The Ndc80 complex of kinetochores doesn't recognize microtubule tips by itself but relies on the Dam1 complex to track microtubule plus ends (see also related paper by Tien et al. in this issue).
Abstract
Kinetochores must remain associated with microtubule ends, as they undergo rapid transitions between growth and shrinkage. The molecular basis for this essential activity that ensures correct chromosome segregation is unclear. In this study, we have used reconstitution of dynamic microtubules and total internal reflection fluorescence microscopy to define the functional relationship between two important budding yeast kinetochore complexes. We find that the Dam1 complex is an autonomous plus end–tracking complex. The Ndc80 complex, despite being structurally related to the general tip tracker EB1, fails to recognize growing ends efficiently. Dam1 oligomers are necessary and sufficient to recruit Ndc80 to dynamic microtubule ends, where both complexes remain continuously associated. The interaction occurs specifically in the presence of microtubules and is subject to regulation by Ipl1 phosphorylation. These findings can explain how the force harvested by Dam1 is transmitted to the rest of the kinetochore via the Ndc80 complex.
Introduction
The ability of spindle microtubules to connect to centromeric DNA is fundamental to ensure faithful transmission of the eukaryotic genome. Kinetochores, large protein complexes, physically tether chromosomes to microtubule plus ends and relay their dynamic instability into directed chromosome movement. To date, the molecular basis of how transitions between microtubule growth and shrinkage are coupled to chromosome movement remains unclear (Westermann et al., 2007; Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009).
Two principal types of kinetochore complexes have been proposed to connect dynamic microtubules to chromosome motion: ring couplers and fibrillary couplers. The yeast Dam1 complex oligomerizes in vitro into a ring that encircles the microtubule plus end like a sleeve, allowing the exchange of tubulin subunits at the end while maintaining a firm grip on the polymer (Cheeseman et al., 2001b; Miranda et al., 2005; Westermann et al., 2005). Processive attachment to depolymerizing ends (Westermann et al., 2006; Gestaut et al., 2008) and force generation in the piconewton range have spotlighted the Dam1 complex as a primary coupler in budding yeast (Asbury et al., 2006; Grishchuk et al., 2008a). These features do not strictly depend on ring formation, leaving open the question of the physiologically active form of the complex in vivo. Interestingly, artificial recruitment of the Dam1 complex to DNA bypasses the requirement for inner kinetochore complexes and is sufficient to drive segregation of yeast mini- or native chromosomes (Kiermaier et al., 2009; Lacefield et al., 2009), providing strong evidence that the complex is a critical attachment factor in vivo.
A paradigm for a fibrillary coupler is the four-protein complex Ndc80, which is part of the ternary KMN network, a central component of the kinetochore–microtubule interface in all eukaryotes (Cheeseman et al., 2006). The Ndc80 complex interacts with microtubules via calponin homology domains located in the Ndc80-Nuf2 head (Wei et al., 2007; Ciferri et al., 2008), and individual Ndc80 molecules have been shown to undergo biased diffusion on depolymerizing microtubule ends, which may in principle allow them to convert microtubule depolymerization into chromosome movement (Powers et al., 2009). The respective contribution of Ndc80 and Dam1 complexes to the plus end–tracking activity of the kinetochore is unknown.
In this study, we present a minimal kinetochore plus end–tracking system. We analyzed the behavior of Dam1 and Ndc80 complexes on dynamic microtubules first individually and then in combination. We find that the Dam1 complex but not the Ndc80 complex has an intrinsic plus end–tracking ability. However, Dam1 mediates continuous Ndc80 tip association with dynamic microtubules, suggesting that in yeast, Dam1 complexes are the prime source of kinetochore plus end–tracking activity, whereas Ndc80 complexes structurally bridge microtubule ends with chromosomes.
Results and discussion
The Dam1 complex tracks microtubule plus ends independently of the general +TIP EB1 in vivo
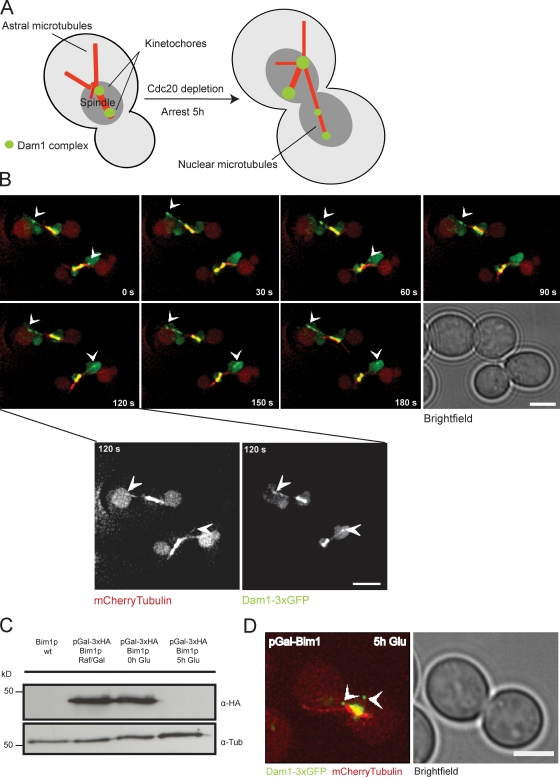
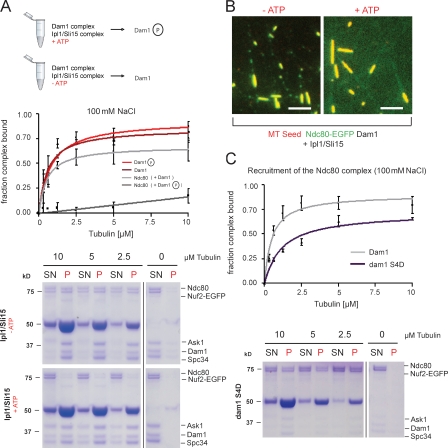
To characterize the interaction of the Dam1 complex with individual microtubule plus ends in vivo, we imaged metaphase-arrested cells expressing Dam1p fused to triple GFP (Dam1-3xGFP) and Tub1p fused to mCherry (Fig. 1 A; Tanaka et al., 2005). We found that the majority of the Dam1 complex is localized at kinetochores, whereas individual signals of the complex accumulate at the tips of growing and shrinking nuclear microtubules. Furthermore, the microtubule lattice is frequently decorated by additional, well-defined Dam1 spots that are collected during microtubule shrinkage consistent with previous data (Tanaka et al., 2007). Interestingly, we observed that the complex continuously tracks microtubule ends during multiple rounds of tubulin polymerization and depolymerization (Fig. 1 B and Videos 1 and 2). Dam1 tip tracking on nuclear microtubules was reminiscent of Bim1p, the budding yeast homologue of the +TIP EB1 (Akhmanova and Steinmetz, 2008; Honnappa et al., 2009). Because interactions between the Dam1 complex and Bim1p have been reported in two-hybrid studies, we considered the possibility that Dam1 tip localization may depend on Bim1p (Cheeseman et al., 2001b; Shang et al., 2003). After 5 h in glucose, Bim1p, placed under the control of the GAL1/10 promoter, was no longer detectable by Western blotting (Fig. 1 C), and nuclear microtubules were extremely short, which is in agreement with previous observations in bim1Δ or bik1Δ strains (Tanaka et al., 2005). However, Dam1 tip localization was retained on short nuclear microtubules, demonstrating that Dam1 plus end–tracking activity is independent of the general +TIP EB1 in vivo (Fig. 1 D and Video 3).
Figure 1.
The Dam1 complex tracks plus ends independently of Bim1p in vivo. (A) Schematic illustration of the live cell imaging strategy (Tanaka et al., 2005). (B) Time-lapse live cell microscopy showing plus end tracking of the Dam1 complex (arrowheads) during phases of microtubule growth and shrinkage (Videos 1 and 2). Insets show higher magnification views of the indicated panel (120 s). (C) Western blot analysis showing depletion of Bim1p-3xHA in a Dam1-3xGFP, mCherry-tubulin background probed with an anti-HA antibody. (D) Still image from a video depicting microtubule plus end localization of the Dam1 complex (arrowheads) in the absence of the +TIP Bim1p (Video 3). Bars, 2 µm.
The Dam1 complex autonomously tracks microtubule plus ends in a minimal in vitro reconstitution system
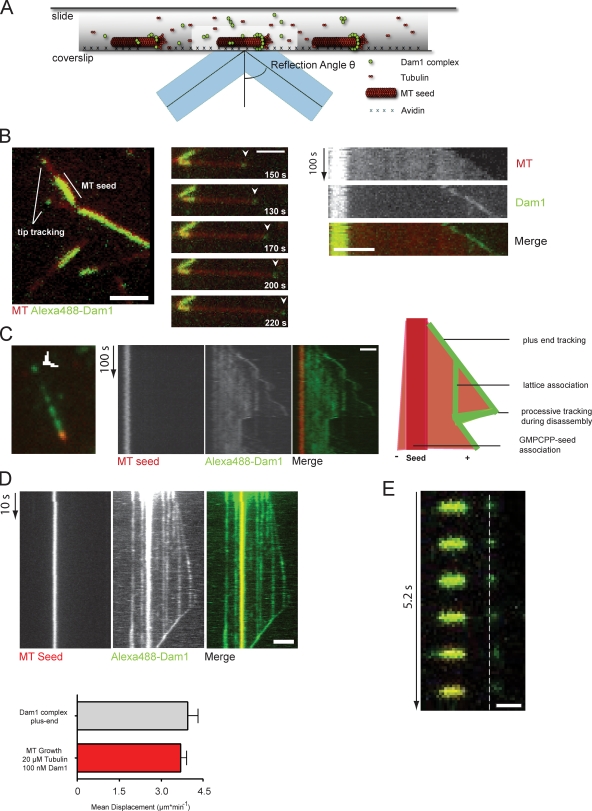
To further establish the minimal requirements for Dam1 plus end tracking, we reconstituted interaction of the complex with dynamic microtubules in vitro using two-color total internal reflection fluorescence (TIRF) microscopy (Fig. 2 A). Previously, the behavior of the Dam1 complex had only been imaged on static or slowly depolymerizing microtubules (Westermann et al., 2006; Gestaut et al., 2008). Visualizing fully dynamic microtubules, we observed the following characteristic features of the complex: the GMPCPP-stabilized seeds were strongly decorated with Dam1 complexes, which is in agreement with previous observations (Westermann et al., 2005; Grishchuk et al., 2008b). In addition, the complex appeared in discrete foci at the plus ends of microtubule extensions, where it persisted during phases of microtubule growth and shrinkage (Fig. 2 B and Video 4). Thus, the Dam1 complex autonomously tracks growing microtubule plus ends in vitro (Fig. 2 C and Videos 5 and 6).
Figure 2.
Reconstitution of Dam1 plus end tracking in vitro using TIRF microscopy. (A) Schematic illustration of the in vitro imaging setup. (B, left) Still image of the Alexa Fluor 488–labeled Dam1 complex (100 nM) on dynamic rhodamine-labeled microtubules (MT). The complex shows preferred association with the plus end and the GMPCPP seed. (middle) A time sequence of Dam1 plus end tracking. (right) Kymographs (time/space plot) of Dam1 plus end tracking are shown (Video 4). (C) Still image and kymograph of a time-lapse video showing Dam1 decoration on unlabeled tubulin extensions. Overlay and individual channels are shown. The Dam1 complex tracks plus ends during phases of growth and shrinkage. The microtubule lattice is decorated by additional Dam1 complexes, which are collected during microtubule disassembly (Videos 5 and 6). (B and C) Arrowheads indicate Dam1 localization at the tip of the microtubule. (D) Kymograph from a streaming video recording continous Dam1 tip tracking. Error bars indicate averaged Dam1 outward displacement at the tip and lattice compared with the mean growth speed of microtubules (n ≥ 20). (E) Still series of a spike experiment depicting an individual Dam1 dot moving away from the stable seed. The dashed line indicates the position of the Dam1 signal at time 0. Bars: (B–D) 2 µm; (E) 1 µm.
As the complex can exist in a variety of concentration-dependent oligomeric states, such as individual complexes, small oligomers, and full rings, we aimed to determine the nature of the tracking species in our assays. We mimicked flow cell conditions in the test tube and examined the decoration of dynamic microtubules (at 14 µM) with different concentrations of Dam1 by EM. We found that at a concentration of 100 nM Dam1, only few rings decorating the microtubules were observed by EM (Fig. S1 A). Quantitative analysis of fluorescent Dam1 signals using TIRF microscopy at the same concentration showed a distribution of various intensities on the microtubule lattice. Most Dam1 spots displayed twice the intensity of the weakest signals, whereas very few spots showed intensities that could correspond to full rings (Fig. S1 B). Thus, under our imaging conditions (100 nM Dam1), the complex predominantly exists in the form of small oligomers.
Dam1 moves continuously with growing microtubule plus ends
The Dam1 complex is only the third molecule for which autonomous tip tracking is reported in vitro. The microtubule polymerase XMAP215 is thought to be processive, persisting at the tip during multiple rounds of tubulin dimer addition (Brouhard et al., 2008), whereas EB1 rapidly turns over on a transiently existing structure at the plus end (Bieling et al., 2007). To distinguish between these possibilities and investigate the mechanism of Dam1 plus end tracking in more detail, we increased the temporal resolution of our videos by recording streaming videos. We observed that those Dam1 foci located most distal to the stable seed continuously moved outward during microtubule growth with a mean speed of 4.02 ± 0.225 µm × min−1. This value is in agreement with the mean growth speed of microtubules in the presence of 100 nM unlabeled Dam1 and 20 µM free tubulin (Fig. 2 D). Moreover, we performed “spike” experiments by reducing the fraction of labeled Dam1 complex at least 10-fold while sustaining the total protein concentration with unlabeled complex. Under these conditions, we were able to visualize individual Dam1 signals that moved away from the stable seed (Fig. 2 E), supporting the notion that the Dam1 complex is an autonomous and continuous plus end tracker.
The Ndc80 complex interacts weakly with dynamic microtubules and is not an autonomous end-binding protein
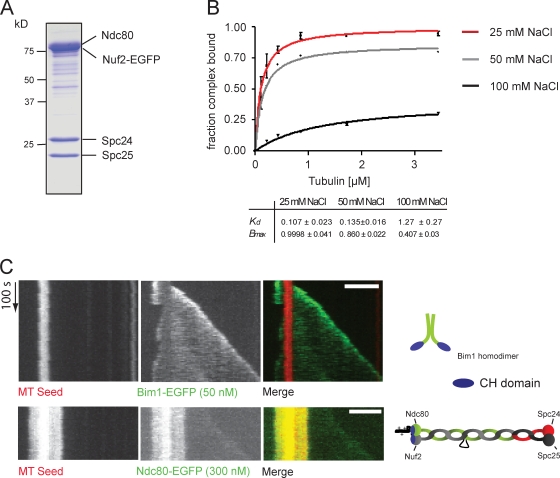
Although a moderate affinity of oligomeric Ndc80 assemblies for depolymerizing microtubule ends has been described in vitro (Powers et al., 2009), it is at present unknown whether the complex can autonomously recognize plus ends and remain attached to them or whether this activity requires additional factors. Using reconstituted Saccharomyces cerevisiae Ndc80 complex with its Nuf2 subunit fused to EGFP (Ndc80–EGFP; Fig. 3 A), we performed microtubule cosedimentation assays and found that the interaction is extremely salt sensitive. Increasing the salt concentration from 25 to 100 mM NaCl lowered the binding affinity by more than one order of magnitude, leading to an apparent dissociation constant of KD (100 mM) = 1.27 ± 0.27 µM compared with KD (25 mM) = 0.107 ± 0.023 µM (Fig. 3 B). This implies that under physiologically relevant salt concentrations, the intrinsic microtubule-binding activity of the Ndc80 complex is significantly impaired.
Figure 3.
Ndc80 is not an autonomous end-binding complex. (A) Coomassie-stained SDS-PAGE showing the reconstituted S. cerevisiae Ndc80 complex. The Nuf2 subunit is fused to EGFP. (B) The Ndc80 complex was tested for its ability to cosediment with taxol-stabilized microtubules in the presence of increasing amounts of salt. The binding affinity curves plot averaged data from three independent experiments. Error bars represent standard error. (C) Kymographs demonstrating 50 nM Bim1-EGFP tip tracking during microtubule (MT) assembly (top) and faint association of 300 nM Ndc80–EGFP with the microtubule lattice (bottom). CH, calponin homology. Bars, 2 µm.
We next studied the interaction of the Ndc80 complex with dynamic microtubules in the TIRF assay. To establish imaging conditions that allow observation of plus end tracking, we first visualized the association of the yeast EB1 protein Bim1p with dynamic microtubules. As expected, Bim1p accumulated at the ends of growing but not shrinking microtubules (Fig. 3 C; Bieling et al., 2007). However, when imaged under the same conditions, the Ndc80 complex was barely visible on the dynamic microtubule extensions, and we did not observe a substantial accumulation on growing or shrinking microtubule ends (Fig. 3 C). Thus, under experimental conditions in which yeast EB1 displays robust plus end tracking in vitro, the Ndc80 complex only weakly interacts with dynamic microtubules and does not accumulate at growing or shrinking ends.
The Dam1 complex is necessary and sufficient to recruit the Ndc80 complex to dynamic microtubule ends
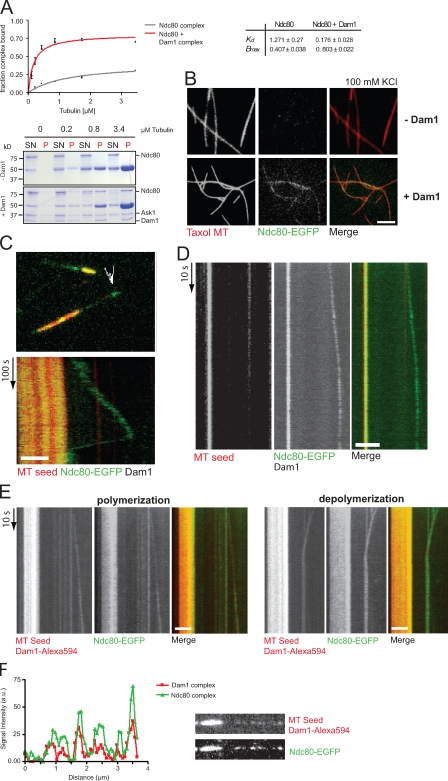
We wondered whether the dynamic properties of the Ndc80 complex would be altered in the presence of the Dam1 complex, which is a tempting idea in the light of autonomous Dam1 plus end–tracking activity and reported two-hybrid interactions between the two complexes (Shang et al., 2003; Wong et al., 2007). We first asked whether inclusion of the Dam1 complex would alter the binding behavior of Ndc80 in cosedimentation assays. At salt concentrations of 100 mM NaCl that strongly impair Ndc80 binding, addition of equimolar amounts of Dam1 drastically increased the affinity of the Ndc80 complex for taxol-stabilized microtubules (Fig. 4 A). Analysis of Coomassie-stained gels revealed that a substantial portion of Ndc80 was now found along with Dam1 in the pellet fraction (Fig. 4 A). The isolated Ndc80-Nuf2 head was sufficient for recruitment (unpublished data). Furthermore, the N-terminal 116 amino acids of Ndc80, critical for the intrinsic microtubule-binding activity of Ndc80, are dispensable for Dam1-dependent recruitment (Fig. S2).
Figure 4.
The Dam1 complex recruits the Ndc80 complex to dynamic microtubule plus ends. (A) Microtubule-binding activity of Ndc80 was tested in the absence and presence of equimolar amounts of Dam1 complex and 100 mM NaCl. (B, top) TIRF micrographs showing little microtubule (MT) decoration of the Ndc80 complex under physiological ionic strength. (bottom) The Dam1 complex efficiently recruits Ndc80 to taxol-stabilized microtubules. SN, supernatant; P, pellet. Bar, 4 µm. (C) Still image and kymograph of a time-lapse video demonstrate that in the presence of 100 nM unlabeled Dam1 complex, Ndc80–EGFP tracks microtubule plus ends (Video 7). Arrowhead indicates Ndc80–EGPF at the tip of the microtubule. (D) Streaming video showing continuous tip tracking of Ndc80–EGFP in the presence of unlabeled Dam1 (Video 8). (E) The Dam1–Alexa Fluor 594 complex and Ndc80–EGFP colocalize at microtubule plus ends during microtubule polymerization and depolymerization (Video 9). (F) Intensity scan demonstrating that Dam1 signals on the microtubule lattice correlate with the signal intensity profile of the Ndc80 complex. Bars, 2 µm.
We next visualized the binding of the Ndc80 complex to rhodamine-labeled, taxol-stabilized microtubules by fluorescence microscopy. In the presence of 100 mM KCl, Ndc80–EGFP was barely visible on the stable microtubules. However, upon addition of 100 nM unlabeled Dam1 complex, Ndc80–EGFP signals were readily observed on the microtubule lattice (Fig. 4 B). We conclude that the Dam1 complex enhances the affinity of the Ndc80 complex for stable microtubules in vitro.
To understand the collective behavior of the complexes on dynamic microtubules, we imaged Ndc80–EGFP in the presence of 100 nM unlabeled Dam1 complex using TIRF microscopy. Strikingly, the Ndc80 complex was now readily visible on the dynamic lattice and displayed features characteristic of Dam1 behavior: it persisted in discrete foci at the end of growing microtubules and tracked the depolymerizing ends of shrinking microtubules (Fig. 4 C and Video 7). To ask whether the association of Ndc80 with growing microtubule ends was continuous, we recorded streaming videos of Ndc80–EGFP in the presence of unlabeled Dam1 complex. We found that Ndc80 signals distal from the stable seed showed continuous outward displacement, and we did not find evidence for turnover at the temporal resolution achievable in our imaging system (Fig. 4 D and Video 8).
Dam1 and Ndc80 colocalize persistently at growing and shrinking microtubule plus ends
We considered two possibilities by which the Dam1 complex may increase the interaction of the Ndc80 complex with dynamic microtubule ends. First, the presence of Dam1 may alter properties of the microtubule lattice, thereby making it more favorable for Ndc80 association. Alternatively, the complexes may physically engage at dynamic microtubule ends. To distinguish between these possibilities, we simultaneously imaged Alexa Fluor 594–labeled Dam1 complex and Ndc80–EGFP complex on dynamic microtubules. We found that Alexa Fluor 594 and EGFP signals colocalized to lattice-associated spots that did not display movement during our observation and to distal spots that showed outward displacement (Fig. 4, E and F; and Video 9). Further experiments using FRAP will be necessary to clarify whether the complexes show turnover at the plus end.
On depolymerizing microtubule ends, Dam1 and Ndc80 spots colocalized, distal signals remained associated with depolymerizing ends, and lattice-associated signals were “picked up,” increasing the fluorescence intensity of the end-tracking species. These observations support the notion that the Ndc80 and Dam1 complexes directly interact on dynamic microtubules. Interestingly, however, we were not able to detect an interaction between the complexes in solution, as they eluted independently in gel filtration experiments (Fig. S3). Thus, the presence of microtubules strongly enhances interaction between these complexes on the lattice (see Tien et al. in this issue).
Kinetochore–microtubule interactions are under the control of the aurora B kinase (Ipl1p in budding yeast), which ensures bipolar attachment of kinetochores to opposite spindle poles. We asked whether the Dam1–Ndc80 interaction is sensitive to Ipl1 phosphorylation by performing microtubule cosedimentation experiments under conditions in which Ndc80 binding to microtubules is dependent on Dam1 (100 mM NaCl). To this end, Dam1 complex and substoichiometric amounts of Ipl1–Sli15 were incubated in the presence or absence of ATP and tested for their ability to recruit Ndc80 to microtubules. Recruitment of the Ndc80 complex to microtubules was diminished in the ATP-containing sample, whereas phosphorylation had little affect on the affinity of the Dam1 complex for microtubules itself (Fig. 5 A). We next tested whether Ipl1 phosphorylation affected Ndc80 recruitment to dynamic microtubules in the TIRF assay. We found that recruitment of the Ndc80 complex to discrete foci on the dynamic microtubule lattice was prevented by inclusion of Ipl1–Sli15 in an ATP-dependent manner (Fig. 5 B). Additionally, we found that a mutant Dam1 complex mimicking Ipl1 phosphorylation on four previously established sites (Dam1 S4D) recruited less Ndc80 than wild-type (Fig. 5 C), but was not as severely affected as the in vitro–phosphorylated complex, suggesting that additional Ipl1 phosphorylation sites on the Dam1 complex may contribute to the inhibition of Ndc80 recruitment. In summary, our results suggest that phosphorylation by Ipl1–Sli15 prevents the Dam1-dependent recruitment of the Ndc80 complex to stable and dynamic microtubules.
Figure 5.
Phosphorylation of Dam1 by Ipl1–Sli15 prevents Ndc80 recruitment to microtubules. (A) The Ndc80 complex was tested for microtubule binding in the presence of phosphorylated and unphosphorylated Dam1 complex. (top) The phosphorylation experiments are outlined. The binding affinity curves plot averaged data from two independent experiments. Error bars represent SEM. (B) TIRF micrographs showing that the Dam1 complex phosphorylated by Ipl1–Sli15 fails to recruit the Ndc80 complex to dynamic microtubules (right). Bars, 2 µm. (C) The ability of the dam1 S4D (S20D, S257D, S265D, and S292) complex to recruit the Ndc80 complex to microtubules was tested in the presence of 100 mM NaCl. The dam1 S4D complex has a decreased ability to recruit Ndc80. The binding affinity curves plot averaged data from two independent experiments. SN, supernatant; P, pellet. Error bars represent SEM.
Conclusions
Our results reveal novel biochemical activities for the Dam1 complex and can explain how it is connected to the rest of the kinetochore. We show that the complex is an autonomous, continuous plus end tracker that recruits Ndc80 in a microtubule-dependent manner to dynamic plus ends. We speculate that the Dam1 complex may exploit its high affinity for GTP-tubulin to diffuse back to the plus end as the microtubule grows.
Furthermore, our results immediately explain several in vivo observations. First, in contrast to human cells, where deletion of the unstructured N terminus of Ndc80 leads to loss of kinetochore–microtubule attachments (Guimaraes et al., 2008; Miller et al., 2008), eliminating the corresponding part in the yeast Ndc80 complex has no effect on viability (Kemmler et al., 2009). Thus, severely crippling the microtubule-binding activity of Ndc80 does not prevent chromosome segregation, suggesting that the microtubule-binding activity provided by Dam1 is sufficient in this system. Second, localization of the Dam1 complex to centromeres, as assayed by chromatin immunoprecipitation, is both dependent on the Ndc80 complex and on microtubules (Li et al., 2002). This can be explained by the microtubule-dependent engagement that we observed in vitro. Our results support the notion that a mature kinetochore–microtubule interface is compositely formed from the centromere and the microtubule side. The role of the Dam1 complex is to provide end recognition and continued association, and the defined geometry with which the calponin homology domains of the Ndc80 complex emerge from microtubule lattice (Wilson-Kubalek et al., 2008) allows the connection to the rest of the kinetochore via the Spc24/25 heads. Future experiments include a detailed structural investigation of the Dam1–Ndc80 interface and a mechanistic understanding of the microtubule-specific interaction between these complexes.
Materials and methods
Yeast genetics
All Westermann laboratory strains are derived from strain S288C. Standard procedures were applied to genetically modify strains.
Live cell imaging
Cells expressing Dam1-3xGFP and mCherry-Tub1 (FLY26: MAT α, lys2-801, ade2-1, leu2, cdc20::TRP1::GAL1/10-CDC20, mCherry-tubulin::URA3, and Dam1-3xGFP::HIS3; or FLY48: MAT a, lys2-801, ade2-1, leu2, cdc20::TRP1::GAL1/10-CDC20, mCherry-tubulin::URA3, Dam1-3xGFP::HIS3, and pGal-3xHA-Bim1::HIS3) were arrested in metaphase for 5 h in synthetic medium containing 2% glucose. Time-lapse images were collected at ambient temperature (21°C) using live cell microscopy (DeltaVision; Applied Precision), a UPlanSApo 100× NA 1.40 oil immersion objective lens (Olympus), and a charge-coupled device (CCD) camera (CoolSnap HQ; Photometrics). Z stacks (0.5 µm) were acquired in 20–30-s intervals, subsequently deconvoluted, projected to two-dimensional images (SoftWoRx software; Applied Precision), and further analyzed by MetaMorph (MDS Analytical Technologies) or ImageJ (National Institutes of Health) software.
Purification of recombinant complexes from bacteria
Expression, purification, and labeling of the Dam1 complex were performed as previously described (Westermann et al., 2005). The two subcomplexes of the Ndc80 complex (Ndc80p–6xHis/Nuf2p–EGFP or ΔN1-116–Ndc80p–6xHis/Nuf2p–EGFP, Spc24p–6xHis/Spc25p) were separately expressed from the pETDuett or pACYCDuet-1 vectors (EMD). Both plasmids were cotransfected into BL21 DE3 (EMD). Bacteria were grown to OD600 = 0.6 at 37°C, induced with 0.2 mM IPTG, and grown for 12–15 h at 18°C. The two subcomplexes were eluted with 150 mM NaCl, 10 mM Hepes, pH 7.0, and 250 mM imidazol from the Ni-NTA beads then subjected to in vitro reconstitution of the full-length Ndc80 complex and further purified on the Superdex 200 HiLoad 16/60 (GE Healthcare). Gel filtration was conducted in 150 mM NaCl and 10 mM Hepes, pH 7.
Microtubule-binding assays
The microtubule cosedimentation assay was performed as described previously (Cheeseman et al., 2001a). Purified Ndc80–EGFP and –Dam1 complexes were precleared and added to a final concentration of 0.4–0.5 µM to taxol-stabilized microtubules. The salt concentration was adjusted to 20, 25, 50, or 100 mM NaCl. Quantification was performed as previously described (Zimniak et al., 2009). To determine apparent KD for Ndc80–EGFP complex binding to microtubules, data points were fitted into the equation Y = Bmax × X/(KD + X) using Prism (version 4.0; GraphPad Software, Inc.).
Analytical gel filtration
Ndc80 and the Dam1 complex were mixed in an equimolar ratio at 5.5 µM and incubated for 10 min at RT. 0.5 ml was loaded onto the Superdex 200 HiLoad 16/60. Proteins were eluted in buffer containing 100 mM NaCl and 10 mM Hepes, pH 7.
In vitro reconstitution of a plus end–tracking system
Flow cells were constructed using silanized glass slides, two double-sided tapes, and precleaned coverslips. Resulting chambers were blocked with 5 mg/ml biotinylated BSA (Vector Laboratories) for at least 3 h and subsequently washed with BRB80 buffer. Afterward, a solution of 0.3 mg/ml avidin DN (Vector Laboratories) in BRB80 was introduced for at least 20 min and exchanged for a blocking solution containing 0.1% pluronic F-127 (Sigma-Aldrich) in BRB80 (Westermann et al., 2006; Zimniak et al., 2009). Rhodamine-labeled short GMPCPP (2'-deoxy-guanosince-5'-[(α,β)-methyleno]triphosphate) microtubule seeds were introduced and incubated for 30 s followed by a wash with wash buffer (BRB80 supplemented with 150 mM KCl, 1 mM GTP, 0.5% [vol/vol] β-mercaptoethanol, 4.5 µg/ml glucose, 200 µg/ml glucose oxidase, and 35 µg/ml catalase). Microtubule growth was induced by introducing a reaction mix composed of 14 µM tubulin (unlabeled or 1 µM labeled with rhodamine), 150 mM KCl, 0.6 mM GTP, 0.13% (wt/vol) methylcellulose, 0.33 mg/ml casein, 0.5% (vol/vol) β-mercaptoethanol, 4.5 µg/ml glucose, 200 µg/ml glucose oxidase, 35 µg/ml catalase, and 100 nM Dam1–Alexa Fluor 488 complex. Time-lapse videos were recorded at 30°C using 4–5-s intervals between frames. Image acquisition was performed using the TIRF3 microscopy system operated with AxioVision software (Carl Zeiss, Inc.), a 100× Plan Apochromat 1.46 NA objective, and an EM CCD camera (C9100-02; Hamamatsu Photonics). For streaming videos, concentration of free tubulin was increased to 20 µM and the time resolution to 400 ms/frame. For Ndc80–EGFP (300 nM) and Dam1–Alexa Fluor 594 (10 nM labeled + 90 nM unlabeled Dam1 complex) experiments, we used coverslips grafted with polyethylene glycol biotin, 100 mM KCl, and an EM CCD camera (Cascade II; Photometrics).
In vitro phosphorylation of the Dam1 complex
0.5 µM recombinant Ipl1–Sli15 complex was incubated with 2.5 µM recombinant Dam1 wild-type complex in kinase buffer (20 mM Hepes, pH 7.5, 100 mM KCl, 10 mM MgCl2, 25 mM β-glycerophosphate, and 1 mM DTT) in the presence or absence of 1 mM ATP. The reaction was performed at 30°C for 40 min.
EM
Short GMPCPP microtubule seeds were incubated for 5 min with a solution containing 14 µM tubulin, 150 mM KCl, 0.6 mM GTP, 0.33 mg/ml casein, 0.5% (vol/vol) β-mercaptoethanol, 4.5 µg/ml glucose, 200 µg/ml glucose oxidase, 35 µg/ml catalase, and variable concentrations of the Dam1–Alexa Fluor 488 complex in BRB80 buffer. The reaction mix was placed on a carbon-coated grid, negatively stained with 2% uranyl acetate, and imaged immediately with an electron microscope operating at 80 kV (Morgagni; FEI) equipped with a CCD camera (Morada SIS; Olympus).
Online supplemental material
Fig. S1 addresses the oligomerization status of the Dam1 complex. Fig. S2 demonstrates that the N-terminal tail of Ndc80p is not essential for Dam1-dependent recruitment to microtubules. Fig. S3 shows that there is no interaction between the Ndc80 complex and the Dam1 complex in solution. Videos 1 and 2 visualize plus end tracking of the Dam1 complex in vivo. Video 3 shows that Dam1 tip localization is independent of the +TIP Bim1p. Videos 4–6 demonstrate autonomous microtubule plus end–tracking activity of the Dam1 complex in vitro. Video 7 displays plus end tracking of the Ndc80–EGFP complex in the presence of unlabeled Dam1 complex. Video 8 shows continuous tip tracking of the Ndc80–EGFP complex in the presence of unlabeled Dam1 complex. Video 9 demonstrates that the Dam1 complex and the Ndc80 complex colocalize on dynamic microtubule plus ends. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912021/DC1.
Acknowledgments
We wish to thank all members of the Westermann laboratory for discussions, Karin Aumayr and Pawel Pasierbek for help with TIRF microscopy and image analysis, and Carrie Cowan, René Ladurner, and Jan-Michael Peters for critical reading of the manuscript.
The research leading to these results has received funding from the European Research Council (ERC) under the European Community's Seventh Framework Program (grant FP7/2007-2013; ERC grant agreement 203499) and by the Austrian Science Fund (FWF; grant SFB F34-B03).
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- TIRF
- total internal reflection fluorescence
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Asbury C.L., Gestaut D.R., Powers A.F., Franck A.D., Davis T.N. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., Surrey T. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Brew C., Wolyniak M., Desai A., Anderson S., Muster N., Yates J.R., Huffaker T.C., Drubin D.G., Barnes G. 2001a. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137–1145 10.1083/jcb.200109063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Enquist-Newman M., Müller-Reichert T., Drubin D.G., Barnes G. 2001b. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212 10.1083/jcb.152.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Efremov A.K., Volkov V.A., Spiridonov I.S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J.R., Ataullakhanov F.I. 2008a. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA. 105:15423–15428 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Spiridonov I.S., Volkov V.A., Efremov A., Westermann S., Drubin D., Barnes G., Ataullakhanov F.I., McIntosh J.R. 2008b. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA. 105:6918–6923 10.1073/pnas.0801811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28:1099–1110 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E., Woehrer S., Peng Y., Mechtler K., Westermann S. 2009. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat. Cell Biol. 11:1109–1115 10.1038/ncb1924 [DOI] [PubMed] [Google Scholar]
- Lacefield S., Lau D.T., Murray A.W. 2009. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat. Cell Biol. 11:1116–1120 10.1038/ncb1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A.A., Wang Y., Qin J., Elledge S.J. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197 10.1101/gad.959402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.A., Johnson M.L., Stukenberg P.T. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 18:1785–1791 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C., Hazbun T.R., Cheeseman I.M., Aranda J., Fields S., Drubin D.G., Barnes G. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 14:3342–3355 10.1091/mbc.E02-11-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 434:987–994 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178:269–281 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., Harrison S.C. 2007. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14:54–59 10.1038/nsmb1186 [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., Barnes G. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 10.1146/annurev.biochem.76.052705.160607 [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek E.M., Cheeseman I.M., Yoshioka C., Desai A., Milligan R.A. 2008. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 182:1055–1061 10.1083/jcb.200804170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Nakajima Y., Westermann S., Shang C., Kang J.S., Goodner C., Houshmand P., Fields S., Chan C.S., Drubin D., et al. 2007. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 18:3800–3809 10.1091/mbc.E07-06-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]