex of She3 and Myo4 (Myo4–She3) to the element through a direct interaction with She3 (Jansen et al., 1996; Bertrand et al., 1998; Münchow et al., 1999; Böhl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000). So far, only Myo4, She3, and She2 have been found to associate with the localization elements in ASH1 mRNA and have been collectively named the locasome.
In vivo imaging of fluorescently tagged RNA in living cells indicates that Myo4 generates continuous transport of RNA to the bud tip at 0.2–0.44 µm/s (Bertrand et al., 1998). To sustain movement of RNA over several micrometers, Myo4 must take many steps along an actin filament before dissociating. Motors walk along filaments by binding and hydrolyzing ATP, and each ATP–hydrolysis cycle includes one phase during which the motor domain detaches from the filament. Processive motors, such as kinesin-1 and myosin Va, maintain contact with filaments through several rounds of ATP hydrolysis because they are dimers and coordinate the enzymatic cycles of their two motor domains, such that when one motor domain detaches from the filament, the second remains bound (Tyska and Mooseker, 2003; Gennerich and Vale, 2009). In vitro assays have shown that processive motors take several steps along a filament before dissociating, making them ideal motors for transporting cargo in vivo. In contrast, Myo4 is a nonprocessive motor and dissociates from filaments before stepping forward (Reck-Peterson et al., 2001; Dunn et al., 2007). One difference between Myo4 and myosin Va is that Myo4 is a monomer that associates with She3 rather than another copy of Myo4, and therefore lacks a second motor domain to coordinate enzymatic cycles (Dunn et al., 2007; Hodges et al., 2008; Bookwalter et al., 2009).
How does Myo4 as a nonprocessive motor generate continuous transport of ASH1 mRNA to the bud tip? One possibility is that the RNA transport machinery includes proteins that increase the processivity of Myo4 when bound to RNA. Genetic and biochemical data have shown that Myo4, She3, and She2 are required for localization of ASH1 mRNA and associate with the localization elements within ASH1 mRNA. She3 copurifies with Myo4 in a 7.6S complex, and purified Myo4–She3 was found to be nonprocessive in vitro (Dunn et al., 2007; Hodges et al., 2008). The effect of She2 on Myo4 processivity has not been tested as it dissociates from Myo4–She3 during purification. Proteins such as Khd1, Puf6, and Loc1 have been shown to regulate the translation or anchoring of ASH1 mRNA at the bud tip, but their role in mediating transport to the bud tip is unclear (Long et al., 2001; Irie et al., 2002; Gu et al., 2004). Whether proteins besides Myo4, She3, and She2 associate with ASH1 localization elements and affect transport of ASH1 mRNA is unknown.
An alternative model is that several copies of Myo4 could associate with ASH1 mRNA, increasing the probability of at least one motor being attached to a filament during transport to the bud tip. As mentioned, ASH1 contains four localization elements, providing four potential sites to recruit Myo4 to the full-length mRNA. However, each element on its own localizes a reporter RNA to the bud tip, which suggests that a single element is sufficient to generate sustained transport to the bud tip (Chartrand et al., 1999; Gonzalez et al., 1999; Jambhekar et al., 2005; Olivier et al., 2005). Whether multiple Myo4 motors could associate with a single element is unclear, as functional elements can be as short as 30 nucleotides (Jambhekar et al., 2005) and would seem to provide limited space to accommodate multiple motors.
In this study, we investigated how Myo4 generates rapid and continuous transport of its RNA cargo. We first developed a novel purification system to identify the protein composition of the RNA transport machinery in S. cerevisiae and determine whether additional regulatory components exist. We show that Myo4, She3, and She2 are the sole, major components that assemble onto a localization element from ASH1 mRNA to form an RNP complex that mediates transport to the bud tip. We demonstrate that Myo4–She3 assembles into a large RNP complex when bound to a single localization element, and further reveal that this RNP complex contains multiple copies of Myo4. In addition, we show that She2 assembles into a dynamic tetramer, and that mutations in She2 that prevent ASH1 mRNA localization also inhibit She2 oligomerization, which suggests that She2 oligomerization facilitates mRNA localization. Finally, we demonstrate that in the absence of She2, increasing the number of Myo4 motors bound to ASH1 RNA increases the efficiency of its transport into the bud. Taken together, our data suggest that continuous transport of ASH1 mRNA to the bud tip is generated by recruiting multiple nonprocessive Myo4 motors to a localization element, possibly through interactions with the tetrameric She2.
Results
Purification of a localized ribonucleoprotein complex
We first explored whether proteins besides Myo4, She3, and She2 associate with ASH1 localization elements and potentially affect the processivity of Myo4. The processivity of the microtubule motor dynein is enhanced by its cofactor dynactin (King and Schroer, 2000), which suggests a possible mechanism to generate processivity from Myo4. To identify proteins that associate with an ASH1 localization element, we modified a tagged RNA system (Takizawa and Vale, 2000) to purify RNA from cytosolic extracts. Our system to image RNA co-expresses an RNA-binding protein, U1Ap, fused to GFP and an NLS (U1Ap-GFP-NLS), along with an RNA that contains four repeats of the U1A aptamer fused to the RNA of interest. To facilitate purification of a specific RNA, we added a tandem affinity purification (TAP) tag (Puig et al., 2001) after GFP in the RNA-binding construct (U1Ap-GFP-TAP-NLS). We chose to purify the U3 localization element from ASH1 mRNA (Fig. 1 A), as the U3 element is sufficient to localize reporter RNAs, such as lacZ, to the bud tip (Chartrand et al., 1999; Gonzalez et al., 1999). In addition, we found that adding a single U3 element to the 3′ end of STE2 mRNA was sufficient to localize STE2 mRNA to the distal tip of buds (Fig. S1). Thus, the U3 element recruits the proteins necessary to generate efficient transport of yeast mRNA to the bud tip. When 4×U1A-tagged U3 RNA expression was induced along with U1Ap-GFP-TAP, bright GFP particles were detected at the bud tip, confirming that U1Ap-GFP-TAP was bound to U3 RNA and that the fusion protein did not interfere with localization (Fig. 1 B).
Figure 1.
Purification of U3 RNP complex. (A) Schematic representation of constructs used for U3 RNP complex purification. The RNA-binding protein is expressed from the constitutive glycerol phosphate dehydrogenase (GPD) promoter, and contains U1Ap, GFP, TAP, and an NLS. The RNA partner is expressed from the inducible galactose (GAL) promoter, and consists of four repeats of U1A aptamer, the U3 localization element, and an ADH1 terminator. (B) GFP-bound U3 RNA localizes to the bud tip using the modified tagged RNA system. Expression of 4×U1A-tagged U3 RNA was induced in cells containing U1Ap-GFP-TAP, and GFP signals in live cells were observed by fluorescence microscopy. The outline was drawn based on the DIC image. The nuclear GFP signal is from U1Ap-GFP-TAP not bound to RNA. Bar, 10 µm. (C) Silver staining of TAP-purified complexes. Extracts prepared from cells expressing U1Ap-GFP-TAP only (lane 1), U1Ap-GFP-TAP and 4xU1A-tagged U3 RNA (lane2), or U1Ap-GFP-TAP and 4xU1A-tagged ADH2 (77 NT) RNA (lane 3) were used for TAP purification. The purified complexes were separated on a 4–15% gradient SDS-polyacrylamide gel, and silver stained. (D) Myo4, She3, and She2 specifically copurify with U3 RNA. Purified complexes from C were separated by 10% SDS-PAGE in the same order as in C and analyzed by Western blotting. U1Ap-GFP was detected by an anti-GFP antibody. (E) Mass spectrometry analysis of U3 RNA copurifying proteins. TAP-purified U3 RNP complexes were separated by SDS-PAGE and stained with Coomassie blue. Protein bands of interest were excised from gels and identified by mass spectrometry analysis (LC-MS/MS).
To identify proteins that bind U3 RNA, extracts were prepared from cells expressing U1Ap-GFP-TAP and 4×U1A-tagged U3 RNA. U1A-GFP-TAP was then purified using the standard TAP purification protocol. As negative controls, we purified U1A-GFP-TAP from cells that were not expressing 4×U1A-tagged U3 RNA and from cells that expressed 4×U1A fused to a 77-nucleotide sequence from ADH2 that resides in the same position as U3 RNA in ASH1. Silver staining of the purified complexes separated by gel electrophoresis revealed several prominent bands (Fig. 1 C). The major proteins that specifically copurified with U3 RNA had sizes similar to the core components of the locasome (Böhl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000; Gonsalvez et al., 2005); Myo4 (169 kD), She3 (47 kD), and She2 (28 kD). She3 has a size similar size to U1Ap-GFP-TAP and was masked in silver-stained gels. To demonstrate that all three locasome proteins were present specifically in the U3 RNA sample, we analyzed the purified complexes by Western blotting using antibodies against the locasome components (Fig. 1 D). The results showed that Myo4, She3, and She2 were present only in complexes that contained U3 RNA. Finally, mass spectrometry analysis (liquid chromatography tandem mass spectrometry [LC-MS/MS]) of the individual protein bands identified the proteins specific to the U3 sample as Myo4, She3, and She2 (Fig. 1 E). Proteins that copurified with both U3 RNA and ADH2 (77 NT) were identified by mass spectrometry as eIF4G1/G2 and poly(A)-binding protein (PABP). These results demonstrated that the modified tagged RNA system purified a U3 ribonucleoprotein complex that contained the three core proteins known to localize ASH1 mRNA: Myo4, She3, and She2.
Myo4, She3, and She2 are the sole components of the minimal, functional U3 RNP complex
To determine whether eIF4G1/G2 and PABP were bona fide components of the locasome, we further isolated U3 RNP by velocity sedimentation on 10–50% sucrose gradients. Silver staining and Western blot analysis of the fractions showed that Myo4, She3, and She2 co-migrated in a complex with an estimated size of 20.61 ± 0.56 S (n = 3), whereas eIF4G1/G2 and PABP migrated further up the gradient (Fig. 2, A and B). These results indicated that eIF4G1/G2 and PABP bind weakly and nonspecifically to U3 RNA and are not integral components of the locasome.
Figure 2.
Myo4, She3, and She2 are the sole, major components of the U3 RNP complex. (A) Myo4, She3, and She2 co-migrate in velocity sedimentation analysis. TAP-purified U3 RNP complexes were resolved on a 10–50% sucrose gradient, and fractions were collected from the bottom (fraction 1). These fractions were analyzed by SDS-PAGE and silver stained. Positions of protein standards thyroglobulin (19S), catalase (11.3S), aldolase (7.3S), and albumin (4.6S) from parallel gradients are indicated. (B) Myo4, She3, and She2 migrate as a 20S RNP complex. TEV-eluted U3 RNP complexes were analyzed as in A, and Western blots were probed with antibodies as indicated. The sedimentation coefficient of U1Ap-GFP-TAP–bound U3 RNP complex was measured as 20.61 ± 0.56S (n = 3). U1Ap-GFP peaks twice, showing proteins bound to U3 RNP complex and unbound free U1Ap-GFP. (C) RNase dissociates the Myo4–She3–She2 complex. TEV-eluted U3 RNP complexes were treated with 0.3 mg/ml RNaseA and further analyzed along with intact U3 RNP complexes in B. The sedimentation coefficient value of Myo4–She3 is reduced to 7.76 ± 0.18S (n = 2) when not bound to RNA cargo.
Next, we tested whether the 20S complex that contained Myo4, She3, and She2 was dependent on RNA by treating the purified U3 RNP with RNaseA and performing velocity sedimentation analysis as previously described. RNaseA treatment shifted all components of the U3 RNP to lighter fractions (Fig. 2 C). Myo4 and She3 migrated in a complex with a size of 7.76 ± 0.18S (n = 2) that is similar to the size of purified Myo4–She3 (7.6 ± 0.3S; Dunn et al., 2007). She2 migrated around 5S, although there was some She2 at the bottom of the gradient. The results suggested that Myo4, She3, and She2 assembled into a 20S complex by associating with U3 RNA.
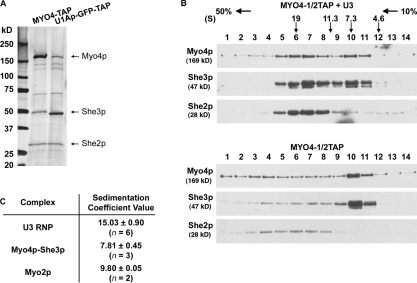
The 20S RNP complex contained not only locasome proteins but also up to eight molecules of U1Ap-GFP-TAP, which increased the size of the complex by an unknown amount. To obtain a more accurate estimate of the size of the U3 locasome, we performed two additional purifications of the U3 locasome. First, we purified U3 RNA from cells that expressed U3 RNA tagged with a single U1A aptamer, reducing the maximum number of U1Ap-GFP-TAP per RNP complex to two. This complex had a similar protein profile as the 20S complex but sedimented at 15.97 ± 0.90S (n = 3; Fig. S3). Next, we isolated the U3 locasome by purifying Myo4-TAP from cells overexpressing U3 RNA. As anticipated, the protein profile of the U3 RNP complex purified via Myo4 was identical to the complex purified via tagged U3 RNA, with She3 more evident due to the absence of U1Ap-GFP-TAP (Fig. 3 A). The size of the complex measured by velocity sedimentation was 15.03 ± 0.90S (n = 6; Fig. 3 B, top). Importantly, both She2 and She3 co-migrated with Myo4 in the complex, and the amount of She2 that copurified with Myo4 was increased, which indicates the assembly of locasome on U3 RNA. To appreciate the relative size of the U3 RNP, we purified Myo4-TAP from cells that did not express U3 RNA and measured its size by velocity sedimentation as 7.81 ± 0.45S (n = 3; Fig. 3 B, bottom), which was similar to previous measurements (Fig. 2 C; Dunn et al., 2007). Some Myo4 was detected further down the gradient and may reflect Myo4 bound to endogenous mRNA. In agreement, Myo4 purified from extracts that were treated with RNaseA eliminated the Myo4 in the heavier fractions (Fig. S2). These results confirm that Myo4, She3, and She2 are the sole, major components that associate with U3 RNA and that these proteins assemble into a 15S RNP complex on U3 RNA.
Figure 3.
Size estimation of the U3 RNP complex. (A) Comparison of U3 RNP protein profiles. U3 RNA was expressed with MYO4-TAP (lane 1) or U1Ap-GFP-TAP (lane 2). TAP-purified complexes from each cell extract were separated by 4–15% SDS-PAGE and silver stained. (B) Myo4–She3 shifts into a 15S RNP complex upon binding U3 RNA. U3 RNA expression was induced (top) or not induced (bottom) in cells expressing MYO4-1/2TAP, and the cell extracts were used for 1/2TAP-purification. The purified complexes were loaded on 10–50% sucrose gradients, and the collected fractions were analyzed by Western blotting. Myo4 bound to U3 RNA purifies as a complex with a size of 15.03 ± 0.90S (n = 6), whereas cargo-free Myo4 purifies at 7.81 ± 0.45S (n = 3). (C) Sedimentation coefficient values of Myo4 associated with U3 RNA and in soluble form. Size determination of U3 RNP complex and Myo4–She3 was based on TAP-purified complexes from cells expressing MYO4-1/2TAP with or without U3 RNA overexpression. Cells expressing MYO2-1/2TAP were used for Myo2p purification.
To determine whether the purified U3 RNP complex contained active Myo4, we analyzed the complex in an actin filament gliding assay. To ensure that the measured activity of Myo4 is from U3 RNP and not free Myo4, we adsorbed the U3 RNP purified via U1Ap-GFP-TAP onto the surface of motility chambers using anti-GFP antibodies. We then tracked the movement of fluorescently labeled actin filaments in the chambers using time-lapse fluorescence microscopy. The maximal velocity of actin filaments was roughly 2.17 ± 0.33 µm/s (n = 14, whereas n is the number of filaments measured; Video 1), which is similar to the velocity produced by purified Myo4 (Dunn et al., 2007). Importantly, U3 RNP complex treated with RNaseA failed to generate actin filament motility when adsorbed into chambers with an anti-GFP antibody. These results demonstrate that the purified U3 RNP contains active Myo4 and suggest that Myo4, She3, and She2 are sufficient to generate a minimal, functional RNP transport unit.
A single U3 RNP complex is composed of multiple motors
The relative size of the purified U3 RNP compared with Myo4–She3 and She2 suggested that the complex contained multiple copies of the locasome proteins. Moreover, as the U3 RNP also largely exceeded the size of the dimeric class V myosin Myo2p (Fig. 3 C), we suspected that multiple motors were assembled on U3 RNA. We first tested whether the U3 RNP complex could cause two differently tagged Myo4 motors to coprecipitate. U3 RNA was overexpressed to induce U3 RNP assembly in cells expressing MYO4-HA and MYO4-MYC. We immunoprecipitated Myo4-myc and measured the amount of Myo4-HA that coprecipitated. Because Myo4 is a monomer, immunoprecipitation of Myo4-myc should not coprecipitate Myo4-HA unless the motors are in the same U3 RNP complex. The results show that formation of the U3 RNP complex increased the amount of Myo4-HA that coprecipitated with Myo4-myc compared with control samples (Fig. 4 A). Small amounts of Myo4-HA and She2 coprecipitated with Myo4-myc even when U3 RNA was not expressed, possibly due to assembly of locasome complexes on native mRNA. In agreement with this, when cell extracts were treated with RNaseA before immunoprecipitation, the amount of Myo4-HA that coprecipitated with Myo4-myc was reduced.
Figure 4.
Multiple Myo4 motors are bound to a single localization element. (A) Myo4-HA coprecipitates with Myo4-myc when bound to U3 RNA. MYO4-HA was expressed in wild-type cells (lanes 1 and 2) or cells expressing MYO4-13xMYC (lanes 3–6), and U3 RNA expression was induced (lanes 1, 2, 5, and 6) or not induced (lanes 3 and 4). RNaseA was added to the indicated cell extracts (lanes 2, 4, and 6). Myo4-myc was immunoprecipitated from each cell extract, and the precipitants were analyzed by a Western blot with anti-HA or anti-She2 antibodies as indicated. Input shows equal amount of initial cell extracts probed with anti-HA antibody. (B) Myo4-myc and Myo4-HA coprecipitation requires She2. MYO4-HA was expressed with MYO4-13xMYC in the presence of SHE2 (lanes 1 and 2) or in she2Δ cells (lane 3), and U3 RNA expression was induced (lanes 2 and 3) or not induced (lane 1). Myo4-myc was immunoprecipitated and analyzed as in A. (C) The 15S U3 RNP complex consists of multiple Myo4 motors. MYO4-HA was expressed with MYO4-TAP, and U3 RNA expression was induced. TAP-purified complexes were loaded on 10–50% sucrose gradients and the collected fractions were analyzed by Western blotting. Note that the Myo4p blot detects both Myo4-TAP and Myo4-HA.
To confirm that the coprecipitation of Myo4-myc and Myo4-HA was caused by formation of U3 RNP complexes, we repeated the immunoprecipitation in she2Δ cells, as She2 is required for the interaction of Myo4 with U3 RNA. Again, overexpression of U3 RNA clearly increased the amount of Myo4-HA that coprecipitated with Myo4-myc, but no increase was detected when She2 was absent (Fig. 4 B). These results suggest that the U3 RNP complex contains more than one copy of Myo4.
We next tested whether the 15S U3 RNP complex contained multiple copies of Myo4. We overexpressed U3 RNA to induce U3 RNP formation in cells expressing MYO4-TAP and MYO4-HA, and purified the U3 RNP complex via Myo4-TAP as previously described. We then tested whether Myo4-HA copurified and co-sedimented with the U3 RNP complex in sucrose gradients. As mentioned, because Myo4 is a monomer, Myo4-HA should not copurify with Myo4-TAP unless they are associated in the same complex. Western blot analysis of the fractions from the sucrose gradients showed that U3 RNA overexpression induced assembly of a 15S U3 RNP complex that contained Myo4, She3, and She2 (Fig. 4 C). Importantly, Myo4-HA co-peaked in the same fractions as the other locasome proteins, which suggests that Myo4-HA, along with Myo4-TAP, was associated with the 15S U3 RNP complex. In parallel experiments without U3 RNA expression, a small amount of Myo4-HA was found to co-migrate with TAP-purified Myo4 in endogenous RNP complexes (Fig. S4 A). When cell extracts were treated with RNaseA, such comigration was abolished, confirming that Myo4-TAP assembled together with Myo4-HA only in the presence of RNA cargo (Fig. S4 B). Together, these results suggest that the locasome that assembles onto U3 RNA contains multiple copies of Myo4, which suggests a potential mechanism to generate continuous transport of ASH1 mRNA.
She2 is assembled into a high-order complex
Our results showing that the U3 RNP contained multiple copies of Myo4 and only three different proteins, Myo4, She3 and She2, raised the question of how multiple motors were recruited to U3 RNA. We focused on She2 because previous structural analysis of She2 indicated that it formed a dimer (Niessing et al., 2004). Because She3 binds directly to She2 (Böhl et al., 2000; Long et al., 2000), a She2 dimer could potentially link more than one Myo4 to a localization element. We first tested the oligomeric state of She2 by cross-linking analysis. SHE2 tagged with 1/2TAP (tobacco etch virus [TEV] protease cleavage site + IgG binding site) was expressed in wild-type cells, and the purified She2 was treated with the zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC). In the absence of EDC, She2 was detected as a doublet, showing that wild-type and TAP-tagged She2 oligomerized (Fig. 5 A). Surprisingly, after EDC treatment, the majority of She2 was found to migrate as a ∼150 kD complex, a size of approximately five She2 molecules (28.3 kD × 5 = 142 kD; Fig. 5 A). To test whether the oligomerization of She2 was concentration-dependent, we treated She2 with EDC over a range of concentrations, from 25 nM to 1 µM. The results showed that She2 was consistently cross-linked into a complex of ∼150 kD (Fig. 5 B). Considering that the cellular concentration of She2 is ∼230 nM (Ghaemmaghami et al., 2003), our results suggest that She2 exists as a high-ordered complex in vivo.
Figure 5.
The oligomeric state of She2. (A) Cross-linking analysis of She2. SHE2-1/2TAP was expressed in wild-type cells, and purified She2 was treated with 40 mM EDC for 1 h at room temperature, separated by 4–15% SDS-PAGE, and probed with anti-She2 antibody. Note that She2 appears as a doublet in the absence of EDC due to oligomerization of wild-type and TAP-tagged She2. (B) She2 oligomerization is not concentration dependent. SHE2-1/2TAP was expressed from a high-copy plasmid in she2Δ cells, and cross-linking analysis was performed with purified She2 diluted to various concentrations. (C) She2 oligomerization does not require RNA binding. SHE2-1/2TAP was expressed in she2Δ, cells and U3 RNA expression was induced. Half of the cell extract was treated with 0.3 mg/ml RNaseA, and purified She2 from each cell extract was cross-linked. (D) Oligomerization is inhibited by mutations in the upper uncharged surface of She2. 1/2TAP-tagged SHE2 mutants (N36S, R63K, T47Y, and L130Y) were expressed in she2Δ cells, and the purified proteins were cross-linked and analyzed as described in A. (E) Analytical ultracentrifugation of She2. Purified wild-type She2 was diluted to 0.15 mg/ml (5.3 µM), 0.53 mg/ml (18.7 µM), 1.13 mg/ml (39.9 µM), and 1.90 mg/ml (67.1 µM), then subjected to analytical ultracentrifugation. A direct boundary modeling program from individual datasets using model-based numerical solutions to the Lamm equation was used to obtain data shown for the normalized continuous sedimentation coefficient, c(s), distribution plot. At 0.15 mg/ml, the weighted mean value of S obtained through integration of the c(s) curve was 5.95S.
We next addressed whether the oligomerization of She2 was dependent on RNA. She2 may be a dimer that assembles into an oligomer upon binding RNA. To test this model, we treated SHE2-1/2TAP cell extracts with RNaseA and then purified She2. Cross-linking of purified She2 from untreated and RNaseA-treated extracts showed no effects of RNaseA on the amount or size of the She2 oligomer (Fig. 5 C). The results indicated that She2 oligomers are stable even when not bound to RNA and suggested that the oligomeric state of She2 was not dependent on RNA binding.
To more accurately determine the size of She2 oligomers, we performed an extensive hydrodynamic analysis of purified She2. Velocity sedimentation in 5–20% sucrose gradients revealed the sedimentation coefficient of She2 to be 5.78 ± 0.00S (n = 2), and gel filtration analysis measured the Stoke’s radius of She2 as 5.35 ± 0.04 nm (n = 2; Fig. S5). Using these numbers, we estimated the molecular weight of She2 as 130 kD (Siegel and Monty, 1966), which suggests that She2 forms a tetramer. We also performed analytical ultracentrifugation to determine more accurately the sedimentation coefficient and size of She2. At 5 µM (0.15 mg/ml), a single species of She2 was detected with a sedimentation coefficient of 5.95S, which is close to our measurement using velocity sedimentation in sucrose gradients (Fig. 5 E). Using the x-ray coordinates of She2 in the HYDROPRO program indicated that a She2 tetramer should have a sedimentation coefficient of 5.6S, whereas a dimer should be 3.7S (García de la Torre et al., 2000; Niessing et al., 2004). These results confirm that purified She2 is most likely a tetramer. Interestingly, increasing the concentration of She2 in the analytical ultracentrifugation resulted in progressively larger sedimentation coefficients up to 8.3S, which is the estimated size of a She2 octamer.
We next investigated the molecular interactions that mediated She2 oligomerization. Several studies have identified mutations in She2 that inhibit binding to RNA and localization of ASH1 mRNA to the bud tip (Gonsalvez et al., 2003; Niessing et al., 2004). Based on a crystal structure of She2, these mutations reside in a positively charged surface or upper uncharged surface; both are functionally important and highly conserved regions. To determine whether these surfaces are required for the oligomerization of She2, we purified She2 that contained single mutations in residues in the charged surface (N36S and R63K) or uncharged surface (T47Y and L130Y). When the oligomeric state of the She2 mutants was examined by EDC cross-linking, mutations in the charged surface did not have any effect, whereas mutations in the uncharged surface strongly reduced oligomerization (Fig. 5 D). The mutations did not prevent dimerization of She2 but inhibited formation of the larger oligomeric structure. To confirm that the mutations in the uncharged surface affected oligomerization of She2, we purified She2 with the L130Y mutation and performed hydrodynamic analysis. Velocity sedimentation analysis in 5–20% sucrose gradients measured a sedimentation coefficient of She2 L130Y as 3.99S (wild-type She2 = 5.78S), and gel filtration analysis estimated the Stoke’s radius of She2 L130Y as 4.14 nm (wild-type She2 = 5.35 nm; Fig. S5). Together, these numbers estimate the molecular mass of She2 L130Y as 69 kD (wild-type She2 = 130 kD), which approximates the molecular mass of a She2 dimer. Thus, the conserved uncharged surface of She2 that was thought to mediate protein–protein interaction proved to be involved in self-oligomerization (Niessing et al., 2004). Together with previous findings, these results suggest that She2 multimerization could be an essential feature that is required for efficient RNA transport and may provide a platform to bind multiple Myo4 motors.
During the preparation of this manuscript, Müller et al. (2009) also concluded that She2 forms a tetramer based on gel filtration and small-angle x-ray scattering analyses. In addition, the authors similarly showed that She2 L130Y is a dimer, and, through reexamination of previously published crystallographic contacts (Niessing et al., 2004), suggest that the uncharged upper surface is involved in dimer–dimer interaction required for the formation of an elongated tetramer. Interestingly, She2 was found to form oligomers larger than tetramers in this study as well. Our results confirm the work of Müller et al. (2009) and suggest that She2 could form a dynamic oligomer in vivo with the tetramer as its base state.
Motor copy number on RNA acts as a criterion for bud localization
Our results indicated that the locasome that assembles onto the U3 localization element from ASH1 contains multiple copies of Myo4 and suggested a possible mechanism by which the nonprocessive Myo4 could efficiently transport ASH1 mRNA to the bud tip. Namely, multiple Myo4 motors bound to the same RNA could be sufficient to generate continuous transport of that RNA. We tested this hypothesis by assessing whether increasing the number of Myo4 motors bound to an RNA improved the localization of that RNA. Because She2 not only links Myo4 to RNA but may also recruit multiple motors to RNA, we engineered an RNA localization system that bypassed She2. Here, one construct expresses the RNA-binding domain from U1Ap fused to the C terminus of She3, and another construct contains zero to eight binding sites for U1Ap added to the 3′ UTR of ASH1 (Fig. 6 A). Both constructs were then expressed in ash1Δ she2Δ cells. Because She2 is absent, Myo4 can only associate with ASH1 mRNA through the interaction between She3-U1Ap and the specific number of U1A sites in ASH1 mRNA.
Figure 6.
Increasing the number of Myo4 motors bound to RNA improves bud localization. (A) Model system for modifying Myo4 motor copy number on ASH1 RNA. She3 is expressed with the RNA-binding domain from U1Ap fused to its C terminus, and ASH1 is expressed with 0, 1, 2, 4, 6, or 8 U1Ap-binding sites positioned between the stop codon and 3′ UTR. Both constructs are expressed in ash1Δ she2Δ cells to ensure that Myo4 binds to ASH1 mRNA only through the interaction between She3-U1Ap and the U1Ap-binding sites tagged to ASH1 RNA. (B) Increasing the number of Myo4 bound to ASH1 RNA enhances bud localization of Ash1. She3-U1Ap and ASH1-HA tagged with the indicated number of U1A aptamers were expressed in ash1Δshe2Δ yeast cells where ADE was under the control of HO promoter. She2 was also coexpressed with She3-U1Ap and ASH1-HA as a control (bottom). Cells were spotted onto selective media plates with or without adenine. (C) U1A tags do not affect Ash1 expression. Protein extracts were prepared from cells expressing ASH1-HA tagged with the indicated number of U1A aptamers. Equal amounts of protein extracts were analyzed by Western blotting with anti-HA antibody. (D) Bud transport of ASH1 mRNA is improved by increasing Myo4 binding sites on ASH1 RNA. She3-U1Ap was expressed with U1A-tagged ASH1-HA in ash1Δshe2Δ cells. wt indicates cells coexpressing She2 with She3-U1Ap and ASH1 as a control. The cells were synchronized and fixed after release from synchronization. ASH1 mRNA was detected by FISH, and the percentage of cells showing bud-localized ASH1 mRNA among ∼100 cells was determined in each cell sample (n = 3). Bud localization was further subdivided to localization confined to the distal tip of buds and that dispersed throughout the bud.
We first tested whether increasing the number of binding sites for Myo4 in ASH1 mRNA improved localization of Ash1 protein. We used a reporter strain in which expression of ADE2 is essential for growth and is under the control of the HO promoter. Because Ash1 represses expression from the HO promoter, the cells grow only if Ash1 localizes to the daughter cell, allowing expression of ADE2 in the mother cell. The results showed that cells lacking U1A sites in the 3′ UTR of ASH1 mRNA failed to grow on media without adenine (Fig. 6 B). Cells that expressed ASH1 mRNA with multiple U1A-binding sites grew more robustly in the absence of adenine, which indicates that Ash1 protein was more efficiently localized to the daughter cell. Western blot analysis confirmed that the U1A repeats in the 3′ UTR did not affect the levels of Ash1 protein in the cell (Fig. 6 C).
To determine whether the increased number of U1A binding sites also improved the localization of ASH1 mRNA, we examined the cells by FISH. As the number of U1A binding sites was increased from one to eight, the proportion of cells with bud-localized ASH1 mRNA gradually increased from 16 ± 2.0% to 49 ± 3.5% (compared with 11 ± 0.5% with 0×U1A, n = 3; Fig. 6 D). However, ASH1 mRNA rarely localized to the distal tip of buds in cells with zero or one U1A binding site. Because localization to the bud tip signifies complete transport of mRNA, we analyzed cells to distinguish between ASH1 mRNA confined to the bud tip and dispersed throughout the bud. Although bud tip localization was not significantly improved by a single U1A binding site (2.4 ± 1.6% with 1×U1A compared with 1.5 ± 2.7% with 0×U1A), increasing the number of U1A binding sites from two to eight increased bud tip localization from 6.4 ± 0.5% to 26 ± 2.5% (Fig. 6 D). Thus, cells expressing ASH1 mRNA with more than one U1A repeat were significantly more likely to have ASH1 mRNA localized to the distal tip of buds. Collectively, these results indicated that increasing the number of Myo4 motors attached to ASH1 mRNA increased the efficiency of localization of ASH1 mRNA and protein, and suggested that yeast generate continuous transport of RNA from a nonprocessive motor by recruiting multiple motors to RNA.
Discussion
The RNP complex
Here we show that Myo4, She3, and She2 are the sole components of an RNP complex that transports RNA to the bud tip. Previous evidence suggested that these proteins were the core components of the locasome (Paquin and Chartrand, 2008), but this paper is the first to define clearly the composition of the RNP complex and describe how these components assemble into an active RNA transport unit. U3 RNA is sufficient to generate bud transport (Fig. S1), and purification of U3 RNA isolated an RNP complex that contained Myo4, She3, and She2 as the major protein components (Fig. 1). Moreover, purification of Myo4-TAP from cells overexpressing U3 RNA copurified only She3 and She2 (Fig. 3 A). Thus, we conclude that Myo4, She3, and She2 are the bona fide components of the U3 RNP complex and that She2 is sufficient to assemble Myo4–She3 into an active RNA transport unit. Other proteins involved in ASH1 mRNA localization could bind transiently for a specific purpose, such as bud-tip anchoring, or be part of a separate complex that associates with a different sequence within ASH1 mRNA.
The U3 RNP complex also copurified with eIF4G1/G2 and PABP (Fig. 1 E). These proteins also copurified with ADH2 (77 NT) RNA and failed to co-migrate with the U3 RNP complex (Figs. 1 C and 2 A), which suggests that they were weakly associated with U3 RNA and are not components of the locasome. eIF4G and PABP are translation initiation factors that directly bind to each other and are both able to bind mRNA (Jackson et al., 2010). Thus, these proteins may have interacted with U3 RNA because there was likely more U3 RNA than She2, She3, or Myo4 in the cells due to overexpression of U3 RNA.
By modifying the tagged RNA system that has been used to visualize RNA in vivo, we purified and identified the proteins associated with an RNA sequence that performs a specific function: mRNA localization. mRNAs are known to be regulated by processes besides localization, including translational control and degradation. Although genetic data has identified the sequences within mRNAs that specify these processes, finding the proteins involved in these different regulatory steps has been more challenging. Our results demonstrate how a tagged RNA system that has been successfully implemented in a wide range of cell types to visualize RNA can be adapted to purify functional RNP complexes, which suggests a way forward to identify proteins that regulate different steps in the life of an mRNA.
Multiple Myo4 motors on a localization element
Because monomeric Myo4 was found to be nonprocessive in its cargo-free state (Reck-Peterson et al., 2001; Dunn et al., 2007), it has been unclear how Myo4 would generate the continuous movement of RNA in vivo. We show that the stable 7S Myo4–She3 complex assembles into a 15S RNP complex containing She2 and a single localization element (Fig. 3). We further demonstrate that this 15S complex contains multiple Myo4 motors and that the assembly of multiple motors is dependent on the interaction with U3 RNA (Fig. 4), providing the first direct evidence that multiple Myo4 motors associate with a single localization element in vivo. Interestingly, single-molecule analysis of Myo4-GFP revealed that ensembles of at least three motors were capable of moving processively (Dunn et al., 2007). Together, these findings suggest that the sustained movement of ASH1 mRNA in vivo is driven by the coordinated activity of multiple Myo4 motors bound to a localization element.
Although we measured the size of the U3 RNP complex as 15S, the complex may be larger. We overexpressed U3 RNA to drive formation of U3 RNP, and cells likely contained an excess of U3 RNA over Myo4, She3, and She2, spreading the locasome proteins across a large amount of U3 RNA. The high ratio of U3 RNA to locasome proteins may have limited the number of locasome proteins that could associate with U3 RNA. Nonetheless, U3 RNA was transported to the distal tip of buds despite its overexpression, which suggests that the 15S RNP complex contained a sufficient number of motors to generate effective transport.
Motors: The more the merrier?
To test whether multiple Myo4 motors attached to an mRNA improved the transport of that mRNA in vivo, we engineered an in vivo RNA transport system that varied the number of motors bound to ASH1 mRNA. In this system, She3 was fused with U1Ap, and a 22-nucleotide RNA aptamer that binds a single U1Ap was used to tag ASH1 RNA (Nagai, 1996). Because U1Ap binds to its target RNA in a 1:1 ratio (Coller and Wickens, 2007), these constructs ensured that the increase in aptamers would correspond with an increase in bound Myo4–She3–U1Ap. The number of aptamers, however, does not necessarily equal the number of motors bound to RNA, as aptamers could be left vacant. In addition, the geometry of the motor complex bound to an aptamer may differ from the native system and could partly explain the reduced efficiency of ASH1 mRNA localization using aptamers to link Myo4 to ASH1 compared with the native system (Fig. 6 D). Nonetheless, we found that increasing the number of motors on RNA improved bud localization of both RNA and protein (Fig. 6). In addition, ASH1 RNA with a single binding site for Myo4–She3 rarely reached the distal tip of the bud (Fig. 6 D), which suggests that ASH1 mRNA must recruit more than one Myo4 motor to generate effective transport. Thus, we conclude that multiple nonprocessive Myo4 motors associated with a single localization element are sufficient to sustain transport of RNA into the bud. We hypothesize that multiple Myo4 motors act in a coordinated manner to ensure that at least one motor is attached to filaments, preventing ASH1 mRNA from dissociating from filaments during transport to the bud tip.
In contrast to our results, previous studies suggested that a single Myo4 motor would be sufficient for RNA bud transport. In one study, bud localization of RNA was described when Myo4 was tethered to a reporter RNA in the absence of She2 (Kruse et al., 2002). However, in this study, the reporter RNA was tagged with MS2 aptamers, and Myo4 was linked to the RNA via She3-MS2p. MS2p exists as a stable dimer but forms multimers even when using mutant forms that prevent aggregation (Johansson et al., 1997; Keryer-Bibens et al., 2008). Thus, it is unclear how many Myo4 motors were associated with MS2-tagged RNA to generate localization to the bud tip. In a more recent paper, a single Myo4 motor fused to a She2 dimer was shown to be sufficient to generate bud transport of RNA in she2Δ cells (Bookwalter et al., 2009). From these observations, the authors suggest that a single motor head per She2 dimer is sufficient for RNA transport. However, based on our results and those from Müller et al. (2009) demonstrating that She2 forms a high-order complex beyond a dimer, a She2 dimer fused to Myo4 would likely cluster Myo4 heads when She2 oligomerizes.
How do multiple, nonprocessive Myo4 motors bound to a single RNA generate sustained transport in vivo? Similar to Myo4, the monomeric Unc104/KIF1A and myosin VI motors contain weak coiled-coil domains that are insufficient to form stable dimers (Okada et al., 1995; Lister et al., 2004; Knight et al., 2005). It has been suggested that binding to cargo clusters these motors and facilitates dimerization, although it is not fully established if such dimerization is required for processivity (Klopfenstein et al., 2002; Tomishige et al., 2002; Park et al., 2006; Phichith et al., 2009; Yu et al., 2009). It is unlikely that a similar mechanism generates Myo4 processivity, as Myo4 stably associates with She3, and this interaction requires the coiled-coil domain in Myo4 (Böhl et al., 2000; Takizawa and Vale, 2000; Hodges et al., 2008). Thus, even if binding to RNA clusters Myo4, the coiled-coil domain in Myo4 may not be accessible to mediate dimerization. Interestingly, a recent report showed that multiple monomeric myosin VI motors cooperate to transport cargo in a similar manner to dimeric motors (Sivaramakrishnan and Spudich, 2009).
Several other studies have also proposed the idea of enhanced cargo transport by increases in motor numbers. In vitro experiments show that run lengths along filaments increase when cargo is bound to more motors (Block et al., 1990; Vershinin et al., 2007; Beeg et al., 2008). The recruitment of additional dynein motors onto RNA has also been suggested to regulate cargo motility in Drosophila embryos in vivo (Bullock et al., 2006). However, this is the first study to manipulate the number of motors bound to an endogenous cargo with full knowledge of the components necessary for transport, and thus directly investigate the role of motor copy number on transport in vivo.
The role of She2 oligomerization in mRNA transport
We found that wild-type She2 forms a tetramer (Fig. 5), and during the preparation of this manuscript, Müller et al. (2009) also come to a nearly identical conclusion. Combined with our results showing that the core components of the locasome are She2 and Myo4–She3 and that multiple copies of Myo4 assemble onto a single localization element, we propose that a She2 tetramer links multiple copies of Myo4–She3 to a localization element through a direct interaction between She2 and She3. In an attempt to demonstrate that tetrameric She2 was recruiting multiple motors, we examined whether the She2 L130Y mutant that fails to form tetramers recruited fewer motors to U3 RNA. However, even with the overexpression of U3 RNA, these mutants rarely formed stable complexes with Myo4–She3 (unpublished data). We speculate this could be caused by reduced affinity between She2 L130Y and U3 RNA (Niessing et al., 2004; Müller et al., 2009), as Myo4–She3 has a stronger association with She2 that is bound to RNA (Fig. 2 C; Böhl et al., 2000).
A She2 tetramer could also indirectly recruit multiple motors to an mRNA by binding to two different mRNAs. Recent work observed that two different mRNAs were co-transported in a common RNP particle in yeast (Lange et al., 2008). A She2 tetramer could potentially function as a cross-linking agent that clusters mRNAs into a single large complex. The presence of multiple mRNAs, each containing a set of localization elements, in one complex would present multiple binding sites for Myo4. However, in our work, the U3 RNP complex was assembled by the overexpression of a single localization element, U3 RNA. Therefore, if a She2 tetramer does bind multiple RNAs, it is probable that in our experiments, U3 RNA occupied all of the potential RNA-binding sites in She2. Thus, multiple Myo4 motors most likely assembled onto U3 RNA through a She2 tetramer that offered multiple, direct binding sites for Myo4–She3. Oligomerization of motor-associated proteins may also cluster other motors. Bicaudal D that binds the dynein–dynactin complex was suggested to form a dimer, and more recently, the RNA-binding protein Staufen, which forms a complex with microtubule motors, was shown to multimerize in vivo (Stuurman et al., 1999; Oh et al., 2000; Hoogenraad et al., 2001; Trucco et al., 2009; Martel et al., 2010).
In summary, we propose that the RNA-binding protein She2p oligomerizes into a tetramer, providing multiple binding sites for Myo4p–She3p, and that the number of motors bound to RNA cargo appears to be a critical factor that determines successful transport to the bud. How multiple Myo4 motors coordinate their activity to generate continuous movement in vivo remains to be elucidated.
Materials and methods
Strains and plasmids
All strains used in this study were derived from W303 and are shown in Table I. MYO4-13xMYC, MYO4-TAP, MYO2-1/2TAP (IgG-binding domain and TEV site), and MYO4-1/2TAP were made by PCR-mediated gene modification by the method of Longtine et al. (1998). pHO-ADE-HO 3′ UTR is described in Irie et al. (2002). STE2-3xHA-U3 was also made using PCR-based modification by inserting U3 zipcode into the AscI site in pFA6a-3HA-His3MX6 (Longtine et al., 1998). Plasmid U1A-GFP-TAP-NLS was generated as in Takizawa and Vale (2000), with a TAP sequence replacing GST. This construct was subcloned into vector pRS304 (PT87), digested with HindIII for integration into trp1 locus, and used to transform ash1Δ (YPT86). Plasmid pGAL-4×U1A-U3-tADH1 (PT185) was made by inserting four repeats of a U1A binding sequence from U1A pre-mRNA 3′ UTR (Allain et al., 1996) into the EcoRI site of pRS423 (2μ) and sequentially inserting a GAL1 promoter into XhoI–EcoRI sites, U3 into BamHI–SacI sites, and ADH1 terminator into a SacI site. To construct plasmid pGAL-4×U1A-ADH2 (77NT)-tADH1 (PT229), U3 in PT185 was replaced by a 77-nucleotide fragment from ADH2 starting seven nucleotides upstream of the stop codon by inserting ADH2 (77NT)-SpeI-tADH1-SacI in place of U3-SacI-tADH1-SacI. Plasmid pGAL-1×U1A-U3-tADH1 (PT204) was derived from PT185 in which 4×U1A was replaced by a single U1A binding sequence with additional restriction sites so that a 36-nucleotide spacer was located in between 1xU1A and U3. Plasmid pGAL-U3-tADH1 (PT244) used for RNA overexpression was derived from PT229 in which EcoRI–4×U1A-ADH2 (77NT)–SpeI was replaced with MfeI–U3–SpeI. The construction of plasmid MYO4-HA has been described previously (Dunn et al., 2007). Cross-linking experiments to examine the effect of RNA binding on She2 oligomerization, and the effect with various She2 mutants, were performed with She2 expressed on CEN/ARS vectors. All other analyses were performed with She2 expressed from high-copy 2μ plasmids. Plasmid pGPD-SHE2-1/2TAP (PT228) was made by inserting XhoI–SHE2–SacI into pRS424 (2μ) and ligating KpnI–pGPD–XhoI upstream of SHE2 start codon (PT94), and inserting XbaI–1/2TAP-SpeI into an NheI site engineered immediately upstream of SHE2 stop codon. To make plasmid SHE2-1/2TAP (PT240), a PCR clone of SHE2 including 1 kb each of the 5′ UTR and 3′ UTR sequences was subcloned into pRS314 (CEN/ARS, PT52), and XbaI–1/2TAP–XbaI was inserted into an NheI site engineered immediately upstream of a SHE2 stop codon. Mutations of SHE2 were made by PCR-mediated mutagenesis. The plasmids SHE2 (N36S)-1/2TAP (AAC mutated to AGC, PT243), SHE2 (T47Y)-1/2TAP (ACG mutated to TAT, PT245), SHE2 (R63K)-1/2TAP (AGA mutated to AAA, PT246), and SHE2 (L130Y)-1/2TAP (TTG mutated to TAC, PT239) were then made by ligating the mutated SHE2, which included 1 kb of 5′ UTR, with 1/2TAP, and inserting XhoI–5′ UTR-SHE2-1/2TAP–XbaI in place of wild-type XhoI–5′ UTR-SHE2–NheI in PT52. For high-level expression of SHE2 L130Y, plasmid pGPD-SHE2 (L130Y)-1/2TAP (PT249) was made from PT94 in which wild-type SHE2 was replaced with SHE2 (L130Y)-1/2TAP. To make plasmid SHE3-U1A (PT248), a PCR clone of SHE3 including 1 kb each of 5′ UTR and 3′ UTR was subcloned into pRS314 (CEN/ARS), and a U1Ap sequence was inserted into an NheI site engineered just upstream of the stop codon. To express ASH1-3×HA with 0 (PT290), 1 (PT291), 2 (PT292), 4 (PT293), 6 (PT294), and 8 (PT295) repeats of the 22-nucleotide U1Ap binding sequence (Nagai, 1996), a PCR clone of ASH1 including 937 bp of 5′ UTR and 563 bp of 3′ UTR was subcloned into pRS313, and an AscI–EcoRI–U1A-GCGC–MfeI–AscI aptamer was generated by PCR. The U1A repeats were made by digesting the aptamer with either EcoRI or MfeI, ligating the two digested products, and performing PCR with 5′–AscI–EcoRI–U1A and 3′–AscI–MfeI–GCGC-U1A primers. The repeats of interest were then digested with AscI and subcloned into pFA6a-3HA-His3MX6 (Longtine et al., 1998) so that the U1A repeats were immediately 3′ of the stop codon. These subclones were used as templates for PCR to obtain BglII–3×HA-(U1A-GCGC)n–MfeI–AscI–BglII and inserted into a BamHI site engineered immediately upstream of the stop codon of ASH1.
Table I.
Strains used in this study
| Strain | Genotype |
| W303 | MATa ura3 trp1 leu2 his3 ade2 can1 GAL |
| YPT4 | she3Δ |
| YPT6 | she2Δ |
| YPT14 | MYO4-13xMYC |
| YPT34 | MYO4-13xMYC she2Δ |
| YPT66 | MYO4-TAP |
| YPT72 | ash1Δ she2Δ HOp-ADE2-HO 3′ UTR |
| YPT86 | ash1Δ GPDp-U1A-GFP-TAP-NLS TRP1 |
| YPT117 | MYO2-1/2TAP |
| YPT118 | MYO4-1/2TAP |
| YPT182 | STE2-3xHA-U3 HIS3 |
Reagents and antibodies
Anti-HA (HA.11) and anti-myc (9E10) antibodies were obtained from Covance. Anti-GFP (JL8) was purchased from Takara Bio Inc. Anti-digoxigenin antibody was obtained from Jackson ImmunoResearch Laboratories, Inc. Anti–mouse Alexa Fluor 488 and a ProLong Antifade kit were obtained from Invitrogen. The preparation of antibodies against Myo4, She3, and She2 has been described in Dunn et al. (2007). IgG Sepharose 6 Fast Flow, protein G Sepharose 4 Fast Flow, Superdex 200 10/300 GL, and high- and low-molecular weight gel filtration calibration kits were obtained from GE Healthcare. Detection reagents for Western blots, EDC, and N-hydroxysuccinimide (NHS) were obtained from Thermo Fisher Scientific. Calmodulin affinity resin was obtained from Agilent Technologies. Anti-sense digoxigenin (DIG)-labeled probes were made using MAXIscript kit from Applied Biosystems.
RNP complex purification
Cells containing pGAL-1x/4×U1A-U3 or pGAL-4×U1A-ADH2 (77 NT) along with U1Ap-GFP-TAP, or those containing pGAL-U3 and MYO4-TAP (or -1/2TAP) were grown overnight in synthetic media containing 2% raffinose. Cultures were adjusted to ∼OD600 0.2 in rich media containing 2% raffinose and incubated at 30°C. At ∼OD600 1.0, galactose was added to 1% and further incubated 1.5 h at 30°C. Cells expressing U1Ap-GFP-TAP only or MYO4-1/2TAP or MYO2-1/2TAP were incubated in rich media containing 2% dextrose at 30°C until OD600 reached ∼1.2–1.5. The cells were harvested by centrifugation, washed with HCB wash buffer (25 mM Hepes-KOH, pH 7.5, 0.15 M KCl, and 2 mM MgCl2), resuspended in 1/2 pellet volume of HCB extract buffer (25 mM Hepes-KOH, pH 7.5, 0.15 M KCl, 2 mM MgCl2, 0.1% NP-40, 1 mM DTT, 5 mM ATP, 0.2 mg/ml heparin, 20 mM vanadyl ribonucleoside complexes, 0.4 mM 4-[2-aminoethyl] benzenesulfonyl fluoride hydrochloride [AEBSF], and 2 µg/ml of aprotinin, leupeptin, and pepstatin), and frozen in liquid nitrogen. The frozen cell extracts were lysed using a mortar and pestle with liquid nitrogen and thawed with added HCB extract buffer (1 ml per culture).
The thawed lysates were centrifuged for 5 min at 4,000 relative centrifugal force (rcf), and supernatants were centrifuged again for 20 min at 16,100 rcf. Supernatants were then protein quantified and incubated with IgG sepharose for 2 h at 4°C. The beads were washed with HCB (25 mM Hepes-KOH, pH 7.5, 0.15 M KCl, 2 mM MgCl2, 0.1% NP-40, and 1 mM DTT) followed by TCB (25 mM Hepes-KOH, pH 7.5, 0.15 M KCl, 0.5 mM EDTA, 0.1% NP-40, and 1 mM DTT), then incubated with AcTEV protease in TCB at 16°C for 2 h. After collecting the TEV-eluted fraction, IgG beads were rinsed with HCB, and this solution was combined with the TEV eluate. The TEV eluate was used for further experiments or adjusted so that the final composition was identical to CCB (25 mM Hepes-KOH, pH 7.5, 0.15 M KCl, 2 mM MgCl2, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, and 1 mM DTT) and incubated with calmodulin resins for 1.5 h at 4°C. The beads were then washed with CCB and the purified complex was either eluted with CEB (25 mM Hepes, pH 7.5, 0.15 M KCl, 2 mM MgCl2, 1 mM imidazole, 10 mM EGTA, 0.1% NP-40, and 1 mM DTT), elution buffer (50 mM Tris-Cl, pH 8.0, 0.15 M NaCl, 12.5 mM EDTA, and 0.1% SDS), or 1× SDS-PAGE sample buffer.
For protein identification, protein bands were cut out from a Coomassie blue–stained SDS-polyacrylamide gel and subjected to LC-MS/MS at the W.M. Keck Foundation Biotechnology Laboratory at Yale University.
Sucrose gradients
RNP complexes eluted from calmodulin resins with CEB or from IgG resins with TEV were added to the top of 10–50% sucrose gradients made with the buffer compositions in the RNP complexes. In some cases, TEV-eluted RNP complexes were incubated with RNaseA (to 0.3 mg/ml) for 1 h before ultracentrifugation to detach RNA cargo from the complex. Purified wild-type She2 and L130Y were loaded on 5–20% sucrose gradients made with 20 mM Hepes-KOH, pH 7.5, 0.1 M NaCl, and protease inhibitors. Protein standards were also loaded on parallel gradients to estimate the S values. The gradients were spun for ∼16 h at 40,000 rpm in a SW50.1 rotor (Beckman Coulter). Fractions were then collected with a peristaltic pump, and TCA precipitated or was directly used for further experiments.
Immunoprecipitation
Yeast lysates were prepared as previously described. Equal amounts of total protein in cell extracts were used in parallel immunoprecipitation experiments. RNaseA (to 0.3 mg/ml) was added to select extracts before further incubation. Cell extracts were precleared by incubating with protein G beads for 1 h at 4°C with gentle rotation. Supernatants were incubated with mouse anti-myc antibody for 1.5 h at 4°C and further incubated with added protein G beads for 1 h at 4°C. The beads were then washed extensively with HCB and incubated with elution buffer.
She2 purification
For analytical ultracentrifugation and concentration-dependent cross-linking analysis of purified wild-type She2, yeast cells containing wild-type SHE2-1/2TAP on a high-copy plasmid (2µ) were grown overnight in synthetic media containing 2% dextrose. The overnight cultures were diluted in rich media containing 2% dextrose to ∼OD600 0.2 and harvested by centrifugation at ∼OD600 2.0. The cell pellet was washed with HB (25 mM Hepes-KOH, pH 7.5, 0.15 M potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 1 mM DTT) without DTT, resuspended in 1/2 pellet volume of HB with protease inhibitors, and frozen with liquid nitrogen. The cell extract was lysed using a mortar and pestle with liquid nitrogen, thawed, and centrifuged for 5 min at 4,000 rcf, 10 min at 48,384 g, and 1 h at 50,000 rpm in a Ti 60 rotor (Beckman Coulter). The clear lysate was then passed over IgG Sepharose columns and washed with 60× column volume of HB and 25× column volume of TB (25 mM Hepes-KOH, pH 7.5, 0.15 M potassium acetate, 1 mM EGTA, 0.5 mM EDTA, and 1 mM DTT) at 4°C. The beads were then mixed with TB containing AcTEV protease and incubated overnight at 4°C with gentle shaking. After the TEV eluate was collected, the beads were rinsed with TB and the eluate was either used for cross-linking or concentrated with a centrifugal filter (Amicon Ultra; Millipore) for analytical ultracentrifugation.
For cross-linking analyses of the She2 mutants, SHE2-1/2TAP was expressed on low-copy plasmids (CEN/ARS), and cell lysates were prepared as previously described but without high speed centrifugation. The mutated She2 were then 1/2TAP purified and the TEV eluates were used for cross-linking experiments.
For gel filtration and sucrose gradient analysis, wild-type SHE2-1/2TAP and SHE2 (L130Y)-1/2TAP were expressed on high-copy plasmids and cultured as previously described. The cell pellets were then resuspended with HNB (20 mM Hepes-KOH, pH 7.5, 0.1 M NaCl, 0.5 mM EDTA, 0.1% NP-40, and 2 mM DTT) with protease inhibitors, and cell lysates were prepared. The clear lysates were incubated with IgG sepharose beads at 4°C for 2 h, washed with HNB, and incubated with HNB containing AcTEV protease at 16°C for 2 h. The TEV eluates were quantified and the protein purity was confirmed by silver staining and Coomassie staining.
EDC/NHS cross-linking
EDC and NHS were dissolved in water to 1 M before usage and added to purified She2 diluted with TB (without DTT) to a final concentration of 40 mM. Cross-linking was performed at room temperature for 1 h and quenched by adding SDS-PAGE sample buffer.
Analytical ultracentrifugation
Wild-type She2 was diluted with TB to 0.15, 0.53, 1.13, and 1.90 mg/ml, and sedimentation velocity analysis was conducted at 20°C and 40,000 rpm using interference optics with an analytical ultracentrifuge (XL-I; Beckman-Coulter) at the Analytical Ultracentrifugation Facility at University of Connecticut. Double sector synthetic boundary cells equipped with sapphire windows were used to match the sample and reference menisci. The rotor was equilibrated under a vacuum at 20°C, and after a period of ∼1 h at 20°C, the rotor was accelerated to 40,000 rpm. Interference scans were acquired at 60 s intervals for ∼5.5 h. Data were processed using the program Sedfit, version 11.71 (National Institutes of Health).
Growth assays
Yeast cells expressing SHE3-U1A, U1A-tagged ASH1-HA, and pHO-ADE (ash1Δ she2Δ) were grown overnight at 30°C in synthetic media. The overnight cultures were pelleted, washed with water, and diluted to ∼OD600 0.2 with water. Three tenfold serial dilutions were made from this culture, and 5 µl of each dilution was spotted on synthetic media plates with or without adenine and incubated at 30°C. Plates with adenine were incubated for 2 d and those without adenine for ∼4 d.
Fluorescence microscopy
Exponentially growing cells were fixed by adding formaldehyde (4% final concentration) to the culture medium and incubating for 1 h at room temperature. The fixed cells were washed, spheroplasted using zymolase 100T (Seikagaku Corporation), and adsorbed onto poly-l-lysine–coated glass coverslips. For FISH, samples were sequentially incubated in hybridization mix (50% formamide, 5× SSC, 1 mg/ml yeast tRNA, 100 µg/ml heparin, 1× Denhardt’s solution, 0.1% Tween 20, 0.1% Triton X-100, and 5 mM EDTA) for 45 min at room temperature; kept in hybridization mix containing anti-sense, DIG-labeled STE2, or ASH1 RNA probes overnight at 37°C; and then were washed in 0.1× SSC at 37°C. For FISH and immunofluorescence, the prepared samples were briefly incubated in blocking buffer (50 mM Tris-Cl, pH 7.5, 0.15 M NaCl, and 5% fetal bovine serum), incubated with mouse anti-DIG antibody or anti-HA antibody in blocking buffer for 30 min at 37°C, and washed at room temperature with washing buffer (50 mM Tris-Cl, pH 7.5, 0.15 M NaCl, and 0.1% Tween 20). Samples were then incubated with goat anti–mouse Alexa Fluor 488 for 45 min at room temperature, washed again with washing buffer, and mounted on glass slides. All samples were inspected with a fluorescence microscope (TE2000; Nikon) equipped with a 100×/NA 1.4 lens (Nikon) and acquired using a charge-coupled device camera (ORCA ER; Hamamatsu Photonics) controlled by IPLab software (Scanalytics, Inc.). For imaging of cells coexpressing tagged U3 RNA and U1Ap-GFP-TAP, GFP fluorescence was observed in cells directly from galactose-induced cultures.
Online supplemental material
Fig. S1 shows that tagging the U3 localization element to STE2 is sufficient to localize STE2 mRNA protein to the distal tip of buds. Fig. S2 shows that RNase treatment eliminates the Myo4 detected in the heavier fractions in Fig. 3 B. Fig. S3 shows silver staining and Western analyses of U3 RNP complex purified with a single U1A aptamer tagged to U3 RNA. Fig. S4 shows that RNase treatment eliminates the small amount of Myo4-HA that co-migrates with TAP-purified Myo4 in the absence of U3 RNA overexpression. Fig. S5 shows gel filtration and sedimentation analyses of wild-type and L130Y mutant She2. Video 1 shows fluorescently labeled actin filaments moving via purified U3 RNP adsorbed to motility chambers with anti-GFP antibodies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912011/DC1.
Acknowledgments
This work was supported by National Institutes of Health grant GM073734.
Footnotes
Abbreviations used in this paper:
- DIG
- digoxigenin
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- NHS
- N-hydroxysuccinimide
- PABP
- poly(A)-binding protein
- rcf
- relative centrifugal force
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- UTR
- untranslated region
References
- Allain F.H., Gubser C.C., Howe P.W., Nagai K., Neuhaus D., Varani G. 1996. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 380:646–650 10.1038/380646a0 [DOI] [PubMed] [Google Scholar]
- Beeg J., Klumpp S., Dimova R., Gracià R.S., Unger E., Lipowsky R. 2008. Transport of beads by several kinesin motors. Biophys. J. 94:532–541 10.1529/biophysj.106.097881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437–445 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Block S.M., Goldstein L.S., Schnapp B.J. 1990. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 348:348–352 10.1038/348348a0 [DOI] [PubMed] [Google Scholar]
- Böhl F., Kruse C., Frank A., Ferring D., Jansen R.P. 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19:5514–5524 10.1093/emboj/19.20.5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookwalter C.S., Lord M., Trybus K.M. 2009. Essential features of the class V myosin from budding yeast for ASH1 mRNA transport. Mol. Biol. Cell. 20:3414–3421 10.1091/mbc.E08-08-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S.L. 2007. Translocation of mRNAs by molecular motors: think complex? Semin. Cell Dev. Biol. 18:194–201 10.1016/j.semcdb.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Nicol A., Gross S.P., Zicha D. 2006. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 16:1447–1452 10.1016/j.cub.2006.05.055 [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X.H., Singer R.H., Long R.M. 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9:333–336 10.1016/S0960-9822(99)80144-4 [DOI] [PubMed] [Google Scholar]
- Coller J., Wickens M. 2007. Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods Enzymol. 429:299–321 10.1016/S0076-6879(07)29014-7 [DOI] [PubMed] [Google Scholar]
- Du T.G., Schmid M., Jansen R.P. 2007. Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18:171–177 10.1016/j.semcdb.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Dunn B.D., Sakamoto T., Hong M.S., Sellers J.R., Takizawa P.A. 2007. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J. Cell Biol. 178:1193–1206 10.1083/jcb.200707080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García De La Torre J., Huertas M.L., Carrasco B. 2000. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78:719–730 10.1016/S0006-3495(00)76630-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A., Vale R.D. 2009. Walking the walk: how kinesin and dynein coordinate their steps. Curr. Opin. Cell Biol. 21:59–67 10.1016/j.ceb.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gonsalvez G.B., Lehmann K.A., Ho D.K., Stanitsa E.S., Williamson J.R., Long R.M. 2003. RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA. 9:1383–1399 10.1261/rna.5120803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez G.B., Urbinati C.R., Long R.M. 2005. RNA localization in yeast: moving towards a mechanism. Biol. Cell. 97:75–86 10.1042/BC20040066 [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Buonomo S.B., Nasmyth K., von Ahsen U. 1999. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9:337–340 10.1016/S0960-9822(99)80145-6 [DOI] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R.H. 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18:1452–1465 10.1101/gad.1189004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A.R., Krementsova E.B., Trybus K.M. 2008. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J. Biol. Chem. 283:6906–6914 10.1074/jbc.M708865200 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Akhmanova A., Howell S.A., Dortland B.R., De Zeeuw C.I., Willemsen R., Visser P., Grosveld F., Galjart N. 2001. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 20:4041–4054 10.1093/emboj/20.15.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K., Tadauchi T., Takizawa P.A., Vale R.D., Matsumoto K., Herskowitz I. 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21:1158–1167 10.1093/emboj/21.5.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Derisi J.L. 2007. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 13:625–642 10.1261/rna.262607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., McDermott K., Sorber K., Shepard K.A., Vale R.D., Takizawa P.A., DeRisi J.L. 2005. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 102:18005–18010 10.1073/pnas.0509229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P., Dowzer C., Michaelis C., Galova M., Nasmyth K. 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 84:687–697 10.1016/S0092-8674(00)81047-8 [DOI] [PubMed] [Google Scholar]
- Johansson H.E., Liljas L., Uhlenbeck O.C. 1997. RNA recognition by the MS2 phage coat protein. Seminars in Virology. 8:176–185 10.1006/smvy.1997.0120 [DOI] [Google Scholar]
- Keryer-Bibens C., Barreau C., Osborne H.B. 2008. Tethering of proteins to RNAs by bacteriophage proteins. Biol. Cell. 100:125–138 10.1042/BC20070067 [DOI] [PubMed] [Google Scholar]
- King S.J., Schroer T.A. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2:20–24 10.1038/71338 [DOI] [PubMed] [Google Scholar]
- Klopfenstein D.R., Tomishige M., Stuurman N., Vale R.D. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358 10.1016/S0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P.J., Thirumurugan K., Xu Y., Wang F., Kalverda A.P., Stafford W.F., III, Sellers J.R., Peckham M. 2005. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280:34702–34708 10.1074/jbc.M504887200 [DOI] [PubMed] [Google Scholar]
- Kruse C., Jaedicke A., Beaudouin J., Böhl F., Ferring D., Guttler T., Ellenberg J., Jansen R.P. 2002. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 159:971–982 10.1083/jcb.200207101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Katayama Y., Schmid M., Burkacky O., Bräuchle C., Lamb D.C., Jansen R.P. 2008. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic. 9:1256–1267 10.1111/j.1600-0854.2008.00763.x [DOI] [PubMed] [Google Scholar]
- Lister I., Schmitz S., Walker M., Trinick J., Buss F., Veigel C., Kendrick-Jones J. 2004. A monomeric myosin VI with a large working stroke. EMBO J. 23:1729–1738 10.1038/sj.emboj.7600180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Singer R.H., Meng X., Gonzalez I., Nasmyth K., Jansen R.P. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 277:383–387 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- Long R.M., Gu W., Lorimer E., Singer R.H., Chartrand P. 2000. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19:6592–6601 10.1093/emboj/19.23.6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Gu W., Meng X., Gonsalvez G., Singer R.H., Chartrand P. 2001. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153:307–318 10.1083/jcb.153.2.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Martel C., Dugré-Brisson S., Boulay K., Breton B., Lapointe G., Armando S., Trépanier V., Duchaîne T., Bouvier M., Desgroseillers L. 2010. Multimerization of Staufen1 in live cells. RNA. 16:585–597 10.1261/rna.1664210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K.C., Ephrussi A. 2009. mRNA localization: gene expression in the spatial dimension. Cell. 136:719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Heuck A., Niessing D. 2007. Directional mRNA transport in eukaryotes: lessons from yeast. Cell. Mol. Life Sci. 64:171–180 10.1007/s00018-006-6286-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Richter K., Heuck A., Kremmer E., Buchner J., Jansen R.P., Niessing D. 2009. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 15:2002–2012 10.1261/rna.1753309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münchow S., Sauter C., Jansen R.P. 1999. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112:1511–1518 [DOI] [PubMed] [Google Scholar]
- Nagai K. 1996. RNA-protein complexes. Curr. Opin. Struct. Biol. 6:53–61 10.1016/S0959-440X(96)80095-9 [DOI] [PubMed] [Google Scholar]
- Niessing D., Hüttelmaier S., Zenklusen D., Singer R.H., Burley S.K. 2004. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 119:491–502 10.1016/j.cell.2004.10.018 [DOI] [PubMed] [Google Scholar]
- Oh J., Baksa K., Steward R. 2000. Functional domains of the Drosophila bicaudal-D protein. Genetics. 154:713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 81:769–780 10.1016/0092-8674(95)90538-3 [DOI] [PubMed] [Google Scholar]
- Olivier C., Poirier G., Gendron P., Boisgontier A., Major F., Chartrand P. 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 25:4752–4766 10.1128/MCB.25.11.4752-4766.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N., Chartrand P. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105–111 10.1016/j.tcb.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Park H., Ramamurthy B., Travaglia M., Safer D., Chen L.Q., Franzini-Armstrong C., Selvin P.R., Sweeney H.L. 2006. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell. 21:331–336 10.1016/j.molcel.2005.12.015 [DOI] [PubMed] [Google Scholar]
- Phichith D., Travaglia M., Yang Z., Liu X., Zong A.B., Safer D., Sweeney H.L. 2009. Cargo binding induces dimerization of myosin VI. Proc. Natl. Acad. Sci. USA. 106:17320–17324 10.1073/pnas.0909748106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Tyska M.J., Novick P.J., Mooseker M.S. 2001. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153:1121–1126 10.1083/jcb.153.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel L.M., Monty K.J. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362 10.1016/0926-6585(66)90333-5 [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S., Spudich J.A. 2009. Coupled myosin VI motors facilitate unidirectional movement on an F-actin network. J. Cell Biol. 187:53–60 10.1083/jcb.200906133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6:363–375 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- Stuurman N., Häner M., Sasse B., Hübner W., Suter B., Aebi U. 1999. Interactions between coiled-coil proteins: Drosophila lamin Dm0 binds to the bicaudal-D protein. Eur. J. Cell Biol. 78:278–287 [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., Vale R.D. 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA. 97:5273–5278 10.1073/pnas.080585897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., Sil A., Swedlow J.R., Herskowitz I., Vale R.D. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 389:90–93 10.1038/38015 [DOI] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D.R., Vale R.D. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267 10.1126/science.1073386 [DOI] [PubMed] [Google Scholar]
- Trucco A., Gaspar I., Ephrussi A. 2009. Assembly of endogenous oskar mRNA particles for motor-dependent transport in the Drosophila oocyte. Cell. 139:983–998 10.1016/j.cell.2009.10.012 [DOI] [PubMed] [Google Scholar]
- Tyska M.J., Mooseker M.S. 2003. Myosin-V motility: these levers were made for walking. Trends Cell Biol. 13:447–451 10.1016/S0962-8924(03)00172-7 [DOI] [PubMed] [Google Scholar]
- Vershinin M., Carter B.C., Razafsky D.S., King S.J., Gross S.P. 2007. Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. USA. 104:87–92 10.1073/pnas.0607919104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Feng W., Wei Z., Miyanoiri Y., Wen W., Zhao Y., Zhang M. 2009. Myosin VI undergoes cargo-mediated dimerization. Cell. 138:537–548 10.1016/j.cell.2009.05.030 [DOI] [PubMed] [Google Scholar]