The Dam1 complex, regulated by aurora B phosphorylation, confers a more stable microtubule association for the Ndc80 complex at kinetochores (see also related paper by Lampert et al. in this issue).
Abstract
The coupling of kinetochores to dynamic spindle microtubules is crucial for chromosome positioning and segregation, error correction, and cell cycle progression. How these fundamental attachments are made and persist under tensile forces from the spindle remain important questions. As microtubule-binding elements, the budding yeast Ndc80 and Dam1 kinetochore complexes are essential and not redundant, but their distinct contributions are unknown. In this study, we show that the Dam1 complex is a processivity factor for the Ndc80 complex, enhancing the ability of the Ndc80 complex to form load-bearing attachments to and track with dynamic microtubule tips in vitro. Moreover, the interaction between the Ndc80 and Dam1 complexes is abolished when the Dam1 complex is phosphorylated by the yeast aurora B kinase Ipl1. This provides evidence for a mechanism by which aurora B resets aberrant kinetochore–microtubule attachments. We propose that the action of the Dam1 complex as a processivity factor in kinetochore–microtubule attachment is regulated by conserved signals for error correction.
Introduction
During mitosis, kinetochores attach to assembling and disassembling microtubule tips while withstanding tensile forces from the mitotic spindle (Skibbens et al., 1993, 1995; Maddox et al., 2003). Kinetochores are able to harness energy from these disassembling microtubule tips to drive movement of chromosomes (for a review see Inoué and Salmon, 1995). Understanding how the kinetochore establishes microtubule attachments under force requires understanding the organization of the kinetochore components and how they bear and transmit load. Recent studies investigated the spatial organization of kinetochore components in vivo and how their arrangement changes throughout mitosis (Joglekar et al., 2009; Wan et al., 2009). Through systematic reconstitution of kinetochore components, we are pursuing a complementary approach with the ultimate goal of mapping the transmission of force across the kinetochore from the dynamic microtubule to the centromere. In this study, we focus on the kinetochore–microtubule interface.
The kinetochores of all eukaryotes contain multiple microtubule-binding elements. The KMN network (KNL-1, Mis12 complex, and Ndc80 complex) and the Ska1 complex both bind microtubules in higher eukaryotic cells (Cheeseman et al., 2006; Gaitanos et al., 2009; Welburn et al., 2009). Yeast also contain the KMN network and the Dam1 complex, possibly the functional homologue of the Ska1 complex (Hanisch et al., 2006; Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009). Cooperation of the three components of the conserved KMN network was shown by cosedimentation with taxol-stabilized microtubules (Cheeseman et al., 2006), but how or whether any of the microtubule-binding components cooperate to achieve attachment to dynamic microtubules is unknown. We show for the first time that cooperation between two kinetochore subcomplexes enhances processive, load-bearing coupling to dynamic microtubule tips.
In the budding yeast kinetochore, all four proteins of the Ndc80 complex and all 10 proteins of the Dam1 complex are essential (Tanaka and Desai, 2008). In vitro, both complexes independently form diffusive attachments to the microtubule lattice and track with disassembling microtubule tips, although the Ndc80 complex requires artificial oligomerization to tip track (Westermann et al., 2006; Gestaut et al., 2008; Powers et al., 2009). The Dam1 complex also tracks robustly with polymerizing microtubules in vitro (Asbury et al., 2006; see Lampert et al. in this issue). When attached to beads, each complex forms load-bearing attachments to dynamic microtubule tips (Asbury et al., 2006; Franck et al., 2007; Grishchuk et al., 2008a,b; Powers et al., 2009). Despite these similarities, the Ndc80 and Dam1 complexes are not redundant. The Ndc80 complex is required in vivo for attachment to microtubules (Kline-Smith et al., 2005), and the Dam1 complex is required for attaching to the tips of microtubules and for establishing biorientation (Tanaka et al., 2005; Shimogawa et al., 2006). Moreover, the Ndc80 complex is required for the assembly of Dam1 complex onto the kinetochore (Janke et al., 2002), and an interaction between the two complexes has been suggested by localization and two-hybrid studies (Shang et al., 2003; Joglekar et al., 2009). Studying the combination of Ndc80 and Dam1 complexes in vitro will allow us to dissect their distinct roles in kinetochore–microtubule binding.
Kinetochores not only serve as physical bridges between chromosomes and spindle microtubules but are also regulatory hubs that ensure chromosome segregation fidelity during mitosis. For example, aurora B kinase is responsible for resetting aberrant kinetochore–microtubule attachments to achieve biorientation (Cheeseman et al., 2002; Tanaka et al., 2002; Hauf et al., 2003; Pinsky et al., 2006). Many of the microtubule-binding components of the kinetochore, including the Ndc80 and Dam1 complexes, are targets of aurora B (Cheeseman et al., 2002, 2006; Shang et al., 2003; DeLuca et al., 2006; Pinsky et al., 2006; Gestaut et al., 2008). In mammalian cells, aurora B phosphorylation of the N-terminal tail of the Ndc80 protein (Hec1 in humans) abolishes kinetochore–microtubule attachment (DeLuca et al., 2006; Guimaraes et al., 2008). Although the budding yeast Ndc80 protein also has an N-terminal tail, it is not essential (Akiyoshi et al., 2009; Kemmler et al., 2009). Previously, we demonstrated that phosphorylation by the yeast aurora B homologue Ipl1 at one target site within the Dam1 complex, Ser20 of Dam1, reduces its affinity for the microtubule lattice (Gestaut et al., 2008). Two-hybrid assays and pull-downs with in vitro–translated proteins using phosphomimetic mutations at Ipl1 target sites in Dam1 also suggested that phosphorylation of the Dam1 complex modulates its interaction with the Ndc80 complex (Shang et al., 2003). Moreover, Ipl1 target sites on Dam1 are dephosphorylated as cells enter metaphase in a cohesin-dependent manner, which could prevent kinetochore–microtubule attachment turnover as biorientation is established (Keating et al., 2009).
In this study, we show that the Dam1 complex is a phospho-regulated processivity factor for the Ndc80 complex in kinetochore–microtubule coupling. Using techniques for tracking and manipulating single molecules in vitro, we demonstrate directly an interaction between the Ndc80 and Dam1 complexes on microtubules. Through this interaction, the Dam1 complex enhances the ability of the Ndc80 complex to maintain attachment to dynamic microtubule tips even in the presence of external load. Finally, this interaction is regulated by Ipl1, further defining the mechanism for aurora B–mediated corrective detachment in vivo.
Results
The Dam1 and Ndc80 complexes interact on microtubules
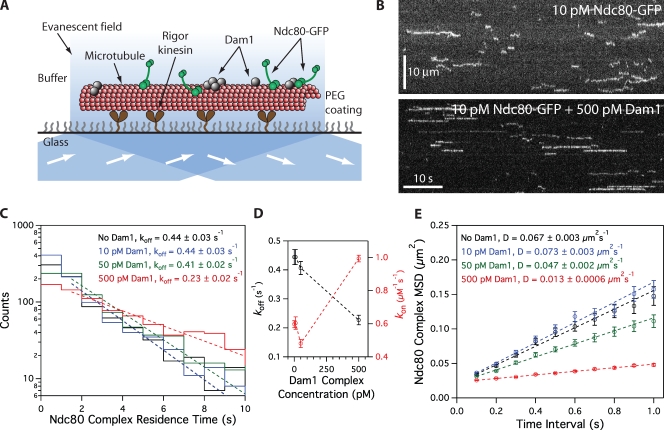
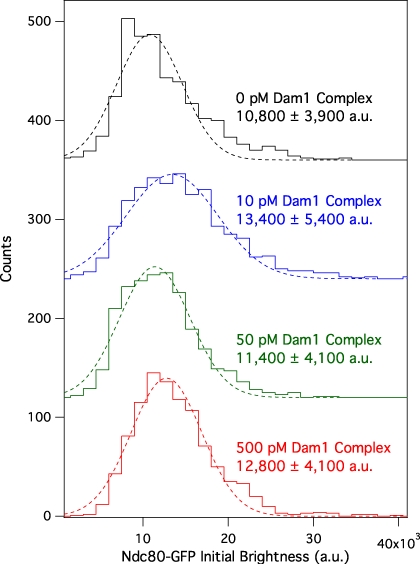
We expressed recombinant Saccharomyces cerevisiae Ndc80 and Dam1 complexes in Escherichia coli and purified each complex by affinity chromatography and gel filtration (Wei et al., 2005; Gestaut et al., 2008; Powers et al., 2009). By velocity sedimentation analysis, we found weak interaction between the Ndc80 and Dam1 complexes free in solution (Fig. S1). Using total internal reflection fluorescence (TIRF) microscopy, we quantified the interaction of GFP-tagged Ndc80 complexes with microtubules in the presence and absence of Dam1 complex (Fig. 1). In the absence of Dam1 complex, individual Ndc80 complexes formed transient and diffusive attachments to microtubules, as reported previously (Powers et al., 2009). We measured a dissociation rate constant (koff) of 0.44 ± 0.03 s−1, an association rate constant (kon) of 0.60 ± 0.02 µM−1 × s−1, and a diffusion constant of 0.067 ± 0.003 µm2 × s−1 (Fig. 1, C–E), which are values comparable with our previous study (Powers et al., 2009). We also simultaneously visualized GFP-tagged Ndc80 complexes and mCherry-tagged Dam1 complexes on microtubules. At concentrations affording single molecule resolution of each complex, interaction events were rare. When the two complexes did associate with each other, they appeared to diffuse more slowly (Fig. S2). However, interaction events between individual Ndc80 and Dam1 complexes were too infrequent to affect population behavior. To increase the frequency of interactions, we raised the concentration of Dam1 complex while maintaining low concentrations (10 pM) of the Ndc80 complex. Overall, Ndc80 complex transitioned gradually to a more persistent and more slowly diffusing behavior as the concentration of Dam1 complex was increased (Fig. 1, C–E). At 500 pM Dam1 complex, the Ndc80 complex dissociated twofold more slowly from the microtubule (koff = 0.23 ± 0.02 s−1) and associated 1.6-fold faster onto the microtubule (kon = 0.99 ± 0.02 µM−1 × s−1) as compared with the Ndc80 complex alone. This corresponds to a threefold decrease in the apparent equilibrium dissociation constant, Kd = koff × kon−1 (0.74 ± 0.06 to 0.23 ± 0.02 µM). At 500 pM Dam1 complex, the Ndc80 complex also diffused fivefold more slowly (0.013 ± 0.0006 µm2 × s−1) as compared with Ndc80 complex alone. The Dam1 complex was unlikely to be acting as a simple barrier to diffusional motility, as the diffusive behavior of the Ndc80 complex was unchanged in the presence of phosphorylated Dam1 complex at the same lattice density (see Ipl1 phosphorylation regulates…). The brightness distribution of the GFP signal remained unchanged across concentrations of the Dam1 complex, demonstrating that oligomerization of the Ndc80 complex did not contribute to its modified behavior in the presence of the Dam1 complex (Fig. 2). Even at 500 pM Dam1 complex, not all Ndc80 complexes bound persistently and diffused slowly. This indicates that not all Ndc80 complexes were associated with Dam1 complexes, so our calculated values describe a mixed population and likely underestimate Dam1 complex–mediated enhancement of Ndc80 complex–microtubule interactions.
Figure 1.
Dam1 complex enhances binding of individual Ndc80 complexes to microtubules. (A) Schematic of the TIRF assay developed to visualize the behavior of GFP-tagged Ndc80 complexes (green rods) in the presence of untagged Dam1 complexes (gray spheres) on microtubules. (B) Representative kymographs showing the binding and one-dimensional diffusion of 10 pM Ndc80 complexes on taxol-stabilized microtubules in the absence or presence of 500 pM Dam1 complex. Positions along the microtubule are shown on the vertical axis, whereas the passage of time is depicted along the horizontal axis. Concentrations are of free complexes in solution. (C) Residence time distributions of 10 pM Ndc80 complex on microtubules without Dam1 complex (black histogram; n = 883 events), with 10 pM Dam1 complex (blue histogram; n = 966), 50 pM Dam1 complex (green histogram; n = 928), and 500 pM Dam1 complex (red histogram; n = 1,003). Dotted lines show the weighted exponential fits used to determine dissociation rate constants, koff. (D) Dissociation rate constants (koff; left axis, black markers) for the Ndc80 complex, calculated from the data in C, are plotted against the concentration of Dam1 complex. Association rate constants (kon; right axis, red markers) of the Ndc80 complex are also plotted (without Dam1 complex, n = 1,103; with 10 pM Dam1 complex, n = 1,426; with 50 pM Dam1 complex, n = 1,179; with 500 pM Dam1 complex, n = 1,412). (E) Mean-squared displacement (MSD) is plotted against time for 10 pM Ndc80 complex on microtubules without Dam1 complex (black markers; n = 803 events), with 10 pM Dam1 complex (blue markers; n = 859), 50 pM Dam1 complex (green markers; n = 883), and 500 pM Dam1 complex (red markers; n = 968). Dotted lines show the weighted linear fits used to determine diffusion constants, D. Markers indicate SEM.
Figure 2.
Dam1 complex does not affect the oligomerization state of the Ndc80 complex on microtubules. Mean initial brightness distributions of 10 pM GFP-tagged Ndc80 complex–binding events on microtubules without Dam1 complex (black histogram; n = 883 events), with 10 pM Dam1 complex (blue histogram; n = 966), 50 pM Dam1 complex (green histogram; n = 928), and 500 pM Dam1 complex (red histogram; n = 1,003). Dotted lines show Gaussian fits used to determine mean values ± SD. These values are similar to the mean brightness from rare single-bleach steps of GFP-tagged Ndc80 complex (9,300 ± 3,200 au; n = 11). For clarity, green, blue, and black histograms are offset vertically by 120, 240, and 360 counts, respectively.
In the presence of the Dam1 complex, diffusion of the Ndc80 complex is slowed far below the reported rate for a single Dam1 complex (Gestaut et al., 2008). Therefore, we hypothesized that at the concentrations required to observe significant changes in the population behavior of the Ndc80 complex, the Dam1 complex forms slowly diffusing oligomers. To test this, we measured the diffusion rate of GFP-tagged Dam1 complex on microtubules (Fig. S3, A and B). At 2 pM, single GFP-tagged Dam1 complexes diffused rapidly, at 0.060 ± 0.003 µm2 × s−1, which is similar to the rates reported previously (Westermann et al., 2006; Gestaut et al., 2008). However, at 20 and 50 pM Dam1 complex, we observed slowly diffusing spots that exhibited fluorescence brighter than individual Dam1 complexes. To maintain single molecule resolution for quantifying the diffusion of Dam1 complex at higher concentrations, we mixed untagged Dam1 complex with a small amount of GFP-tagged Dam1 complex. At 500 pM, Dam1 complex diffused at least 60-fold more slowly than at 2 pM (Fig. S3 B). These observations indicate that oligomerization of the Dam1 complex slows its diffusion rate, as reported previously (Grishchuk et al., 2008a). Moreover, they imply that the enhanced binding of Ndc80 complex to microtubules that we have quantified here (Fig. 1, C–E) occurs via interaction with Dam1 complexes that are primarily in an oligomeric state.
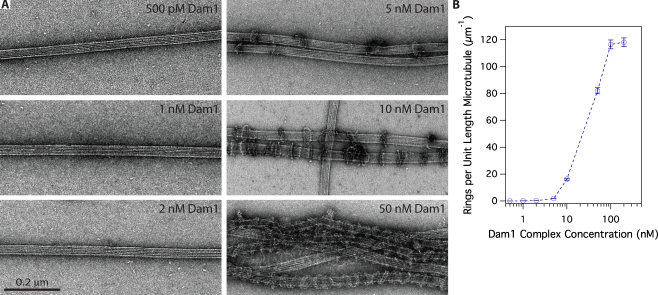
In vitro, the Dam1 complex forms rings of 16–25 complexes that encircle microtubules (Miranda et al., 2007; Wang et al., 2007). To investigate whether rings are important for interaction with the Ndc80 complex, we used negative-stain EM to quantify ring formation on taxol-stabilized microtubules (at 36 nM tubulin) across a range of Dam1 concentrations (Fig. 3). At 500 pM Dam1 complex, the highest concentration used in our TIRF assays, rings were absent. Instead, we observed small particles scattered around or attached to the filaments. The dimensions of these particles were consistent with Dam1 complex dimers (Wang et al., 2007). Rings first appear on microtubules at 1 nM Dam1 complex, substantially increase in density between 5 and 10 nM, and saturate at 100 nM (Table I). These findings are consistent with a strong and cooperative binding of the Dam1 complex to microtubules, as reported previously (Gestaut et al., 2008).
Figure 3.
Assembly of oligomeric rings of the Dam1 complex around microtubules. (A) Negative-stain electron micrographs of oligomeric rings formed by the Dam1 complex around taxol-stabilized microtubules. (B) The number of rings observed per unit length (micrometers) of microtubule was quantified (statistics shown in Table I) and plotted against the total concentration of Dam1 complex. Error bars represent counting uncertainties.
Table I.
EM of ring formation on microtubules at different Dam1 complex concentrations
| Dam1 complex concentration | No. of microtubules | Total microtubule length | No. of rings | Ring density |
| nM | µm | µm−1a | ||
| 0.5 | 27 | 259 | 0 | 0 |
| 1 | 16 | 280 | 20 | 0.1 ± 0.02 |
| 2 | 26 | 308 | 76 | 0.2 ± 0.03 |
| 5 | 26 | 256 | 450 | 2 ± 0.08 |
| 10 | 18 | 33 | 535 | 17 ± 0.7 |
| 50 | 13 | 16 | 1,312b | 82 ± 3 |
| 100 | 11 | 11 | 1,282b | 120 ± 3 |
| 200 | 9 | 11.5 | 1,358b | 120 ± 3 |
Errors represent counting uncertainties.
Rings stacked together in pairs to form doublets and/or coils.
Although 500 pM Dam1 complex did not assemble into rings on microtubules at 36 nM tubulin, reducing the amount of tubulin could promote ring formation by increasing the density of Dam1 complex bound to microtubules. To explore the magnitude of this effect, we imaged 500 pM Dam1 complex on microtubules at fivefold lower tubulin (7 nM). Rings were again absent (n = 8 microtubules; 101 µm total). Further reductions in tubulin concentration were impractical because the microtubules became too sparse on the EM grids. Because the effective concentration of tubulin polymer in our TIRF assays was lower still (∼1 nM), it remains possible that Dam1 complex rings contributed to the observed alterations in behavior of the Ndc80 complex. However, we note that two observations suggest that ring formation is not required for the initial interaction between the Ndc80 and Dam1 complexes. First, the Dam1 and Ndc80 complexes interact during velocity sedimentation, where the Dam1 complex is primarily in dimeric form (Fig. S1). Second, interactions between individual Ndc80 and Dam1 complexes can be observed directly in TIRF assays (albeit rarely; Fig. S2).
The Dam1 complex enhances attachment of the Ndc80 complex to dynamic microtubule tips
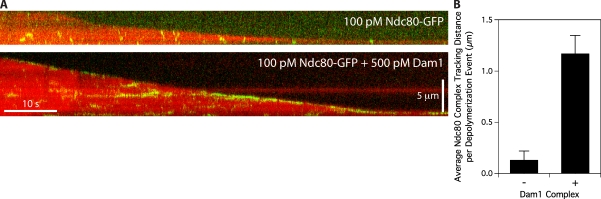
The Ndc80 complex has been shown to track efficiently with disassembling microtubule tips in vitro, but only when it is bound to beads or to antibodies (Powers et al., 2009). In contrast, the Dam1 complex tracks robustly with disassembling tips without artificial oligomerization (Westermann et al., 2006; Gestaut et al., 2008). Therefore, we tested whether the Dam1 complex enhances tip tracking by the Ndc80 complex. We grew microtubules from nonhydrolyzable GMPCPP seeds in the presence of free fluorescent-labeled tubulin and GTP. We visualized the behavior of GFP-tagged Ndc80 complex as microtubules disassembled after the free tubulin was removed. By itself, the Ndc80 complex localized only briefly to microtubule tips during disassembly (Fig. 4 A). Most binding events were transient and diffusive, which is similar to those seen on taxol-stabilized microtubules (Powers et al., 2009). In contrast, the addition of Dam1 complex, which accumulates at the disassembling microtubule tip (Fig. S3 C), substantially increased the tip tracking behavior of the Ndc80 complex (Fig. 4 A). Ndc80 complexes bound preferentially at the microtubule tip were more persistently attached and moved with the disassembling tip.
Figure 4.
Ndc80 complex tracks with disassembling tips in the presence of Dam1 complex. (A) Representative two-color kymographs showing the tip tracking ability of 100 pM Ndc80 complex in the presence or absence of 500 pM Dam1 complex. Movement of GFP-tagged Ndc80 complex (green) is shown on disassembling microtubules (red). Concentrations are of free complexes in solution. (B) Mean tracking distance of Ndc80 complex per depolymerization event in the absence of Dam1 complex (n = 19) or in the presence of 500 pM Dam1 complex (n = 62). Error bars indicate SEM.
For quantification, we defined tip tracking as the colocalization of GFP-tagged Ndc80 complex with disassembling microtubule tips. In the presence of Dam1 complex, Ndc80 complex tracked with 78% (62/80) of disassembling microtubule tips over a mean distance of 1.2 ± 0.2 µm compared with only 27% (19/71) of tips over a mean distance of 0.13 ± 0.09 µm in the absence of Dam1 complex (Fig. 4 B). In the presence of the Dam1 complex, tip tracking events by the Ndc80 complex often continued until the tips reached the microtubule seeds. Therefore, we likely underestimate the effect of the Dam1 complex to enhance the ability of the Ndc80 complex to track disassembling tips.
We then used an optical trapping-based force clamp (Asbury et al., 2006; Franck et al., 2007, 2010; Powers et al., 2009) to test whether the Dam1 complex enhances the tip-tracking ability of Ndc80 complex while under load. We attached beads decorated with Ndc80 complex to the tips of assembling microtubules in the presence and absence of free Dam1 complex. We applied constant tensile force until the attachment broke, the microtubule switched to disassembly, or, in a few cases, the event was terminated by other causes (e.g., the bead became stuck to the coverslip). In the absence of Dam1 complex, bead-bound Ndc80 complex formed persistent load-bearing attachments to assembling and disassembling microtubule tips (Fig. 5) as reported previously (Powers et al., 2009). While bearing 1.8 ± 0.4 pN (mean ± SD) of continuous load, travel distances during assembly were broadly distributed with a mean of 350 nm (n = 115). Detachment from assembling tips occurred at a rate of 0.026 ± 0.003 s−1 (Fig. 5 B). To mimic the likely arrangement in vivo, we added free Dam1 complex lacking an affinity tag so that it interacted with the beads only via its interaction with Ndc80 complex (i.e., direct Dam1 complex–bead interactions were prevented; see Materials and methods). In the presence of the Dam1 complex, the mean travel distance increased threefold to 1,100 nm (n = 42; P = 3 × 10−8 by Kolmogorov-Smirnov test), and the detachment rate decreased fivefold to 0.005 ± 0.0008 s−1 (Fig. 5 B). Accordingly, plots of survival probability versus distance show that the couplers remained more persistently attached when Dam1 complex was present (Fig. 5 C).
Figure 5.
Dam1 complex enhances the coupling of bead-bound Ndc80 complex to assembling microtubule tips under fixed load. (A) Representative records of bead position versus time for microtubule tip attachments by bead-bound Ndc80 complex in the absence (blue traces) or presence (red traces) of free Dam1 complex during continuous application of tensile load. Increasing position represents assembly-coupled movement in the direction of applied force. Arrows mark transitions from assembly to disassembly. Decreasing position represents disassembly-driven movement against the applied force. Circles indicate detachment. For clarity, each record is offset vertically by an arbitrary amount. (B) Rates of bead detachment from assembling microtubule tips are estimated by counting the number of detachment events and dividing by total observation time. Error bars represent uncertainty based on Poisson statistics. (C) Survival probability versus distance for attachments composed of bead-bound Ndc80 complex in the absence (blue) or presence (red) of free Dam1 complex. The survival probability is the number of events that persisted beyond a given distance divided by the total number of events.
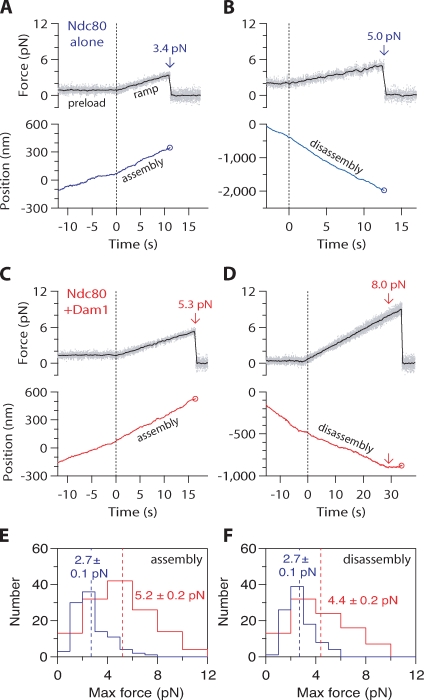
We also developed a force ramp assay to test the coupling performance of bead-bound Ndc80 complex across a broader range of forces on both assembling and disassembling microtubule tips (Franck et al., 2010). After an initial preload period at ∼1 pN constant force, we gradually increased the force on a tip-attached bead at a constant rate (0.25 pN × s−1) until the bead detached from the microtubule tip, the load limit of the trap (10–12 pN) was reached, or, in the case of disassembling filaments, the microtubule switched from shortening to growth (Fig. 6). The maximum force achieved before any one of these termination points was recorded for each event. Without Dam1 complex present, all events recorded during microtubule assembly ended in detachment. Most events during disassembly also ended in detachment (93/96), but a few ended with a shortening to growth transition (2/96) or when the trap load limit was reached (1/96). The resulting maximum forces were distributed narrowly, with means of 2.7 ± 0.1 pN (n = 101) during assembly and 2.7 ± 0.1 pN (n = 96) during disassembly (Fig. 6, E and F). The addition of Dam1 complex resulted in a clear improvement in the load-bearing capacity of the Ndc80 complex–coated beads. Most events recorded during assembly ended in detachment (112/131), but some persisted until the trap load limit was reached (19/131). Of the events recorded during disassembly, only about half ended in detachment (43/92). The remainder terminated when the microtubule switched to assembly (43/92), or, in a few cases, when the load limit was reached (6/92). The high frequency of shortening to growth transitions indicates that tension applied through linkages composed of both Ndc80 and Dam1 complexes promotes microtubule rescue, a phenomenon we saw previously using bead-bound Dam1 complex alone (Franck et al., 2007). The resulting maximum forces were distributed broadly with means of 5.2 ± 0.2 pN during assembly (n = 131) and 4.4 ± 0.2 pN during disassembly (n = 92), values that are twofold higher than in the absence of Dam1 complex (assembly, P < 1 × 10−8; disassembly, P = 1 × 10−8). These observations, together with the force clamp results, show that interactions between Dam1 and Ndc80 complexes enhance coupling to both assembling and disassembling microtubule tips under load. This enhancement persists across a range of loads (up to 10 pN), and it occurs under conditions in which the entire load is ultimately transmitted to the cargo through the Ndc80 complex.
Figure 6.
Dam1 complex enhances the coupling of bead-bound Ndc80 complex to assembling and disassembling microtubule tips across a range of loads. (A–D) Representative records showing tensile force (top) and bead position (bottom) versus time for bead-bound Ndc80 complexes attached to assembling and disassembling microtubule tips in the absence (A and B) or presence (C and D) of free Dam1 complex. The instrument was programmed to automatically increase the force at a constant rate (0.25 pN × s−1) after ∼500 nm of movement occurred. Arrows mark maximum forces, recorded either at rupture or when the microtubule switched from disassembly to assembly. Circles mark ruptures. (E) Distributions of maximum force for bead-bound Ndc80 complexes attached to assembling tips in the absence (blue histogram; n = 101) or presence (red histogram; n = 131) of free Dam1 complex. (F) Distributions of maximum force for bead-bound Ndc80 complexes attached to disassembling tips in the absence (blue histogram; n = 96), or presence (red histogram; n = 92) of free Dam1 complex. Dotted vertical lines indicate the mean for each distribution. Uncertainties represent standard errors.
Ipl1 phosphorylation regulates the interaction between Ndc80 and Dam1 complexes
We asked whether Ipl1 phosphorylation of the Dam1 complex regulates its interaction with the Ndc80 complex on microtubules. Phosphorylation of Ser20 on the Dam1 protein weakens the interaction of the Dam1 complex with microtubules (Gestaut et al., 2008). To determine how phosphorylation at sites other than Ser20 affects the interaction between the Dam1 and Ndc80 complexes, we used a modified Dam1 complex with a Ser20 to Ala mutation (S20A). With the S20A substitution, the Dam1 complex interacts with microtubules in a manner that is indistinguishable from the wild-type complex except that the interaction is insensitive to Ipl1 phosphorylation (Fig. S4, A and B). The phosphorylated S20A Dam1 complex also tracks with disassembling microtubule tips and is less diffusive at high concentrations, as expected for oligomers (Fig. S3). Phosphorylated S20A Dam1 complex also slows the disassembly of microtubules, as reported for wild-type Dam1 complex (Westermann et al., 2006; Franck et al., 2007; Grishchuk et al., 2008a).
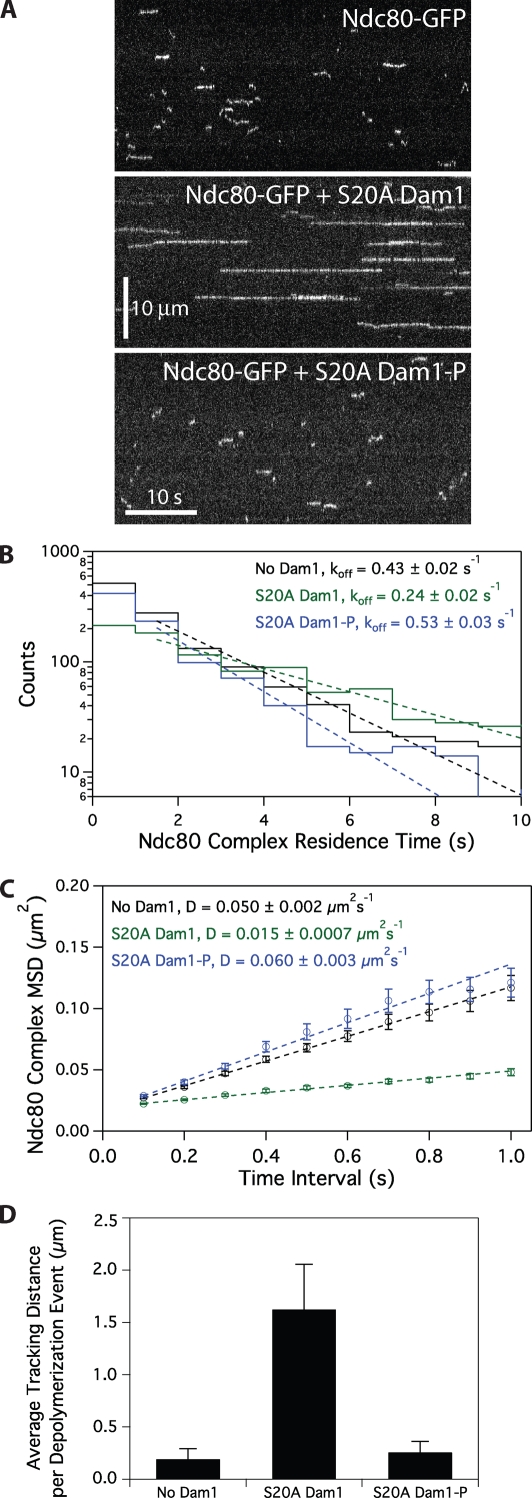
In the presence of unphosphorylated S20A Dam1 complex, diffusion of the Ndc80 complex on microtubules is slowed, dissociation rate constant is decreased, and tip tracking is enhanced, as described for the wild-type Dam1 complex (Fig. 7). However, Ipl1 phosphorylation of the S20A Dam1 complex abolished the ability of Dam1 complex to slow the diffusion and decrease the dissociation rate constant of the Ndc80 complex (Fig. 7, B and C). Moreover, phosphorylated S20A Dam1 complex did not enhance the tip-tracking ability of the Ndc80 complex (Fig. 7 D). Control experiments were performed to ensure that after the initial Ipl1 phosphorylation reaction with the S20A Dam1 complex, residual Ipl1 activity was negligible (see Materials and methods; Fig. S5). Furthermore, the 10 proteins of the Dam1 complex do not dissociate from one another when the complex is phosphorylated by Ipl1 (Fig. S4 C). Because phosphorylation of the S20A Dam1 complex does not alter the behavior of the Dam1 complex alone but abolishes its ability to change the behavior of the Ndc80 complex, we conclude that Ipl1 phosphorylation of the Dam1 complex inhibits its interaction with the Ndc80 complex.
Figure 7.
Ipl1 phosphorylation of the Dam1 complex regulates its interaction with the Ndc80 complex. (A) Representative kymographs showing changes in behavior of 10 pM Ndc80 complex with the addition of 500 pM S20A Dam1 complex with or without Ipl1 phosphorylation. Concentrations are of free complexes in solution. (B) Residence time distributions of 10 pM Ndc80 complex on microtubules without Dam1 complex (black histogram, n = 1,266 events), with 500 pM S20A Dam1 complex (green histogram, n = 1,081), and 500 pM Ipl1-phosphorylated S20A Dam1 complex (blue histogram, n = 974). Dotted lines show the weighted exponential fits used to determine dissociation rate constants, koff. (C) Mean-squared displacement (MSD) is plotted against time for 10 pM Ndc80 complex on microtubules without Dam1 complex (black markers, n = 1,102), with 500 pM S20A Dam1 complex (green markers, n = 1,030), and with 500 pM Ipl1-phosphorylated S20A Dam1 complex (blue markers, n = 860). Dotted lines show the weighted linear fits used to determine diffusion constants, D. (D) Mean tracking distance of 100 pM Ndc80 complex per depolymerization event in the absence of Dam1 complex (n = 19), in the presence of 500 pM S20A Dam1 complex (n = 28), or in the presence of 500 pM Ipl1-phosphorylated S20A Dam1 complex (n = 39). Error bars indicate SEM.
Discussion
The Dam1 complex acts as a processivity factor for the Ndc80 complex
Many molecular machines require factors that enhance their processivity. For example, the proliferating cell nuclear antigen sliding clamp is required for efficient DNA replication by DNA polymerase-ε (Kelman, 1997). Likewise, dynactin is required for long-distance movement of cytoplasmic dynein along microtubules (King and Schroer, 2000). Kinetochores are processive and form persistent attachments to dynamic microtubule tips over the times and distances required for chromosome biorientation and segregation. However, the contribution of individual components to the processivity of kinetochore–microtubule attachments is poorly understood. In this study, we show that the Dam1 complex enables the Ndc80 complex to track with disassembling microtubule tips over distances in excess of the length of the entire yeast spindle. We also show that the Dam1 complex strengthens the attachment of the Ndc80 complex to dynamic microtubule tips. In vivo, assembly of the Dam1 complex onto the kinetochore requires the Ndc80 complex (Janke et al., 2002). In our optical trap experiments, bead-bound Ndc80 complex was assayed with the Dam1 complex free in solution to mimic this arrangement in vitro. The increased ability of bead-bound Ndc80 complexes to bear load in the presence of free Dam1 complex indicates that tensile force can be transmitted through an Ndc80 complex–based linkage in a physiologically relevant arrangement.
We propose that the Dam1 complex acts as a processivity factor for the Ndc80 complex and that the two complexes cooperate to form load-bearing kinetochore–microtubule attachments. In vivo, the Ndc80 complex forms lateral attachments to spindle microtubules before kinetochore association of Dam1 complex and biorientation (Tanaka et al., 2005; Shimogawa et al., 2006). Our results are consistent with a model in which the Ndc80 complex initially mediates kinetochore attachment to microtubules. The Dam1 complex is later loaded onto the kinetochore to maintain attachment to dynamic microtubule tips. Association of the Dam1 complex is particularly important for these attachments to withstand the tensile forces required for biorientation. The existence of a distinct and separable processivity factor also provides a point of regulation for corrective detachment.
A mechanism for aurora B–mediated corrective detachment
The regulatory mechanism that ensures chromosome biorientation has been proposed to respond to the level of tensile force on the kinetochore (Kelly and Funabiki, 2009). When kinetochores make attachments that generate little tension, such as monotelic or syntelic attachments, progression to anaphase is blocked. Key to this regulation, the conserved aurora B kinase is responsible for the release of aberrant kinetochore–microtubule attachments (Biggins et al., 1999; Tanaka et al., 2002; Hauf et al., 2003; Pinsky et al., 2006). We showed previously that phosphorylation by the yeast aurora B kinase Ipl1 at Ser20 of Dam1 decreases the affinity of the Dam1 complex for the microtubule lattice (Gestaut et al., 2008). We show in this study that Ipl1 phosphorylation of the Dam1 complex at sites other than Ser20 weakens its interaction with the Ndc80 complex. Collectively, these observations suggest that Ipl1 phosphorylation of the Dam1 complex promotes corrective detachment of kinetochores via two distinct mechanisms, decreasing the affinity of the Dam1 complex for both the Ndc80 complex and for microtubules. Regulation by aurora B kinase is a conserved feature of kinetochore function in all eukaryotes. Therefore, we propose that regulation at both the kinetochore–microtubule interface and between components of the kinetochore itself will extend to mechanisms of corrective detachment in higher eukaryotes.
Materials and methods
Protein expression and purification
The S. cerevisiae Ndc80 and Dam1 complexes were expressed from polycistronic vectors in E. coli as described previously (Wei et al., 2005; Gestaut et al., 2008, 2010; Powers et al., 2009). For TIRF microscopy, the Ndc80 complex Nuf2 subunit was tagged with GFP, and the Dam1 complex Dad1 subunit was tagged with GFP or mCherry. Complexes were purified by affinity chromatography and gel filtration as previously described (Asbury et al., 2006; Franck et al., 2007; Gestaut et al., 2008; Powers et al., 2009).
For optical trap bead assays, a tobacco etch virus (TEV) cleavage site was inserted adjacent to the His6 affinity tag within the GFP-tagged Dam1 complex. The complex was purified by affinity chromatography and gel filtration as previously described (Gestaut et al., 2008). The cleavage reaction was performed in 50 mM phosphate buffer and 350 mM NaCl, pH 6.9, with 1 mM DTT, 0.5 mM EDTA, and recombinant TEV protease for 2 h at 4°C. TEV-cleaved Dam1 complex was isolated by gel filtration, and cleavage was verified by immunoblot analysis.
Phosphorylation of the Dam1 complex
Dam1 complex was phosphorylated with purified GST-Ipl1 and GST-Sli15 as described previously (Gestaut et al., 2008). The 50 µl reaction contained 4 µM GFP- or mCherry-tagged S20A Dam1 complex, 0.5 µM GST-Ipl1, 0.5 µM GST-Sli15 (residues 554–698), 200 mM NaCl, 10 mM ATP, 25 mM MgCl2, and 50 mM Hepes buffer, pH 7.2. Reactions were incubated at 30°C for 90 min. Control reactions lacked GST-Ipl1 and GST-Sli15. Control reactions lacking ATP were also performed and gave similar results as previously reported (Gestaut et al., 2008). Ipl1 activity was not eliminated after the phosphorylation reaction. Therefore, to ensure that residual Ipl1 from the reaction did not affect our assays, we performed mock phosphorylation reactions using BSA in place of the Dam1 complex. The components of this mock reaction had no effect on the diffusion and dissociation rate constants of the Ndc80 complex either in the absence or presence of the Dam1 complex (Fig. S5).
TIRF microscopy
A custom TIRF illumination system was constructed for simultaneous excitation of Alexa Fluor 647 and GFP (Gestaut et al., 2008, 2010; Powers et al., 2009). Total internal reflection of a far-red laser (FTEC-635-0-25-PFQ; Blue Sky Research) and a blue laser (Sapphire 488–75; Coherent) was achieved using a through the objective arrangement with a 100× 1.4 NA Plan Apochromat lens (CFI; Nikon). Images from the far-red and green channels were projected side by side onto a cooled EM charge-coupled device camera (iXon 887-BI; Andor Technology).
A custom flow cell construction method was used (Gestaut et al., 2008, 2010; Powers et al., 2009). Glass slides (Gold Seal) were drilled with two holes along the short axis. Double-sided sticky tape (Scotch) was placed on either side of the holes to produce the walls of the flow channel. Silanized coverslips (Corning) were pressed firmly onto the tape, and the ends of the channel were sealed with vacuum grease. To draw fluid through the channel, a peristaltic pump was used via a custom adaptor attached above one of the holes on the glass slide with adhesive transfer tape (3M).
Flow cells were washed with three 100 µl vol dH2O. To bind taxol-stabilized microtubules, we flowed in a modified “rigor” kinesin (G234A) lacking motor activity (Rice et al., 1999) diluted in BRB80 containing 8 mg × ml−1 BSA (BB80). Flow cells were washed with two 50 µl vol BB80, the second of which contained 10 µM taxol (BB80T). Alexa Fluor 647–labeled microtubules were diluted in BB80T and incubated in flow cells for 5 min. Flow cells were washed with two 50 µl vol BB80T. Proteins were then introduced, diluted in BB80T containing 0.02–0.1 mg × ml−1 κ-casein, 200 µg × ml−1 glucose oxidase, 35 µg × ml−1 catalase, 25 mM glucose, and 5 mM DTT. When assayed in combination, Ndc80 and Dam1 complexes were premixed before their introduction into flow cells. After flowing in the protein mixture, 2,000-frame videos were taken at 10 frames per second with iXon software (Andor Technology). All assays were performed at 26°C.
For disassembling microtubule assays, “rigor” kinesin was bound to flow cells and washed with 50 µl BB80 followed by 50 µl BB80 containing 0.1 mg ml−1 κ-casein and 1 mM GTP (growth buffer [GB]). Alexa Fluor 647–labeled GMPCPP microtubule seeds were bound and washed with two 50 µl vol GB. Microtubules were grown by incubating for ∼15 min in GB containing 2 mg × ml−1 tubulin (1:100; Alexa Fluor 647 labeled), 200 µg × ml−1 glucose oxidase, 35 µg ml−1 catalase, 25 mM glucose, and 5 mM DTT. Microtubule depolymerization was induced by buffer exchange removing free tubulin and simultaneously introducing proteins diluted in BB80 containing 0.1 mg × ml−1 κ-casein, 200 µg × ml−1 glucose oxidase, 35 µg × ml−1 catalase, 25 mM glucose, and 5 mM DTT. Videos were started concomitantly with induction of depolymerization and taken at 10 frames per second for 2,000 frames.
TIRF microscopy data analysis
Software analysis of TIRF microscopy data was performed using Labview (National Instruments) as previously described (Gestaut et al., 2008, 2010; Powers et al., 2009). The software generated the position and brightness of individual GFP-tagged complexes on microtubules over time. Custom Igor Pro (WaveMetrics) programs were used to generate histograms of Ndc80 complex residence times on microtubules. A weighted single exponential fit was applied to determine the mean residence time, τ, and to calculate the dissociation rate constant, koff = τ−1. Association rate constants, kon, were estimated as the number of observed Ndc80 complex–binding events per tubulin dimer per second divided by the free concentration of Ndc80 complex. Standard diffusion plots of mean-squared displacement versus time were generated in Igor Pro. A weighted linear fit was used to calculate the one-dimensional diffusion constant, D, of GFP-tagged complexes on microtubules.
To quantify Ndc80 complex tip tracking, brightness profiles along disassembling tips were created in Labview. Fluorescent signals at the tips were averaged across seven frames (0.7 s), and we required a minimum intensity threshold of 20% above background to score a tip-tracking event. For each individual frame, the instantaneous depolymerization rate was calculated as the change in tip position over 50 frames (5 s). A microtubule disassembly event was defined to start at the first appearance of GFP-tagged Ndc80 complex at the tip and to end when the rate of depolymerization dropped <0.03 µm × s−1. Microtubule tips without tracking as defined by this criterion were omitted from further analysis. The total tracking distance for each individual tip was determined, and the mean tracking distance per depolymerization event was calculated.
To quantify binding to microtubules, we created brightness profiles of 500 pM mCherry-tagged Dam1 complex using our TIRF assay. After a 5-min incubation with taxol-stabilized microtubules, an image was recorded (six or seven images per condition). For each microtubule in the image, the integrated intensity of mCherry was measured in ImageJ (National Institutes of Health), and the brightness per unit length was calculated. Brightness per unit length values were averaged across all microtubules within one image and reported as means from multiple images.
EM
Taxol-stabilized microtubules were made by polymerizing cleared tubulin in a total volume of 40 µl BRB80 containing 1.75 mM GTP, 1 mM MgCl2, and 3.5% DMSO at 37°C for 30 min. Various concentrations of Dam1 complex were mixed with taxol-stabilized microtubules to a final concentration of 36 nM tubulin in BRB80 containing 10 µM taxol and incubated for 15 min. Samples were prepared for analysis by EM as follows: carbon-coated copper grids were positively charged in a glow discharge device (Electron Microscopy Sciences) for 2 min. A 2-µl drop of sample was applied onto a freshly discharged grid and incubated for 20 s. Excess solution was blotted off, and the grid was washed twice with water and once with 0.075% uranyl formate before staining with uranyl formate. The stain was blotted off, and the grid was air dried. The preparations were viewed on a transmission electron microscope (Spirit T12; FEI) operating at 120 kV, and images were recorded on a 1,000 × 1,000 bottom-mount slow-scan charge-coupled device camera (Gatan) at a nominal magnification of either 15,000 or 52,000× at the specimen level. For each preparation, the total number of Dam1 complex rings on microtubules was counted and divided by the total length of microtubules to generate a mean number of Dam1 complex rings per microtubule micron. In control experiments performed in the presence of blocking proteins (8 mg × ml−1 BSA and 0.02 mg × ml−1 κ-casein), rings were still absent at 500 pM Dam1 complex.
Optical trap instrumentation and bead preparation
Our optical trap has been described previously (Franck et al., 2007, 2010; Powers et al., 2009). The instrument is built around an inverted microscope (TE2000; Nikon) equipped for video-enhanced differential interference contrast imaging. Custom-mounted optics direct the infrared trapping laser (J20-BL10–106Q; Spectra Physics) through a 100× 1.4 NA oil infrared Plan Apochromat objective lens (CFI; Nikon), through a high NA oil immersion condenser, and onto a position-sensitive detector. During force clamp experiments, a computer feedback–controlled piezo specimen stage (P-517.3CL; Physik Instrumente) was programmed (Labview) to maintain a fixed offset between the tip-attached bead and the trap center by moving to accommodate changes in microtubule length, thereby keeping the tensile force constant. During force ramp experiments, the bead trap separation was increased at a fixed rate, 0.25 pN × s−1, up to a preset maximum of 10–12 pN (just below the escape force of the trap). For both force ramp and force clamp experiments, the stage position was updated and stored to disk at 50 Hz. Bead trap separation was sampled at 40 kHz but decimated to 200 Hz for storage.
Beads were prepared as previously described (Powers et al., 2009). Ndc80 complex was linked to 0.44-µm-diameter streptavidin-coated beads (Spherotech) with biotinylated His5 antibody (QIAGEN). Ndc80 complex was diluted to 13–15 nM in BB80 with 1 mM DTT and incubated with 6 pM beads at 4°C for ∼90 min. In some experiments, recombinant His6-tagged GFP was used as a blocking agent. In this case, Ndc80 complex was diluted to 30 nM in BB80 with 1 mM DTT and incubated with 12 pM beads at 4°C for ∼90 min. These beads were mixed 1:1 with 6 µM GFP and incubated for an additional 30 min before use. The amount of complex per bead and the final bead concentration was the same in both protocols. Both protocols yielded a molar ratio of Ndc80 complexes to beads of 2,200–2,500. Based on simple geometric considerations (Powers et al., 2009), we estimate that <100 Ndc80 complexes could simultaneously interact with the filament. The Ndc80 complex/bead ratio was chosen to create tip attachments of moderate strength, so the full force range of the optical trap could be used to assess the contribution of Dam1 complex to the Ndc80 complex–based attachments. Results obtained with and without the GFP block were statistically indistinguishable (Ndc80 complex during assembly, P = 0.3010; Ndc80 complex during disassembly, P = 0.5518; Ndc80 complex + Dam1 complex during assembly, P = 0.1663; Ndc80 complex + Dam1 complex during disassembly, P = 0.8597), so they were pooled and analyzed together.
Optical trap bead assays, data collection, and analysis
Flow chambers were constructed and functionalized as previously described (Powers et al., 2009; Franck et al., 2010). In brief, two lengths of double-sided sticky tape (Scotch) were placed across the width of a microscope slide (Gold Seal) to form an inverted chamber of 2–3-mm width. A cleaned coverslip (Corning) longer than the slide width was pressed firmly onto the tape to form the chamber bottom, the overhanging edges acting as reservoirs for pipetting and aspirating solutions through the chamber. The chamber was functionalized by introducing 1 vol 1 mg × ml−1 biotinylated BSA (Vector Laboratories) and incubating for >10 min at room temperature. The chamber was washed with ∼20 vol BRB80 followed by ∼20 vol 0.33 mg × ml−1 avidin DN (Vector Laboratories). After a second wash with ∼20 vol BRB80, stable biotinylated microtubule seeds were introduced and washed with a growth and blocking buffer, BRB80 containing 1 mM GTP, 2 mg × ml−1 κ-casein, and 2% pluronic F-187. Subsequently, we introduced Ndc80 complex–coated beads that were diluted eightfold into GB, BB80 containing 1 mM GTP, 1.4 mg × ml−1 tubulin, 1 mM DTT, 250 µg × ml−1 glucose oxidase, 30 µg × ml−1 catalase, and 4.5 µg × ml−1 glucose. In assays with Dam1 complex, His6-cleaved GFP-tagged Dam1 complex was used at a final concentration of 9–15 nM and added to the bead mixture just before introduction into the flow chamber. Microtubule disassembly events either occurred by a spontaneous switch from assembly to disassembly or were induced by laser scission (Franck et al., 2010). All trap assays were performed at 26°C.
Records of bead position versus time were analyzed using custom software written in Igor Pro. Periods of microtubule assembly and disassembly were visually identified in the records. Detachments were scored when the force on a bead under load suddenly dropped to zero and the stage exhibited open-loop (“run away”) movement. The maximum force was taken as the mean of the final 10 data points before event termination (detachment or microtubule assembly/disassembly state switching). The survival probability was calculated by dividing the number of events that persisted beyond a given distance by the total number of events.
A bead–microtubule-binding assay was used to verify that His6-cleaved GFP-tagged Dam1 complex did not bind directly to the GFP-blocked beads. Taxol-stabilized microtubules were introduced into a flow chamber and given 1 min to adhere nonspecifically to the coverslip. After a wash and 10-min incubation with surface block (BRB80 with 2 mg × ml−1 κ-casein and 10 µM taxol), free beads were introduced. After waiting 10 min to allow beads to bind, the number of microtubule-attached beads was counted across many fields of view (each 822 µm2). Beads decorated with Ndc80 complex bound microtubules at a density of 7,800 beads × mm−2 (1,355 beads in 210 fields of view), whereas beads blocked with GFP in the presence of free Dam1 complex bound at only 24 beads × mm−2 (five beads in 250 fields of view) under identical conditions.
Online supplemental material
Fig. S1 shows a weak interaction between the Ndc80 and Dam1 complexes in solution by velocity sedimentation. Fig. S2 shows kymographs of single-molecule Ndc80 and Dam1 complexes interacting on taxol-stabilized microtubules. Fig. S3 shows that the Dam1 complex oligomerizes on microtubules and tracks with disassembling microtubule tips. This behavior is unaffected by the S20A mutation and subsequent phosphorylation by Ipl1. Fig. S4 further shows that phosphorylation of S20A Dam1 complex does not affect its microtubule binding. In addition, phosphorylation does not cause disassembly of the wild-type Dam1 complex. Fig. S5 shows that the behavior of the Ndc80 complex on microtubules is unaffected by residual components of the Dam1 complex phosphorylation reaction. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910142/DC1.
Acknowledgments
We thank V. MacKay and B. Kennedy for help with velocity sedimentation experiments. We also thank A. Powers, M. Shimogawa, B. Graczyk, and M. Wargacki for helpful scientific discussion.
We thank the Murdock Charitable Trust and the Washington Research Foundation for generous support of our electron cryomicroscopy facility. This work was supported by a National Sciences and Engineering Research Council of Canada scholarship (to J.F. Tien), a National Institutes of Health traineeship (grant T32 GM008268 to N.T. Umbreit), a National Science Foundation Integrative Graduate Education and Research traineeship (grant DGE-0504573 to A.D. Franck), a Searle Scholar award (grant 06-L-111 to C.L. Asbury), a Packard Fellowship for Science and Engineering (grant 2006-30521 to C.L. Asbury), and by the National Institute of General Medical Sciences (grants R01GM40506 and R01GM79373 to T.N. Davis and C.L. Asbury, respectively). T. Gonen is a Howard Hughes Medical Institute early career scientist.
Footnotes
Abbreviations used in this paper:
- GB
- growth buffer
- TEV
- tobacco etch virus
- TIRF
- total internal reflection fluorescence
References
- Akiyoshi B., Nelson C.R., Ranish J.A., Biggins S. 2009. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 183:1591–1595 10.1534/genetics.109.109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C.L., Gestaut D.R., Powers A.F., Franck A.D., Davis T.N. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Franck A.D., Powers A.F., Gestaut D.R., Gonen T., Davis T.N., Asbury C.L. 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9:832–837 10.1038/ncb1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A.D., Powers A.F., Gestaut D.R., Davis T.N., Asbury C.L. 2010. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. doi:10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., Nigg E.A. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Cooper J., Asbury C.L., Davis T.N., Wordeman L. 2010. Reconstitution and functional analysis of kinetochore subcomplexes. Methods Cell Biol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Efremov A.K., Volkov V.A., Spiridonov I.S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J.R., Ataullakhanov F.I. 2008a. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA. 105:15423–15428 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Spiridonov I.S., Volkov V.A., Efremov A., Westermann S., Drubin D., Barnes G., Ataullakhanov F.I., McIntosh J.R. 2008b. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA. 105:6918–6923 10.1073/pnas.0801811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A., Silljé H.H., Nigg E.A. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué S., Salmon E.D. 1995. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 6:1619–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortíz J., Tanaka T.U., Lechner J., Schiebel E. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193 10.1093/emboj/21.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating P., Rachidi N., Tanaka T.U., Stark M.J. 2009. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J. Cell Sci. 122:4375–4382 10.1242/jcs.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21:51–58 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. 1997. PCNA: structure, functions and interactions. Oncogene. 14:629–640 10.1038/sj.onc.1200886 [DOI] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28:1099–1110 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.J., Schroer T.A. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2:20–24 10.1038/71338 [DOI] [PubMed] [Google Scholar]
- Kline-Smith S.L., Sandall S., Desai A. 2005. Kinetochore-spindle microtubule interactions during mitosis. Curr. Opin. Cell Biol. 17:35–46 10.1016/j.ceb.2004.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Straight A., Coughlin P., Mitchison T.J., Salmon E.D. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382 10.1083/jcb.200301088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J., King D.S., Harrison S.C. 2007. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol. Biol. Cell. 18:2503–2510 10.1091/mbc.E07-02-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Kung C., Shokat K.M., Biggins S. 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8:78–83 10.1038/ncb1341 [DOI] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.A., Tanenbaum M.E., Maia A.F., Medema R.H. 2009. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J. Cell Sci. 122:2436–2445 10.1242/jcs.051912 [DOI] [PubMed] [Google Scholar]
- Rice S., Lin A.W., Safer D., Hart C.L., Naber N., Carragher B.O., Cain S.M., Pechatnikova E., Wilson-Kubalek E.M., Whittaker M., et al. 1999. A structural change in the kinesin motor protein that drives motility. Nature. 402:778–784 10.1038/45483 [DOI] [PubMed] [Google Scholar]
- Shang C., Hazbun T.R., Cheeseman I.M., Aranda J., Fields S., Drubin D.G., Barnes G. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 14:3342–3355 10.1091/mbc.E02-11-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa M.M., Graczyk B., Gardner M.K., Francis S.E., White E.A., Ess M., Molk J.N., Ruse C., Niessen S., Yates J.R., III, et al. 2006. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr. Biol. 16:1489–1501 10.1016/j.cub.2006.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Rieder C.L., Salmon E.D. 1995. Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 108:2537–2548 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Desai A. 2008. Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 20:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 434:987–994 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Ramey V.H., Westermann S., Leschziner A.E., Welburn J.P., Nakajima Y., Drubin D.G., Barnes G., Nogales E. 2007. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol. 14:721–726 10.1038/nsmb1274 [DOI] [PubMed] [Google Scholar]
- Wei R.R., Sorger P.K., Harrison S.C. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R., III, Cheeseman I.M. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., Barnes G. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]