Phosphorylation of a dynactin subunit is important for its release from mitotic spindles.
Abstract
Aurora A is a spindle pole–associated protein kinase required for mitotic spindle assembly and chromosome segregation. In this study, we show that Drosophila melanogaster aurora A phosphorylates the dynactin subunit p150glued on sites required for its association with the mitotic spindle. Dynactin strongly accumulates on microtubules during prophase but disappears as soon as the nuclear envelope breaks down, suggesting that its spindle localization is tightly regulated. If aurora A's function is compromised, dynactin and dynein become enriched on mitotic spindle microtubules. Phosphorylation sites are localized within the conserved microtubule-binding domain (MBD) of the p150glued. Although wild-type p150glued binds weakly to spindle microtubules, a variant that can no longer be phosphorylated by aurora A remains associated with spindle microtubules and fails to rescue depletion of endogenous p150glued. Our results suggest that aurora A kinase participates in vivo to the phosphoregulation of the p150glued MBD to limit the microtubule binding of the dynein–dynactin complex and thus regulates spindle assembly.
Introduction
The Drosophila melanogaster aurora A gene was first identified through mutations resulting in either female sterility or maternal effect lethality. In both cases, mutations were associated with mitotic defects including failure for centrosomes to separate (Glover et al., 1995). Subsequently, it was shown to encode a protein kinase that is widely present in eukaryotic genomes (Giet and Prigent, 1999). Aurora A is an essential gene in mammals and is expressed at elevated levels in a wide variety of tumor cells. Its overexpression is sufficient to trigger genetic instability and transformation in NIH3T3 mouse fibroblasts but not in normal cells, suggesting that this protein might behave as an oncogene under specific genetic backgrounds (Giet et al., 2005; Cowley et al., 2009). The multiple roles of aurora A protein kinase in centrosome function and mitotic spindle assembly in Drosophila, Caenorhabditis elegans, Xenopus laevis, and human cells have been extensively studied (Roghi et al., 1998; Hannak et al., 2001; Giet et al., 2002; Barr and Gergely, 2007). In many systems, the phenotypes caused by aurora A loss of function suggest that aurora A regulates the dynamics of astral microtubules. To do so, it has been shown that aurora A phosphorylates several microtubule-associated proteins, including the D-TACC subunit of the D-TACC–Msps microtubule-stabilizing complex. Indeed, after phosphorylation by aurora A, the D-TACC–Msps complex is targeted to the centrosome component, centrosomin. The Msps subunit of the complex (XMAP215 homologue) binds directly to microtubules to promote microtubule growth. It is thus proposed that phosphorylation of the D-TACC–Msps complex favors stabilization of newly nucleated microtubules at the centrosome (Giet et al., 2002; Terada et al., 2003; Barros et al., 2005; Zhang and Megraw, 2007). In mitotic X. laevis egg extracts, aurora A phosphorylates the kinesin-related protein Eg5, hepatoma up-regulated protein, and its coactivators, TPX2. These proteins are required for bipolar mitotic spindle assembly and can be found in a complex with XMAP215 (Giet and Prigent, 1999; Wong et al., 2008). Furthermore, phosphorylation of the mitotic centromere-associated kinesin by aurora A induces its redistribution onto spindle microtubules, where it facilitates the establishment of spindle bipolarity (Zhang et al., 2008). Finally, the aster-associated protein, required for spindle assembly, is protected from degradation by the proteasome during mitosis after aurora A phosphorylation (Saffin et al., 2005; Venoux et al., 2008).
In this study, we show that aurora A can phosphorylate the p150glued component of the dynein–dynactin complex (DDC) at the microtubule-binding domain (MBD) to prevent the accumulation of dynactin and its associated protein, dynein, on the spindle microtubules.
Results and discussion
Most known aurora A substrates are associated with centrosomes and spindle microtubules (Barr and Gergely, 2007). Thus, to identify new aurora A substrates, we decided to ask whether they could be enriched in microtubule preparations. To this end, we prepared microtubule-associated proteins (MAPs) from Drosophila embryos (Fig. 1 A). We used these preparations as substrates for an aurora A in vitro kinase assay (see Materials and methods). We observed a prominent labeled band of 150 kD, which was analyzed by mass spectrometry (Fig. 1 B). This protein was identified as p150glued, a subunit of the dynactin complex required for several aspects of mitosis (Goshima and Vale, 2003; Morales-Mulia and Scholey, 2005; Delcros et al., 2006).
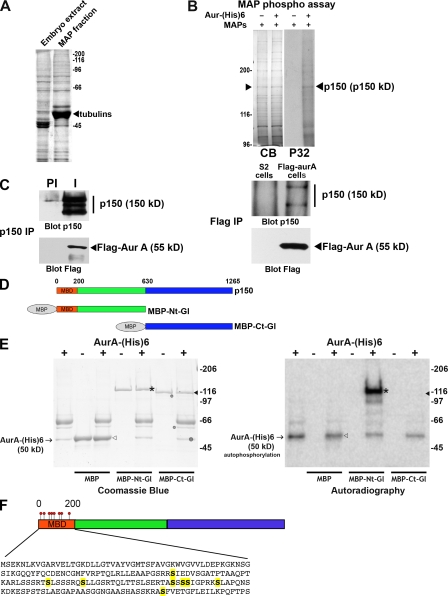
Figure 1.
p150glued is an aurora A substrate in vitro. (A) Coomassie blue–stained gel of the total embryonic extract (left) and the MAPs fraction obtained after sedimentation of taxol-polymerized microtubules (right). The strong band corresponds to the tubulins (arrowhead). (B) A kinase assay with (+) or without (−) aurora A–(His)6 was performed using 20 µg MAPs preparation. (left) The proteins were separated by SDS-PAGE and stained by Coomassie blue (CB). (right) The discrete phosphorylated band (P32) was excised and identified by mass spectrometry as p150glued. The white line indicates that intervening lanes have been spliced out. (C, left) Extracts from S2 cells stably expressing 3xFlag–aurora A were subjected to IP with preimmune (PI) or affinity-purified immune (I) anti-p150glued antibodies. (right) Extracts from wild-type S2 cells (control) or S2 cells stably expressing 3xFlag–aurora A were subjected to anti-Flag IP. The precipitates were revealed with anti-p150glued (top) or anti-Flag antibodies (bottom). Note the presence of p150glued in 3xFlag–aurora A precipitates and, conversely, the presence of aurora A in p150glued immunoprecipitates. (D) Scheme of the p150glued fusion proteins used in the kinase assay. N- and C-terminal fragments of p150glued are displayed in green and blue, respectively. (E) Recombinant MBP, MBP-Ct-Gl, and MBP-Nt-Gl were used for in vitro kinase assays using (+) or not using (−) aurora A–(His)6 protein kinase in the presence of radio-labeled γ-[32P]ATP. The position of the aurora A–(His)6 band is indicated by arrows (+). MBP and MBP-Ct-Gl, indicated by open and closed arrowheads, respectively, are not phosphorylated, whereas MBP-Nt-Gl (asterisks) is strongly phosphorylated by aurora A. The Coomassie blue–stained gel (left) and the corresponding autoradiography (right) are shown. (F) Position of the eight phosphorylated Ser residues (yellow) in the p150glued MBD (amino acids 0–200).
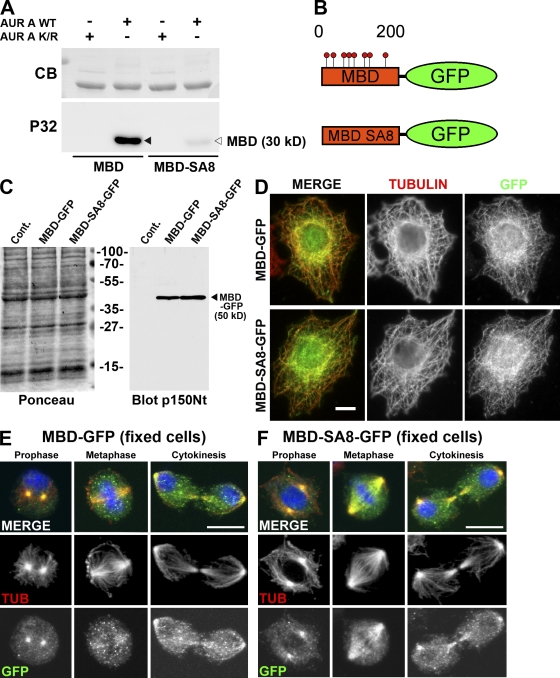
To determine whether aurora A and dynactin might be physically associated in vivo, we performed immunoprecipitation (IP) experiments in Drosophila S2 cells stably expressing a tagged aurora A protein kinase (see Materials and methods). Endogeneous p150glued was able to pull down tagged aurora A (Fig. 1 C, left) and was found in tagged aurora A immunoprecipitates (Fig. 1 C, right), indicating the ability of aurora A and p150glued to interact in S2 cells. We wanted to check whether aurora A was able to directly phosphorylate p150glued. For this purpose, we produced recombinant N-terminal and C-terminal fragments of p150glued in fusion with the maltose-binding protein (MBP; named MBP-Nt-Gl and MBP-Ct-Gl, respectively; Fig. 1 D) in Escherichia coli to use in an in vitro kinase assay (Fig. 1 E). MBP alone and MBP-Ct-Gl were not phosphorylated by aurora A in vitro, whereas MBP-Nt-Gl was a good substrate. Phosphomapping by mass spectrometry revealed phosphorylation of eight Ser residues (Ser85, -109, -117, -135, -137, -138, -144, and -183) within a 200–amino acid N-terminal domain corresponding to the MBD (Fig. 1 F). Most of these sites (except Ser137 and Ser138) matched the yeast and mammalian aurora phosphorylation known consensus site RXS/TΦ (where Φ is a hydrophobic residue; Cheeseman et al., 2002; Ferrari et al., 2005). These phosphorylated sites were also detected with other sites after proteomic analyses of p150glued isolated from Kc Drosophila cultured cells in vivo (http://www.sbeams.org/dev1/sbeams/cgi/Glycopeptide//peptideSearch.cgi). A purified recombinant MBD in which all phosphorylated Sers were mutated into Alas had lost its ability to be phosphorylated by aurora A in vitro (Fig. 2 A). Interestingly, these sites contribute to ∼60% of total MBD phosphorylation by S2 cell extracts, confirming that other phosphorylation sites exist on p150glued MBD in vivo (Fig. S1 A). Furthermore, phosphorylation assays using an aurora A–depleted extract showed that aurora A by itself contributes to ∼25% of total MBD phosphorylation observed in S2 cells (Fig. S1 B). p150glued MBD is responsible for the microtubule-binding properties of dynactin, and its deletion affects mitotic but not interphase functions of the protein (Kim et al., 2007). To analyze the relevance of those phosphorylation events in vivo, we examined the consequences of expressing either a GFP-tagged wild-type (MBD-GFP) or nonphosphorylatable variant of the p150glued MBD (MBD-SA8-GFP) in S2 cells (Fig. 2 B). We established stable cell lines in which such constructs were expressed at equivalent levels (Fig. 2 C). By immunostaining, both proteins were able to bind microtubules during interphase when aurora A was not active (Fig. 2 D). During early mitosis, MBD-GFP and MBD-SA8-GFP were strongly associated with centrosomal microtubules. However, the nonphosphorylatable MBD-SA8-GFP mutant protein was retained on the spindle microtubules during mitosis in all cells examined during metaphase but not MBD-GFP (n > 200; Fig. 2, compare E [middle] with F [middle]). We assessed the binding of recombinant MBD and MBD-SA8 to taxol-polymerized microtubules in vitro. The p150glued MBD affinity for microtubules was five times weaker after incubation with aurora A protein kinase and ATP. In these conditions, we found that the mutagenesis of the eight Sers into Alas restored the ability of p150glued MBD to bind to the microtubules (Fig. S1 C). Thus, the direct interaction of the p150glued MBD with the microtubules appears to be negatively regulated by aurora A phosphorylation in vitro and in vivo during mitosis.
Figure 2.
The MBD of p150glued is phosphorylated by aurora A after nuclear envelope breakdown. (A) Wild-type (WT) or mutant MBD fragment in which the eight Sers (phosphorylated by aurora A) were mutated into Alas (MBD-SA8) were subjected to a kinase assay in the presence of active (AUR A WT) or inactive (AUR A K/R) recombinant aurora A kinase. The Coomassie blue (CB)–stained gel (top) was subjected to autoradiography (bottom). The p150glued MBD fragment (closed arrowhead) is strongly phosphorylated in the presence of active (but not inactive) aurora A protein kinase, whereas the MBD-SA8 fragment (open arrowhead) remains unphosphorylated. (B) Scheme of the wild-type (MBD-GFP) or mutant (MBD-SA8-GFP) proteins stably expressed in S2-cultured cells. (C) Control (cont) or S2 cell extracts expressing MBD-GFP or MBD-SA8-GFP were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and stained by Ponceau S as a loading control (left). The membrane was blotted for p150glued. The position of the ∼50-kD MBD-GFP and MBD-SA8-GFP proteins is indicated (arrowhead). (D–F) Interphase S2 cells expressing MBD-GFP (top) or MBD-SA8-GFP (bottom) were fixed and stained for tubulin (red; middle in monochrome) or GFP (green; right in monochrome). Both GFP fusions associate with the interphase microtubule network. S2 cells expressing either MBD-GFP (E) or MBD-SA8-GFP (F) were methanol fixed and stained for DNA (blue), tubulin (red; middle in monochrome), and GFP (green; bottom in monochrome). During prophase, the GFP signal was strong at the centrosome region for both proteins. Unlike MBD-GFP, the mutant MBD-SA8-GFP protein remains strongly associated with spindle microtubules during all mitotic steps. Bars, 10 µm.
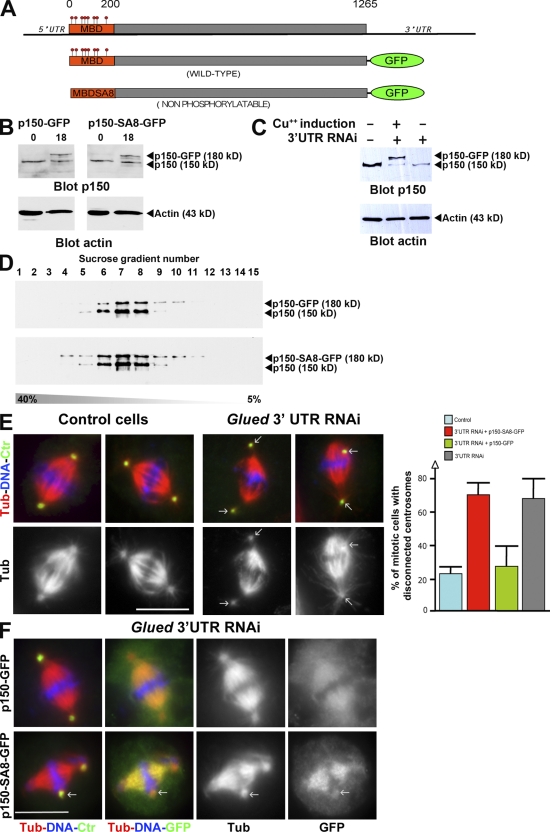
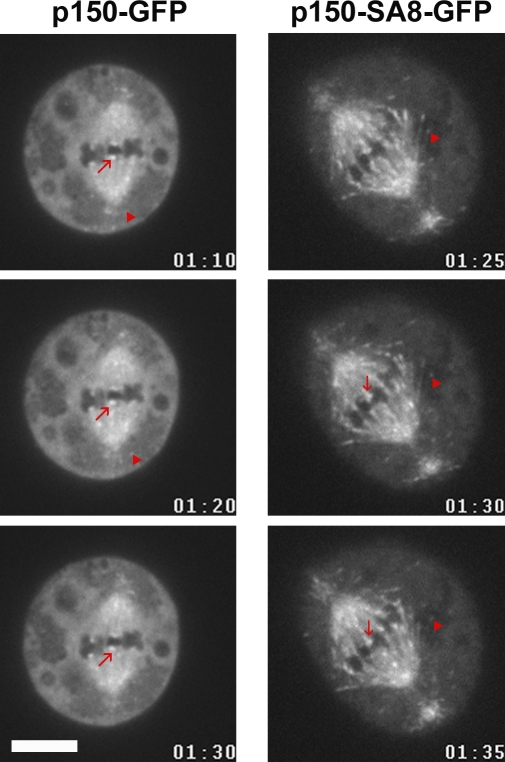
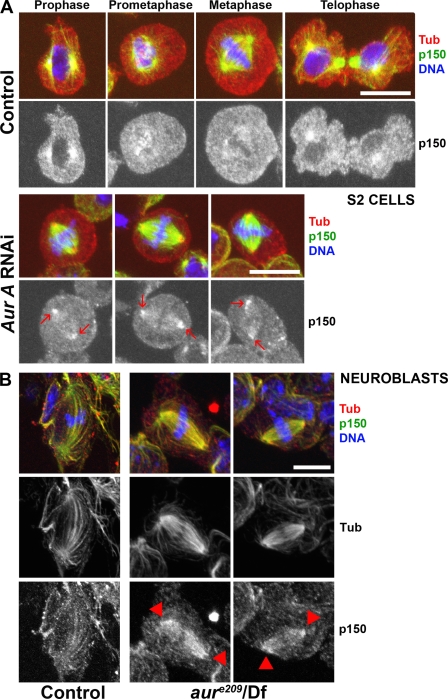
As p150glued MBD is required for mitotic spindle assembly (Kim et al., 2007), it was tempting to speculate that phosphorylation at the sites we have mapped is required to control the overall dynactin accumulation on the mitotic spindle. To test this hypothesis, we checked whether a full-length p150glued GFP fusion protein in which the eight aurora A phosphorylation sites (Sers) had been replaced by Alas (p150-SA8-GFP) would (a) accumulate on spindle microtubules and (b) be able to complement p150glued depletion. For this purpose, we induced the expression of either the GFP-tagged p150glued wild-type protein (p150-GFP) or a mutant variant (p150-SA8-GFP; Fig. 3 A). After several hours of induction of their metallothionein promoter with copper, both GFP-fused proteins were detected by Western blot analysis (Fig. 3 B). Using a double-stranded RNA (dsRNA) specific to the glued 3′UTR gene (Fig. 3 A; Kim et al., 2007), we were able to deplete the endogenous p150glued but not the exogenous GFP-tagged protein (Fig. 3 C). In addition, sucrose gradient separation analyses revealed that both GFP fusion proteins were sedimenting together with endogenous p150glued in the 19S dynactin complex (Fig. 3 D). As reported previously in S2 cells, p150glued-depleted mitotic cells showed an obvious disconnection of centrosomes from the mitotic spindle in 67.3 ± 11.6% of the metaphase cells (Fig. 3 E, compare left with right), whereas only 22.4 ± 4% of the cells showed this phenotype in control cells (Goshima and Vale, 2003; Goshima et al., 2005b; Siller et al., 2005). Interestingly, we completely rescued this defect by expression of p150-GFP (26.7 ± 12%) but not p150-SA8-GFP (69.6 ± 6.9%; Fig. 3 E). In addition to that, the wild-type protein was weakly localized on spindle microtubules compared with the mutant, which showed strong association with these microtubules (Fig. 3 F). This localization of p150-GFP and p150-SA8-GFP was confirmed by high resolution time-lapse video microscopy. As seen in Fig. 4 (left; and Video 1), the p150-GFP protein weakly localized on the spindle region and microtubule plus ends (Fig. 4, triangles), whereas p150-SA8-GFP strongly decorated mitotic spindle fibers and microtubule plus ends (Fig. 4, right; and Video 2). The kinetochore localization (Fig. 4, arrows) remained identical for both constructs. All of these results led us to investigate whether aurora A was responsible for the dynamics of dynactin during mitosis. To do so, we generated a new p150glued antibody against the C-terminal fragment of p150glued (Fig. S2 A) to localize the p150glued protein during mitosis in Drosophila S2-cultured cells. Using this antibody, we observed a staining of the centrosomal microtubules during prophase and the early stages of spindle formation. The signal decreased during spindle maturation before becoming enriched once more at telophase, suggesting that spindle pole localization of p150glued was inhibited after nuclear envelope breakdown (Fig. 5 A). We also found a transient staining of the kinetochore regions of chromosomes during prometaphase that was replaced after chromosome congression by a weak staining of spindle fibers, which is similar to the findings of others (Siller et al., 2005). After aurora A RNAi, 60% of the metaphase cells (n = 3; 200 cells) showed stronger staining of the spindle pole regions (Fig. 5 A, arrows; and Fig. S3). Interestingly, identical results were obtained after aurora A knockdown in human HeLa cells (unpublished data), suggesting that DDC regulation is conserved from Drosophila to humans. We further showed the same behavior for the dynactin-interacting protein dynein, which accumulated at the spindle poles after aurora A RNAi in S2-cultured cells (Fig. S2 B). We also asked whether dynactin could become mislocalized in larval neuroblasts from aurora A mutants. As previously described (Siller et al., 2005), dynactin was faintly distributed along the spindle fibers of wild-type cells in metaphase. In contrast, ∼60% of the aurora A mutant cells with a bipolar spindle showed an accumulation of dynactin in the spindle pole regions or all over the spindle (six brains; 60 metaphase cells; Fig. 5 B, arrowheads). In these mutant cells, dynein strikingly followed the same behavior (n = 3; 35 metaphase cells; Fig. S2 C). Together, these data strongly suggest that aurora A participates in the control of the dynactin binding to microtubules, which is an event required for DDC function.
Figure 3.
A full-length p150glued that can no longer be phosphorylated by aurora A binds more strongly to microtubules but cannot rescue glued spindle assembly defect. (A) Scheme of the GFP fusion proteins expressed in S2 cell lines under the control of the metallothionein promoter (inducible by Cu2+). The constructs lack the glued gene 3′UTR (top) targeted by RNAi. (B) p150glued (top) or actin (bottom) Western blot showing p150-GFP (left) and p150-SA8-GFP (right) protein levels after 0 and 18 h induction on the corresponding cell line. (C) p150glued, p150-GFP, p150-SA8-GFP, and actin protein levels after glued 3′UTR RNAi (lanes 2 and 3). Note the knockdown of endogenous p150glued, whereas p150-GFP protein level remains stable. (D) p150glued Western blot analysis of the different sucrose fractions after the sedimentation assay (from 5 to 40%) of S2 cell extracts expressing either p150-GFP (top) or p150-SA8-GFP (bottom). (E, left) Fixed control or p150glued-depleted metaphase cells are stained for DNA in blue, microtubules in red (monochrome in bottom), and centrosomes in green. The arrows point to disconnected centrosomes. (E, right) Quantification of the centrosome disconnection phenotype in control or glued 3′UTR dsRNA-treated S2-cultured cells. Note the rescue obtained after induced expression of p150-GFP protein but not p150-SA8-GFP. (F) Spindle morphology in p150glued cells after p150-GFP (top) or p150-SA8-GFP (bottom) expression. DNA (blue), microtubules (red; monochrome in the third column), centrosomes (green), and GFP (green in merge; monochrome on the right) are displayed. p150-SA8-GFP interacts more strongly with spindle microtubules than its wild-type counterpart. The arrows point out the disconnected centrosomes. Error bars indicate mean ± SD. Bars, 10 µm.
Figure 4.
p150-SA8-GFP displays higher affinity for microtubules than wild-type p150-GFP protein during mitosis. S2 cells expressing equivalent levels of p150-GFP (left) or p150-SA8-GFP (right) were imaged by time-lapse video microscopy (Videos 1 and 2). The p150-SA8-GFP protein is strongly associated with spindle fibers and microtubule plus ends, whereas p150-GFP shows moderate association with these structures (arrowheads). The kinetochore localization (arrows) is not affected. Bar, 10 µM.
Figure 5.
p150glued accumulates at spindle poles of aurora A RNAi-deficient S2 cells and aurora A mutant neuroblasts. (A) Control S2 cells (top) were fixed and stained for tubulin (red), p150glued (green), and DNA (blue). The mitotic phases are displayed at the top. p150glued localization at spindle poles is clearly detected during prophase and cytokinesis, whereas the signal is very weak during prometaphase and metaphase. In parallel, ∼60% of aurora A–depleted cells (bottom) exhibit p150glued accumulation at spindle poles (arrows). (B) Wild-type (left) or aurora Ae209/Df(3R)T61 (right) mutant neuroblasts were fixed and stained for tubulin (green; monochrome in middle), p150glued (red; monochrome in bottom), and DNA (blue). In control metaphase cells, p150glued is faintly detected along spindle fibers. In aurora A mutant neuroblasts, p150glued accumulates on spindle fibers and spindle poles (arrowheads). Bars, 10 µm.
The dynactin complex, composed of p150glued and at least 10 other polypeptides, cooperates with dynein to fulfill multiple roles during mitotic progression (Karki and Holzbaur, 1999; Goshima and Vale, 2003; Morales-Mulia and Scholey, 2005; Delcros et al., 2006). In Drosophila S2 cells, DDC is required for the centrosome connection to the mitotic spindle and for the metaphase/anaphase transition (Goshima and Vale, 2003; Morales-Mulia and Scholey, 2005). Our RNAi experiments in S2 cells confirm these previous studies, as centrosomes were disconnected from the poles in ∼70% of the p150glued-depleted mitotic cells. Both RNAi experiments and mathematical models concur in the notion that kinetochore fibers are captured and focused at the poles by the DDC, which connects these fibers to the centrosome-nucleated microtubules. Thus, a lack of DDC can explain the fact that centrosomes are disconnected from the spindle (Maiato et al., 2004; Goshima et al., 2005a). Our study revealed that aurora A participates in the phosphoregulation of p150glued, but other kinases can contribute to this process on p150glued MBD itself (Fig. S1) or other DDC subunits (Huang et al., 1999; Vaughan et al., 2001, 2002). Interestingly, the variant that can no longer be phosphorylated by aurora A remains associated with spindle microtubules and cannot complement the glued loss of function. This suggests that DDC levels on spindle microtubules and at the poles need to be tightly controlled in Drosophila cells to avoid centrosome disconnection, a phenotype sometimes also observed in aurora A mutant embryos (unpublished data), but not in other cell types in which other kinases possibly participate in dynactin limitation. The reason why DDC localization at spindle poles needs to be modulated remains unclear. It is possible that DDC function requires a moderate association of dynactin with spindle microtubules. It is also possible that overaccumulation of DDC (and cargoes) could create a traffic obstruction at spindle poles to compete with other MAPs. Consequently, this would prevent the connection between kinetochore fibers and astral microtubules. Further studies will be required to elucidate the complete DDC regulation during mitosis.
Materials and methods
Preparation of microtubules from Drosophila embryos for aurora A phosphorylation assays
500 µl Drosophila early embryos (0–4 h) were collected and lysed in 500 µl BRB80 buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) supplemented with protease inhibitors (Roche) and 0.5% NP-40. The crude extract was centrifuged at 10,000 g for 15 min at 4°C. The supernatant was centrifuged a second time at 100,000 g for 20 min at 4°C. 0.4 ml supernatant containing the soluble proteins was supplemented with 20 µM taxol and 1 mM GTP. Microtubule polymerization was induced for 20 min at 25°C. The microtubules were sedimented on a 0.4 ml BRB80 cushion supplemented with 20 µM taxol, 1 mM GTP, and 40% glycerol. The microtubule pellet containing the MAPs was washed twice in BRB80 and resuspended in BRB80. The MAP fractions were stored at −80°C.
Phosphorylation assay
20 µg MAPs fraction or 3 µg recombinant substrate proteins was incubated with 200 ng recombinant aurora A–(His)6 kinase, 1 µCi radio-labeled γ-[32P]ATP (GE Healthcare), and 100 µM ATP in kinase buffer (Giet et al., 2002).
The samples were separated by SDS-PAGE and stained by Coomassie blue. p150glued was identified by mass spectrometry on the excised band from the gel. To map the phosphorylation sites on the MBP-Nt-Gl protein, the kinase assay was performed without radio-labeled ATP, and the phosphorylated band was excised and sent for phosphopeptide identification to the Proteomic Platform of the Quebec Genomic Center (http://proteomique.crchul.ulaval.ca/en/equipment.html).
For in vivo phosphorylation assay, 3 × 106 control or aurora A–depleted cells were resuspended in 500 µl of lysis buffer containing protease inhibitors (10 mM Na2VO4 and 50 mM β-glycerophosphate; Giet et al., 2002). 5 µl of the extract was used to phosphorylate 5 µg MBD or MBD-SA8 in the presence of 2.5 µCi radio-labeled γ-[32P]ATP (GE Healthcare) and 100 µM cold ATP in kinase buffer. MBD and MBD-SA8 phosphorylation signals were quantified using a phosphoimager (Molecular Dynamics).
dsRNA production and constructs
The aurora A dsRNA production was described previously (Giet and Glover, 2001; Giet et al., 2002). In brief, an ∼1,000-bp PCR product containing a T7 sequence at each end was used as a template to generate RNA using the Megascript kit (Promega). To obtain the glued template (for the 3′UTR region), an ∼450-bp PCR product using the primers 5′-AATAATACGACTCACTATAGGGAAAGGATCDTGTATCGTGGCA-3′ and 5′-AATAATACGACTCACTATAGGGGAGTTATACAACATCAGCAAA-3′ was produced. After isolation, the RNAs were boiled for 20 min and annealed by slow cooling overnight at room temperature. dsRNAs were analyzed by agarose gel electrophoresis and aliquoted at −80°C before use in RNAi experiments. To generate the expression construct encoding the full-length aurora A in fusion with 3xFlag at its N-terminal end, aurora A ORF was first cloned into the pENTR vector (to create an entry clone) using a cloning kit (TOPO; Invitrogen) and recombined using the gateway system (Invitrogen) into the pHFW containing the 3xFlag tag and a heat shock promoter. An aurora A kinase-dead mutant (K193/R) was generated by mutation of the conserved lysine of the kinase subdomain II into an arginine.
For the MBD expression constructs, each Ser to Ala mutation in the p150glued MBD was obtained by sequential PCRs. The wild-type (MBD) or nonphosphorylatable (MBD-SA8) domains were cloned directly into pET102/TOPO bacterial expression vector or pENTR to generate entry clones. The latter clones were recombined into pDEST17 to generate bacterial expression constructs and in pAWG to allow S2 cell expression of either MBD-GFP or MBD-SA8-GFP under the control of the actin 5c promoter. Both pAWG and pHFW were obtained from the Carnegie Institution of Washington.
N- and C-terminus domains of p150glued were amplified by PCR and cloned into the pMal-C2E expression vector using KpnI and BamHI restriction sites. All constructs were verified by sequencing.
Full-length p150glued was cloned into pMT-GFP-Ct (provided by M. Savoian, University of Cambridge, Cambridge, England, UK) using the gateway system (through an entry clone) to generate p150-GFP expression construct. p150-SA8-GFP was subsequently obtained with the QuickChange Mutagenesis kit (Agilent Technologies). The pAct5C-Cherry–α-tubulin construct was provided by G. Hickson (University of California, San Francisco, San Francisco, CA).
Production of the recombinant proteins and antibody purification
Aurora A–(His)6, aurora A–K/R-(His)6, MBD-(His)6, or MBD-SA8-(His)6 were purified as described previously (Giet et al., 2002). MBP-Nt-Gl and MPB-Ct-Gl were expressed in E. coli BL21(DE3) pLysS (EMD) for 4 h at 25°C, purified on an amylose column, and dialysed against PBS and stored at −80°C before use for aurora A kinase assay. The purified MBP-Ct-Gl protein was also used to immunize rabbits. Obtained anti-p150glued antibodies were affinity purified on a nitrocellulose membrane, and the purified antibodies were stored at −80°C as described previously (Montembault et al., 2007).
In vitro microtubule-binding assay
1 µg purified recombinant p150glued MBD or MBD-SA8 proteins in fusion with a hexahistidine tag was incubated with either 200 ng aurora A or aurora A–K/R proteins in kinase buffer containing 1 mM ATP in a total volume of 10 µl for 20 min. The kinase reaction was then incubated with 25 µl (100 µg) of taxol-stabilized microtubules for 10 min at 37°C (Roghi et al., 1998). The microtubule pellet and the supernatant containing microtubule-unbound proteins were separated by sedimentation at 100,000 g for 20 min at 37°C (TLA-100; Beckman Coulter). The proteins present in the pellets and supernatants were analyzed by Western blotting or Coomassie blue staining.
RNAi, transfections, drug treatment, stable line generation, and rescue experiments
Drosophila S2 cells were grown and processed for RNAi as described previously (Clemens et al., 2000). In brief, 106 cells were incubated with 10 µg/ml dsRNA in media without serum. Alternatively, 10 µg transfection reagent (Transfast; Promega) was added together with 3 µg dsRNA following the manufacturer's instructions. After 1 h, fresh media were added to the cells. At 4 d after transfection, the cells were fixed and analyzed for mitotic defects (Giet and Glover, 2001). 100–200 mitoses were scored and analyzed per experiment, and each experiment was repeated at least three times.
To make S2 stable lines expressing 3xFlag–aurora A, MBD-GFP or MBD-SA8-GFP, p150-GFP, and p150-SA8-GFP, 5 µg of each expression construct was cotransfected with 1 µg pIB/V5-His/CAT (Invitrogen). Alternatively, 5 µg pAct5c-Cherry–α-tubulin was cotransfected in those cells. Transfection reagent (Effectene; QIAGEN) was used for transfection. Stable lines were selected and expanded in media containing 25 µg/ml blasticidin S (Invitrogen). For rescue experiments, expression of either p150-GFP or p150-SA8-GFP proteins was induced 18 h before fixation by supplementing media with CuSO4 (300 µM final concentration).
Immunofluorescence analysis
S2 cells were fixed in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgCl2) containing 3.7% formaldehyde and 0.1% Triton X-100. In some cases, the cells were plated on concanavalin A–coated coverslips for 1 h and fixed in methanol at −20°C for 10 min. The fixed cells were briefly washed in PBS and blocked for 1 h in PBS containing 0.1% Triton X-100 and 1% BSA (PBST-BSA). Primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated for 1 h at room temperature in PBST-BSA. DNA was stained with Hoechst 33258. Slides were mounted in ProLong gold (Invitrogen) and observed with a microscope (DMRXA2; Leica) using a 63× 1.3 NA objective. Images were acquired with a camera (CoolSnap HQ; Photometrics) and processed with MetaMorph software (Universal Imaging). Alternatively, images were acquired with an inverted confocal microscope (SP2; Leica).
Time-lapse imaging
S2 cells expressing p150-GFP or p150-SA8-GFP were incubated with CuSO4 (300 µM final concentration) for 18 h and plated for 1 h on concanavalin A–coated incubation chambers (Labtek; Sigmatek) before imaging. Images were aquired using a spinning-disk system mounted on an inverted microscope (Elipse Ti; Nikon) using a 100× 1.4 NA objective. Images were acquired every 5 or 10 s with a camera (CoolSnap HQ2; Photometrics) controlled by the MetaMorph acquisition software.
Antibodies and Western blotting
The YL1/2 rat anti-tyrosinated tubulin antibody (1:1,000) was obtained from Millipore, and the GTU-88 mouse anti–γ-tubulin antibody (1:1,000) was obtained from Sigma-Aldrich. The mouse anti-dynein clone 1H4 antibody (1:100) was provided by T. Hays, and the anti-GFP monoclonal antibodies (1:1,000) were obtained from Roche or Invitrogen. Affinity-purified antibodies against aurora A were used at 0.5 µg/ml (Giet et al., 2002). The affinity-purified anti-p150glued antibody was purified as described previously (Montembault et al., 2007) and used at 2 µg/ml. Secondary peroxidase-conjugated antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc., and Alexa Fluor–conjugated secondary antibodies were obtained from Invitrogen. For Western blotting, ECL reagent was purchased from Thermo Fisher Scientific.
Online supplemental material
Fig. S1 shows the contribution of aurora A and other unknown kinases to p150glued MBD phosphorylation in vivo. It also shows that aurora A kinase inhibits MBD binding to microtubules in vitro. Fig. S2 shows p150glued antibody Western blotting in control or aurora A–depleted cells together with dynein localization in control or aurora A–depleted S2 cells. The colocalization of dynein and dynactin is also displayed in wild-type or aurora A mutant neuroblasts. Fig. S3 shows the distribution (line scans) of p150glued and tubulin in control or aurora A–depleted cells. Videos 1 and 2 show the behavior of wild-type and nonphosphorylatable p150glued proteins fused to GFP during metaphase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001144/DC1.
Acknowledgments
We thank the laboratory members for stimulating discussions, Clotilde Petretti, Thomas Hays, Pier Paolo d'Avino, Gilles Hickson, and Arnaud Echard for reagents, and Stéphanie Dutertre for microscopy facilities of the IFR140. R. Giet thanks Kathryn Lilley for initial proteomic studies and the members of David Glover's group in Cambridge, where this project was initiated (Department of Genetics, University of Cambridge, Cambridge, England, UK). Special thanks are due to Claude Prigent for his full support and to Alex Holmes, Emeric Sevin, and M. Savoian for their critical reading of the manuscript.
R. Giet and N. Franck are funded by the Agence Nationale de la Recherche (programme Jeune Chercheur) and the Ligue Nationale Contre le Cancer (Equipe Labellisée). E. Montembault and P. Romé are doctoral fellows of the French Ministère de la Recherche.
Footnotes
Abbreviations used in this paper:
- DDC
- dynein–dynactin complex
- dsRNA
- double-stranded RNA
- IP
- immunoprecipitation
- MAP
- microtubule-associated protein
- MBD
- microtubule-binding domain
- MBP
- maltose-binding protein
References
- Barr A.R., Gergely F. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barros T.P., Kinoshita K., Hyman A.A., Raff J.W. 2005. Aurora A activates D-TACC–Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170:1039–1046 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503 10.1073/pnas.110149597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D.O., Rivera-Pérez J.A., Schliekelman M., He Y.J., Oliver T.G., Lu L., O'Quinn R., Salmon E.D., Magnuson T., Van Dyke T. 2009. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol. Cell. Biol. 29:1059–1071 10.1128/MCB.01062-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcros J.G., Prigent C., Giet R. 2006. Dynactin targets Pavarotti-KLP to the central spindle during anaphase and facilitates cytokinesis in Drosophila S2 cells. J. Cell Sci. 119:4431–4441 10.1242/jcs.03204 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Marin O., Pagano M.A., Meggio F., Hess D., El-Shemerly M., Krystyniak A., Pinna L.A. 2005. Aurora-A site specificity: a study with synthetic peptide substrates. Biochem. J. 390:293–302 10.1042/BJ20050343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Prigent C. 1999. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 112:3591–3601 [DOI] [PubMed] [Google Scholar]
- Giet R., McLean D., Descamps S., Lee M.J., Raff J.W., Prigent C., Glover D.M. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156:437–451 10.1083/jcb.200108135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Petretti C., Prigent C. 2005. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15:241–250 10.1016/j.tcb.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Glover D.M., Leibowitz M.H., McLean D.A., Parry H. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 81:95–105 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., Vale R.D. 2005a. Mechanisms for focusing mitotic spindle poles by minus end–directed motor proteins. J. Cell Biol. 171:229–240 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., Vale R.D. 2005b. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Hannak E., Kirkham M., Hyman A.A., Oegema K. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109–1116 10.1083/jcb.200108051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Chang C.P., Huang C.L., Ferrell J.E., Jr 1999. M phase phosphorylation of cytoplasmic dynein intermediate chain and p150(Glued). J. Biol. Chem. 274:14262–14269 10.1074/jbc.274.20.14262 [DOI] [PubMed] [Google Scholar]
- Karki S., Holzbaur E.L. 1999. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 11:45–53 10.1016/S0955-0674(99)80006-4 [DOI] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., Gelfand V.I. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., Khodjakov A. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembault E., Dutertre S., Prigent C., Giet R. 2007. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 179:601–609 10.1083/jcb.200703133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J.M. 2005. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 16:3176–3186 10.1091/mbc.E04-12-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C., Giet R., Uzbekov R., Morin N., Chartrain I., Le Guellec R., Couturier A., Dorée M., Philippe M., Prigent C. 1998. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 111:557–572 [DOI] [PubMed] [Google Scholar]
- Saffin J.M., Venoux M., Prigent C., Espeut J., Poulat F., Giorgi D., Abrieu A., Rouquier S. 2005. ASAP, a human microtubule-associated protein required for bipolar spindle assembly and cytokinesis. Proc. Natl. Acad. Sci. USA. 102:11302–11307 10.1073/pnas.0500964102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Serr M., Steward R., Hays T.S., Doe C.Q. 2005. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 16:5127–5140 10.1091/mbc.E05-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Uetake Y., Kuriyama R. 2003. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 162:757–763 10.1083/jcb.200305048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan P.S., Leszyk J.D., Vaughan K.T. 2001. Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 276:26171–26179 10.1074/jbc.M102649200 [DOI] [PubMed] [Google Scholar]
- Vaughan P.S., Miura P., Henderson M., Byrne B., Vaughan K.T. 2002. A role for regulated binding of p150Glued to microtubule plus ends in organelle transport. J. Cell Biol. 158:305–319 10.1083/jcb.200201029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venoux M., Basbous J., Berthenet C., Prigent C., Fernandez A., Lamb N.J., Rouquier S. 2008. ASAP is a novel substrate of the oncogenic mitotic kinase Aurora-A: phosphorylation on Ser625 is essential to spindle formation and mitosis. Hum. Mol. Genet. 17:215–224 10.1093/hmg/ddm298 [DOI] [PubMed] [Google Scholar]
- Wong J., Lerrigo R., Jang C.Y., Fang G. 2008. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell. 19:2083–2091 10.1091/mbc.E07-10-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Megraw T.L. 2007. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell. 18:4037–4049 10.1091/mbc.E07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., Walczak C.E. 2008. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 19:2752–2765 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]