Abstract
Profilin-1 (Pfn1), a ubiquitously expressed actin-binding protein, has gained interest in epithelial-derived cancer because of its downregulation in expression in various adenocarcinoma. Pfn1 overexpression impairs tumorigenic ability of human breast cancer xenografts thus suggesting that Pfn1 could be a tumor-suppressor protein. The objective of the present study was to determine how Pfn1 overexpression affects cell-cycle progression of breast cancer cells. We show that Pfn1 overexpression in MDA-MB-231 breast cancer cells causes cell-cycle arrest in G1 phase and dramatically reduced proliferation in culture. Pfn1 overexpression results in increased protein stability of p27kip1 (p27 – a major cyclin-dependent kinase inhibitor) and marked elevation in the overall cellular level of p27. Proliferation defect of Pfn1 overexpressers can be partly rescued by silencing p27 expression thus suggesting a critical role of p27 in Pfn1-induced growth inhibition of MDA-MB-231 cells. Finally, Pfn1 overexpression was found to sensitize MDA-MB-231 cells to apoptosis in response to cytotoxic stimulus thus suggesting for the first time that survival of breast cancer cells can also be negatively influenced by Pfn1 upregulation. These findings may provide novel insights underlying Pfn1’s tumor-suppressive action.
Keywords: Profilin-1, MDA-MB-231, breast cancer cells, proliferation, apoptosis, p27kip1
INTRODUCTION
Profilins (Pfns) belong to a class of small G-actin-binding proteins comprising of four members identified to date: Pfn1 (ubiquitously expressed in almost all cell types), Pfn2 (mainly expressed in nervous system in vertebrates), Pfn3 and 4 (expression restricted to kidney and testis). Besides binding to actin, Pfns also interact with a multitude of other ligands including various phosphoinositides and proteins containing proline-rich motifs that are involved in actin cytoskeletal regulation, endocytosis and gene transcription. Pfn1, the founding member of the protein family, promotes actin polymerization in cells by virtue of its ability to i) catalyze nucleotide exchange factor (ADP-to ATP) on G-actin, ii) shuttle G-actin to the barbed ends of actin filaments, and iii) interact with almost all major protein families that are known to be involved in nucleation and/or elongation of actin filaments (Jockusch et al., 2007; Witke, 2004). Although Pfn1 has been most thoroughly studied for its role in actin polymerization at the leading edge during cell migration, other findings including early stage embryonic lethality of Pfn1−/− mice, reduced survival of Pfn1 −/+ mice (Witke et al., 2001), and slower growth of vascular endothelial cells in culture after Pfn1 depletion (Ding et al., 2006) suggest that Pfn1 also plays a role in cell proliferation. In the context of cell proliferation, Pfn1 has been mainly linked to cytokinesis for its essential role in actomyosin-based contractile ring formation in lower organisms such as yeast and C. Elegans (Balasubramanian et al., 1994; Severson et al., 2002). A recent study using a conditional knockout mouse model has shown that Pfn1 function is dispensable for actomyosin-based contractile ring formation in chondrocytes, but abscission of daughter cells in the final stage of cytokinesis still requires Pfn1. Actin-binding mutant of Pfn1 fails to rescue the cytokinesis defect in Pfn1-null chondrocytes thus suggesting importance of Pfn1-actin interaction in cytokinesis, at least, in those cell types (Bottcher et al., 2009).
Tumor cells acquire gains-of-function in proliferation and survival over their normal counterparts. Given the importance of Pfn1’s function in normal cell proliferation, it is not intuitively clear why Pfn1 expression is significantly downregulated in various types of epithelial-derived tumors including those originating in breast, pancreas and liver (Gronborg et al., 2006; Janke et al., 2000; Wu et al., 2006). Consistent with reduced expression of Pfn1 in breast cancer, overexpression of Pfn1 has been shown to inhibit tumorigenicity of breast cancer cell lines (CAL-51, MDA-MB-231) in xenograft models therefore suggesting that Pfn1 is a tumor-suppressor protein (Janke et al., 2000; Zou et al., 2007). How Pfn1 overexpression affects cell cycle progression of breast cancer cells is not known, and addressing this gap was the overall goal of the present study.
Cell cycle progression is tightly regulated by coordinated activities of cyclin/cyclin-dependent kinase (CDK) complexes. The interactions of cyclins with their partner CDKs are negatively regulated by CDK inhibitors (CDKIs). Two families of CDKIs, namely the CIP/KIP (p21Cip1/Waf1 (p21), p27Kip1 (p27) and p57Kip2 (p57)) and INK4 (p16INK4a, p15INK4b, p18INK4c, and p19INK4d) cause cell cycle arrest at G1 phase (reviewed in (Sherr, 1994; Sherr and Roberts, 1995)). Tumor-suppressor proteins often cause G1 arrest of cancer cells through upregulating expression levels and/or activities of CDKIs. Among these various CDKIs, p27 (binds to CDK2 and inhibits CDK2-cyclin E complex) has drawn major attention in cancer because of its downregulation in a substantial number of human epithelial malignancies (breast, prostate, lung, colon, head and neck). There is also clinical evidence of reduced p27 level correlating with tumor aggressiveness and poor patient survival (reviewed in (Chu et al., 2008)). In this study, we establish a novel link between Pfn1 overexpression and suppression of proliferation of breast cancer cells via p27 upregulation.
MATERIALS AND METHODS
Antibodies and Reagents
Monoclonal GAPDH antibody is a product of Abd Serotec (Raleigh, NC). Monoclonal p21, p27, and histone H1 antibodies were obtained from BD Biosciences (San Diego, CA). Polyclonal cyclin-D1 was purchased from Neomarker (Freemont, CA). Polyclonal PARP (poly-ADP ribose polymerase) antibody is a product of Cell Signaling (Danver, MA). Polyclonal Phospho-S10-p27 antibody was obtained from Santa Cruz (Santa Cruz, CA). Campothecin was purchased from Sigma (St. Louis, MO). All cell culture reagents, propidum iodide (PI), RNAse are products of Invitrogen (Carlsbad, CA).
Cell culture and siRNA transfection
Generation and culture of MDA-231 stably expressing GFP and GFP-Pfn1 have been previously described (Zou et al., 2007). Smart-pool control siRNA and p27 siRNA (also a pool of 3-4 individual siRNAs targeting different regions of mRNA) are commercially available from Dharmacon Inc (Chicago, IL). SiRNA transfection was done at a working concentration of 25 nM using Dharmacon transfection reagent according to the manufacturer’s instructions.
RT-PCR
Total RNA was extracted from cells cultured in normal serum-containing growth medium using the RNeasy mini kit commercially available through Qiagen (Valencia, CA). RT-PCR reactions were performed using a commercial kit also available through Qiagen. The primer sequences of p27 were 5′-TGCAACCGACGATTCTTCTACTCAA-3′ (sense), and 5′-CAAGCAGTGA TGTATCTGATAAACAAGGA-3′ (antisense). The primer sequences for GAPDH were 5′-CGGAGTCAACGGATT TGGTCGTAT-3′ (sense) and 5′- AGGCTTCTCCATGGTGGTGAAGAC-3′ (antisense). The PCR cycle conditions of p27 and GAPDH were 94°C (1 min), 55°C (1 min), and 72°C (1 min) with a total of 40 cycles. The PCR products were run on a 1.5% agarose gel. For qRT-PCR experiments, SYBR green method was employed and fold-change in transcript level was quantified by the standard delta-delta-CT method.
Cell proliferation assay
To assess serum-induced cell growth in culture, 30,000 cells were plated overnight in triplicates in the wells of a 12-well plate, serum-starved for 24 hours and then stimulated with the complete growth media for additional 24 and 48 hours. At those indicated time-points, cells were trypsinized and counted after performing trypan blue exclusion assay. The number of cells, based on the average count of all the wells, was compared between the different groups from a total of 3 independent experiments. The experimental protocol for assessing the effect of p27 knockdown on cell proliferation was essentially similar except in this case equal number of control- and p27-siRNA transfected cells were replated 24 hours after transfection and growth kinetics was followed for 2 additional days by counting cells as described before.
Apoptosis assay
For TNF-induced apoptosis experiments, cells were pre-treated with cycloheximide (CHX; 10 or 50 μM) for 30 minutes before being exposed to TNFα (40ng/ml) for 16-18 hours. Both adherent and floating cells were harvested, pooled and then processed for further FACS analyses of DNA content (following PI staining) and immunoblotting analyses of PARP cleavage. Apoptosis was additionally confirmed by nuclear condensation and fragmentation from DAPI staining. For campothecin-induced apoptosis experiments, cells were treated with 15 μM campothecin 15 μM for 24 hours before fixing and staining with DAPI to detect nuclear condensation and fragmentation of apoptotic cells.
FACS analyses of DNA content
Cells were trypsinized and washed with PBS twice before fixing with ice-cold 70% ethanol at 4°C overnight. After removing ethanol by centrifugation, cells were washed with PBS and treated with PI solution (50 μg/ml PI combined with 100 μg/ml DNAse free RNAse A, 0.1% Ttiton X-100 and 0.1mM EDTA in PBS) for 45 minutes in dark. After washing with PBS, PI-stained cells were subjected to FACS analyses and data were analyzed with FlowJo software.
Protein extraction and immunoblotting
Total cell lysate was prepared by extracting cells with either warm 1X sample buffer or modified RIPA buffer (50 mM Tris-HCl -pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.3% SDS, 2 mM EDTA) supplemented with 50 mM NaF, 1mM sodium pervanadate, and protease inhibitors. For subcellular fractionation, cells were first extracted in buffer A (10mM HEPES, pH7.9, 10mM KCl, 0.1mM EDTA, 0.1% NP-40, 1mM DTE with protease inhibitors) for 15 minutes and centrifuged at 5000 rpm for 5 minutes. The supernatant was saved as cytoplasmic fraction. Cell pellet was then washed with buffer A, resupended in buffer B (20mM HEPES, pH7.9, 0.4M NaCl, 1mM EDTA, 10% glycerol and 1 mM DTE with protease inhibitors) and vortexed for one hour at 4°C before centrifuging at 13000 rpm for 5 minutes. The supernatant was collected as the nuclear fraction (the purity of the nuclear fraction was confirmed by positive and negative immunoreactitivity of histone-H1 and GAPDH antibodies, respectively). For immunoblotting, the following concentrations of the antibodies were used: GAPDH (1:1000), p27 (1:500), cyclin-D1(1:200), phospho-S10-p27 (1:200) and PARP (1:1000), histone-H1 (1:500).
RESULTS
Pfn1 overexpression in MDA-231 cells causes cell-cycle arrest at G1 phase
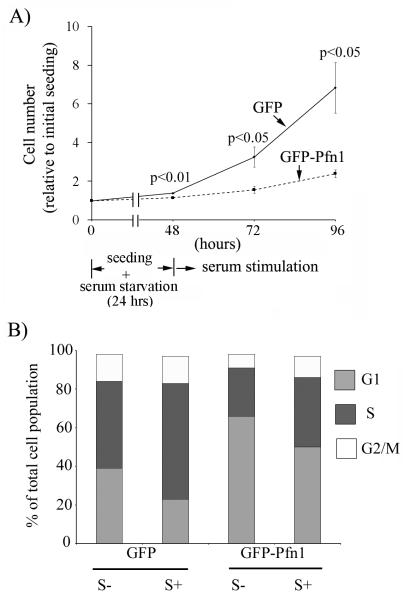
We previously reported generation of stable sublines of MDA-231 cells expressing either GFP-fused Pfn1 (GFP-Pfn1) at a modest level of overexpression (< 2-fold) or GFP as control, and in vivo studies showed that GFP-Pfn1 expressers of MDA-231 cells fail to form tumor when xenografted in nude mice (Zou et al., 2007). GFP-expressers serve as an appropriate control since we confirmed that the proliferation kinetics of GFP-expressers is nearly identical to that of the parental cell line (unpublished data) and xenografts of GFP expressers form robust tumors in mice. Since tumor-suppressor genes inhibit cell proliferation, we first assessed the effect of Pfn1 overexpression on serum-stimulated growth of MDA-231 cells in culture. The average cell counts of GFP expressers were approximately ~2-and ~3-fold higher than the corresponding readouts of GFP-Pfn1 expressing cells at 24 and 48 hours after serum stimulation thus demonstrating growth inhibition of MDA-231 cells induced by Pfn1 overexpression (Fig 1A). Similar results were obtained when growth rates of these two cell lines were compared in continuous serum-based culture without involving serum-withdrawal in the experimental protocol (data not shown). We did not find any evidence of cell death in the growing culture for either of the experimental groups suggesting that the differential growth rate is strictly due to proliferation differences between the two cell lines.
Fig 1. Pfn1 overexpression causes G1 arrest of MDA-231 cells.
A) Relative growth kinetics of GFP and GFP-Pfn1 expressers of MDA-231 cells in response to serum stimulation B) Summary of cell-cycle status of GFP and GFP-Pfn1 expressers in serum-starved (S−) and 48 hrs post serum-stimulation (S+) conditions (data summarized from 3 independent experiments).
We next compared the cell-cycle profiles of GFP and GFP-Pfn1 expressers in serum-starved vs serum-stimulated conditions by FACS-based analyses of their DNA content, the results of which are summarized in the form of a bar graph in Fig 1B. Differences in the cell cycle profiles between the two cell lines were most pronounced for G1 and S phases. For serum-starved condition, the average % of GFP-expressing cells in G1 and S phases were equal to (39.5±2.4) and (45.5±2.8), respectively; the corresponding readouts for GFP-Pfn1 expressers were equal to (66.1.5±5.4) and (25.6±7.0), respectively. In response to serum-stimulation, % of GFP cells in G1 phase reduced to (23.3±4.8) with a concomitant increase in S-phase population to (60.4±3.5); the corresponding readouts of Pfn1 overexpressers were equal to (50.0±0.22) and (36.6±1.5), respectively. Although both cell lines displayed a general trend of G1-to-S phase progression in response to serum stimulation as expected, our data are indicative of a general G1 arrest of MDA-231 cells induced by Pfn1 overexpression. We did not find any significant difference in G2/M phase population between the two cell lines therefore suggesting that at least a moderate level of overexpression of Pfn1 does not lead to cytokinesis defect in MDA-231 cell line.
Pfn1 overexpression inhibits MDA-231 cell proliferation partly via upregulating p27
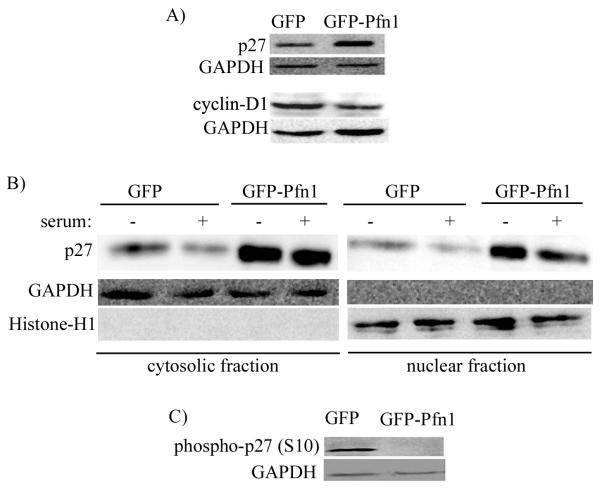
G1-S transition of cell cycle is inhibited by cyclin-dependent kinase inhibitors [CDKIs]. Among different CDKIs, p27 is frequently downregulated in various cancers including breast cancer. We therefore examined the relative expression level of p27 in actively growing cultures of GFP and GFP-Pfn1 expressers by immunoblot analyses of total cell extracts which showed a dramatic elevation of p27 level in Pfn1 overexpressing cells (Fig 2A). Also, consistent with the G1-arrest phenotype, Pfn1 overexpressing cells showed substantially lower cyclin-D1 levels compared to control GFP cells (Fig 2A).
Fig 2. Pfn1 overexpression upregulates p27 level in MDA-231 cells.
A) Relative expression profile of various cell-cycle markers (p21, p27 and cyclin-D1) between GFP and GFP-Pfn1 expressers of MDA-231 cells. B) P27 immunoblot of cytoplasmic and nuclear fractions isolated from GFP and GFP-Pfn1 expressers in serum-starved (starvation time – 24 hrs) and serum-stimulated (stimulation time - 12 hrs) conditions. Histone H1 and GAPDH blots serve as the loading controls for nuclear and cytosolic fractions, respectively. C) Relative levels of S10 phosphorylated form of p27 between GFP and GFP-expressers (GAPDH blots serves as the loading control).
P27 inhibits CDK2-cyclinE complex (a necessary step for CDK2 activation leading to E2F transcription factor activation and DNA replication) and elicits its anti-proliferative effect when it is in the nucleus. There are also several examples of cancer including certain carcinomas of the breast, thyroid, esophagus and colon where p27 level is maintained at a normal level, but is mislocalized to cytoplasm (Blain and Massague, 2002). In essence, nuclear content of p27 strongly correlates with its growth inhibitory action. We therefore performed subcellular fractionation to examine the relative cytosolic and nuclear contents of p27 between the two sublines of MDA-231 cells, either in serum-starved state or following 12 hrs of serum-stimulation. Although both cell lines displayed a general trend of decrease in nuclear p27 level in response to serum stimulation as expected, Pfn1 overexpressing cells were found to have higher p27 level in both cytoplasmic and nuclear fractions of cell lysate when compared to control GFP cells (Fig 2B).
Various phosphorylation events are thought to impinge on the biological functions of p27 through either localization and/or proteolysis control. Several phosphorylation sites of p27 have been identified which include serine 10, threonine 157 (not conserved in mouse), 187 and 198; and tyrosine 74, 88, and 89. Serine 10 (S10) is the major phosphorylation site of p27 (accounts for an estimated 75% of total phosphate incorporation) (Grimmler et al., 2007; Kaldis, 2007). Phosphorylation of p27 at S10 residue not only promotes its nuclear export but can also indirectly regulates p27 level through its proteolysis by kip1-ubiquitylation-promoting complex (KPC) in the cytoplasmic compartment. Since Pfn1 overexpressing cells showed p27 elevation in both nuclear and cytoplasmic compartment, we examined the relative levels of S10-phosphorylated p27 between our two sublines of MDA-231 cells by phosphospecific immunoblot analyses of total cell extracts. Indeed, we found a dramatic inhibition in S10 phosphorylation of p27 as a result of Pfn1 overexpression (Fig 2C). Also, consistent with nuclear export of S10-phosphorylated form of p27, immunostaining data further revealed that S10-phosphorylated p27 localized primarily in the cytosolic compartment (supplemental Fig 1 - weaker fluorescence signal of phospho-S10-p27 in Pfn1 overexpressing cells compared to GFP cells also conformed to the immunoblot data).
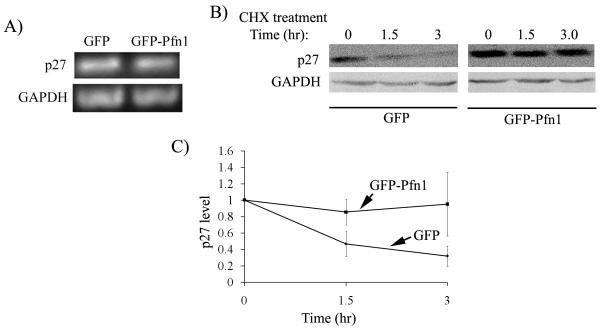
We next asked how Pfn1 overexpression might lead to elevated p27 level in MDA-231 cells. P27 expression is subjected to transcriptional (by Forkhead family of transcription factors), translational (through microRNAs, binding of proteins to its IRES site) and post-translational (through proteolysis) control mechanisms (Chu et al., 2008; le Sage et al., 2007; Vervoorts and Luscher, 2008). To determine if there is any Pfn1-dependent change in p27 transcription, we first performed semi-quantitative RT-PCR experiments which showed comparable levels of p27 mRNA between GFP and GFP-Pfn1 overexpressers (Fig 3A). This data was further validated by real-time qRT-PCR experiments (supplemental Fig 2) thus suggesting that Pfn1 overexpression does not transcriptionally upregulate p27 in MDA-231 cells.
Fig 3. Pfn1 overexpression enhances protein stability of p27 in MDA-231 cells.
A) RT-PCR data showing relative levels of p27 and GAPDH (loading control) mRNAs in GFP and GFP-Pfn1 expressers. B) P27 immunoblot of total cell extracts prepared from GFP and GFP-Pfn1 expressers at different time-points (0, 1.5, and 3 hours) after CHX treatment (GAPDH blot serves as the loading control). C) Kinetics of p27 degradation of GFP and GFP-Pfn1 expressers in response to CHX treatment (these data are based on p27 band intensity normalized to internal control (GAPDH or actin) and are expressed relative to the value calculated at time t=0; data summarized from 3 independent experiments).
Expression level of p27 is most often regulated at a post-translational stage by proteolysis. Therefore, to determine if Pfn1 overexpression alters the protein stability of p27 in MDA-231 cells, we inhibited new protein synthesis in our cell lines with cycloheximide (CHX) treatment and performed quantitative immunoblot analyses of p27 from cell extracts prepared at different time-points (0, 1.5, 3 hrs) after CHX addition. Fig 3B shows a set of representative p27 immunoblots from extracts of our two cell lines after different lengths of exposure to CHX. Densitometric analyses of these immunoblot data are summarized in Fig 3C which clearly shows that CHX-induced p27 decline was significantly inhibited in GFP-Pfn1 expressing cells when compared to control GFP cell thus indicating that Pfn1 overexpression enhances the protein stability of p27 in MDA-231 cells.
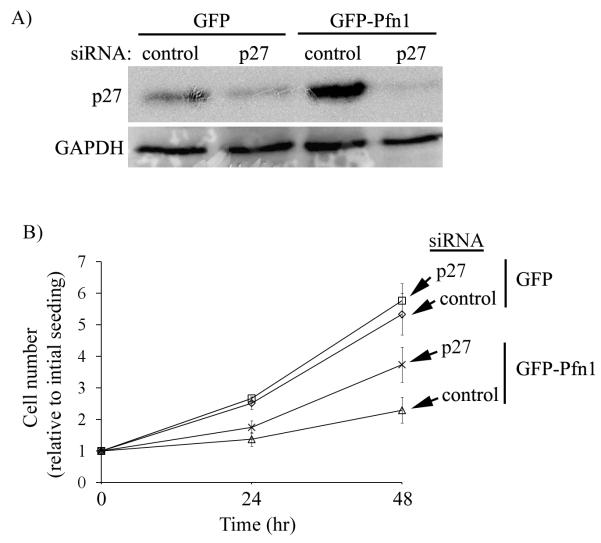
Next to determine whether elevated p27 level is, at least, partially responsible for Pfn1-induced growth suppression of MDA-231 cells, we silenced p27 expression in GFP and GFP-Pfn1 expressers by transient transfection of a pool of p27-specific siRNAs (as control, cells were transfected with a pool of non-targeting control siRNAs), and then followed the growth of transfected cells up to 48 hours after cell-seeding (corresponds to 72 hours after transfection). Immnublot data in Fig 4A confirms that p27 expression can be strongly suppressed by p27-siRNA treatment, while control siRNA treatment preserves the p27 differential between GFP and GFP-Pfn1 expressers as expected. We did not find any significant effect of p27 knockdown on the growth kinetics of GFP cells at least for the two time-points we tested (Fig 4B). By contrast, silencing p27 expression led to substantial faster growth of Pfn1 overexpressing cells (1.3−and 1.7−folds increase at 24 and 48 hour time-points, respectively). The cell counts of GFP-Pfn1 expressers at both time points were still lower than the corresponding readings for GFP expressers suggesting that p27 upregulation is partly responsible for growth inhibition of MDA-231 cells induced by Pfn1 overexpression.
Fig 4. P27 upregulation is partly responsible for Pfn1-induced growth suppression of MDA-231 cells.
A) P27 immunoblot demonstrating strong suppression of p27 expression 72 hours after 25 nM siRNA treatment (GAPDH blot serves as the loading control). B) Effect of p27 silencing on the proliferation of GFP and GFP-Pfn1 expressers.
Pfn1 overexpression negatively influences MDA-231 cell survival
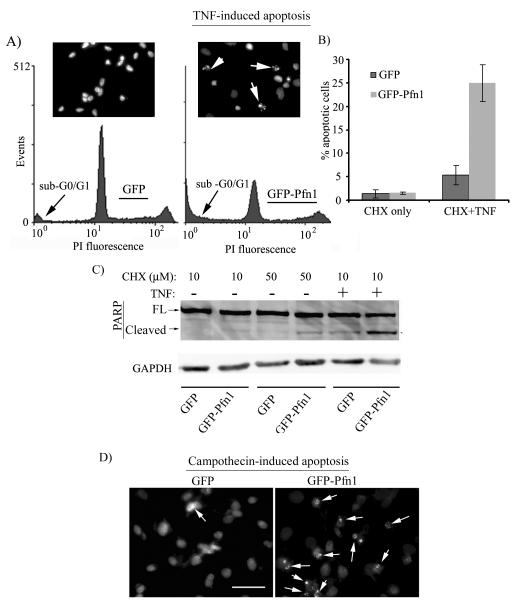
Finally, in addition to acquiring unrestrained proliferative capacity, tumor cell also develop resistance to apoptosis. Because of tumor suppressive action of Pfn1, we therefore asked whether Pfn1 overexpression has any effect on the survival of MDA-231 cells. To address this question, we subjected GFP and GFP-Pfn1 expressers to co-treatment of TNF (tumor necrosis factor – a pro-apoptotic factor) and CHX for 18 hours (addition of CHX was necessary to suppress induction of survival genes in response to the apoptotic stimulus). As a control, cells were subjected to CHX treatment only. Apoptotic sensitivity of the two cell lines was determined by FACS based analyses of sub-G0/G1 population of cells, extent of PARP cleavage (a caspase-3-mediated event and a hallmark of apoptosis) and nuclear fragmentation/condensation (another hallmark of apoptotic cells). Fig 5A shows a typical set of FACS profiles representing the DNA content of our two cell lines in response to TNF treatment. Quantitative analyses of FACS data showed a ~3 fold higher sub-G0/G1 population of cells in Pfn1 overexpressers compared to control GFP group (Fig 5B). Conforming to this FACS data, PARP immunoblot and nuclear staining by DAPI show stronger TNF-induced cleaved PARP product (Fig 5C) and enhanced nuclear condensation/fragmentation (inset of Fig 5A), respectively in Pfn1 overexpressing cells. These data demonstrate that Pfn1 overexpression sensitizes MDA-231 cells to TNF-induced apoptosis. To further examine whether Pfn1 overexpression enhances apoptotic susceptibility of breast cancer cells to chemotherapeutics, we treated GFP and GFP-Pfn1 expressers with campothecin, a commonly used chemotherapeutic drug. Conforming to our observation in TNF-challenge experiments, we found that campothecin also invoked a much stronger apoptotic response in Pfn1 overexpressing cells compared to GFP cells (Fig 5D). Collectively, our data suggest for the first time that breast cancer cell survival can also be modulated by Pfn1 upregulation.
Fig 5. Pfn1 overexpression sensitizes MDA-231 cells to apoptosis.
A) FACS profile of TNF-treated cells show a larger fraction of GFP-Pfn1 cells with sub G0/G1 nuclear content compared to GFP cells (DAPI staining in the inset reveals nuclear condensation and fragmentation of apoptotic cells (arrows)). B) A bar graph comparing % apoptotic cells for the two cell lines following 18 hours of either co-treatment of TNF and CHX or CHX only. C) Corresponding PARP immunoblot shows stronger cleaved PARP band in the case Pfn1 overexpressers (GAPDH blot serves as the loading control). D) DAPI staining shows increased sensitivity of Pfn1-overexpressing cells to campothecin-induced apoptosis (arrows mark nuclear condensation and fragmentation of apoptotic cells).
DISCUSSION
Although tumor-suppressive action of Pfn1 on breast cancer cell lines has been previously reported (Janke et al., 2000; Zou et al., 2007), the molecular pathways by which Pfn1 upregulation suppresses growth of breast cancer cells were not elucidated in those studies. Previous studies performed with various normal cells of eukaryotic origins have implicated Pfn1’s role in cell proliferation mostly in the context of cytokinesis, either during ingression of cleavage furrow and/or the final stage of abscission of daughter cells (Balasubramanian et al., 1994; Bottcher et al., 2009; Severson et al., 2002). This is not surprising given that cytokinesis is an actin-driven process and Pfn1 is an important regulator of actin polymerization in cells. However, given that overexpression of actin-binding deficient mutant of Pfn1 has no negative effect on mammary tumor growth in vivo (Wittenmayer et al., 2004), an indispensable role of Pfn1-actin interaction in cell proliferation mainly during cytokinesis may be, at least, not generalizable for breast cancer cells. In this study we investigated for the first time how Pfn1 upregulation alters cell-cycle progression of MDA-231, a highly aggressive breast cancer cell line that previously showed evidence of tumor-suppression in response to Pfn1 overexpression (Zou et al., 2007). We here report a novel finding that Pfn1 overexpression in MDA-231 cells is associated with a dramatic upregulation of p27 level, cell-cycle arrest at G1 phase (consistent with p27 upregulation) and reduced cell proliferation in vitro (also congruent with Pfn1’s act of tumor-suppression in vivo). We, however, failed to see any apparent defect in cytokinesis of Pfn1 overexpressers.
We showed that transient silencing of p27 expression leads to a substantial, although not complete, rescue of proliferation defect of Pfn1-overexpressing cells thus establishing for the first time a direct causal relationship between Pfn1-induced p27 upregulation and growth inhibition of MDA-231 cells. Partial restoration of growth inhibition by silencing p27 expression is not surprising for at least two reasons. First, our experiments were performed in a transient transfection setting and only followed short-term, and therefore long-term effect of p27 suppression is not known. Second, it is possible that more than just p27 upregulation contributes to decreased mitogenesis of Pfn1 overexpressing cells (for example, we also observed a significant decrease in cyclin-D1 level in MDA-231 cells as a result of Pfn1 overexpression).
We demonstrated that Pfn1-induced p27 upregulation does not occur at the transcriptional level and showed further evidence of post-translational regulation via conferring enhanced protein stability. P27 degradation can occur in both nuclear (a Skp2 mediated process that is dependent on p27 phosphorylation at T187 residue by CDK2) and cytosolic (a KPC-dependent event that is preceded by S10 phosphorylation and subsequent nuclear export of p27) (Ishida et al., 2002; Kamura et al., 2004; Lee and Kay, 2008). We have shown that S10-phosphorylation of p27 is inhibited in Pfn1 overexpressing cells. Although we have not shown directly that inhibition of S10 phosphorylation primarily accounts for higher nuclear content with an overall reduction in degradation of p27 in Pfn1 overexpressing cells, our data is nevertheless consistent with the literature. There are several kinases that are capable of phosphorylating p27 at S10 residue including AKT, KIS (kinase interacting stathmin) and MAPK (mitogen activated protein kinase). AKT can also phosphorylate other sites of p27 and there is strong experimental evidence of AKT’s involvement in downregulating p27 through transcriptional, translational and post-translational regulation (Liang and Slingerland, 2003; Liang et al., 2002). We have recently shown that Pfn1 overexpression upregulates PTEN (phosphatase and tensin homolog deleted on chromosome 10 – a negative regulator of PI3-kinase/AKT pathway) and suppresses AKT activation in both MDA-231 (Das et al., 2009). Therefore, one likely pathway by which Pfn1 overexpression could upregulate p27 level in breast cancer cells is through modifying lipid turnover and suppression of AKT pathway. It has been shown that cell-cell adhesion reduces the expression level of skp2 (a ubiquitin ligase responsible for p27 proteolysis) and hence increases p27 level in cells (Migita et al., 2008; Motti et al., 2005; Polyak et al., 1994; St Croix et al., 1998). There is also experimental evidence that cell-cell adhesion stabilizes and upregulates PTEN in carcinoma cells, and this is another mechanism by which p27 level can be potentially elevated by intercellular adhesion in an AKT-linked pathway (Subauste et al., 2005; Uegaki et al., 2006). We have recently shown that Pfn1 overexpression restores adherens junctions in mesenchymal MDA-231 cells (Zou et al., 2009).Therefore, Pfn1 overexpression could also elevate p27 level in MDA-231 cells secondary to changes in cell-cell adhesion. These various but not mutually exclusive possibilities will need to be explored in the future.
Finally, AKT pathway plays a critical role in regulating cell survival. Particularly, PTEN status is critical determinant of apoptotic sensitivity of cancer cells to chemotherapeutic agents. Cancer cells with reduced PTEN expression and therefore having hyperactivated AKT signaling are resistant to chemotherapy and conversely, PTEN overexpression increases apoptotic susceptibility of cancer cells to chemotherapeutics (Frattini et al., 2007; Jiang et al., 2007; Yan et al., 2006). Therefore, suppressed AKT activation could also form the basis of our observation relating reduced survival of Pfn1 overexpressing cells in the face of apoptotic insult. It will be interesting to determine if expression of constitutively active AKT rescues the survival defect of Pfn1 overexpressing MDA-231 cells
In conclusion, we here highlight for the first time a molecular mechanism involving p27 underlying Pfn1-induced growth inhibition of MDA-231 breast cancer cells. It will be therefore interesting to further validate our observation in vivo as to whether p27 plays a role in the actual tumor-suppressive action of Pfn1 in MDA-231 breast cancer cells or in even other adenocarcinoma in which Pfn1 expression is downregulated.
Supplementary Material
Supplemental Fig 1: Immunostaining of phospho-S10-p27 of GFP and GFP-Pfn1 expressers (bar – 30 μm).
Supplemental Fig 2: SYBR-green fluorescence tracings of GAPDH and p27 RT-PCR products in real-time qRT-PCR experiments.
Acknowledgement
This study was funded by a grant from the National Institute of Health (CA108607) to P.R. We thank Tuhin Das for technical assistance.
References
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. Journal of Cell Biology. 1994;125(6):1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain SW, Massague J. Breast cancer banishes p27 from nucleus. Nat Med. 2002;8(10):1076–1078. doi: 10.1038/nm1002-1076. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Wiesner S, Braun A, Wimmer R, Berna A, Elad N, Medalia O, Pfeifer A, Aszodi A, Costell M, Fassler R. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. Embo J. 2009;28(8):1157–1169. doi: 10.1038/emboj.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218(2):436–443. doi: 10.1002/jcp.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119(Pt 19):4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97(8):1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128(2):269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- Ishida N, Hara T, Kamura T, Yoshida M, Nakayama K, Nakayama KI. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J Biol Chem. 2002;277(17):14355–14358. doi: 10.1074/jbc.C100762200. [DOI] [PubMed] [Google Scholar]
- Janke J, Schluter K, Jandrig B, Theile M, Kolble K, Arnold W, Grinstein E, Schwartz A, Estevez-Schwarz L, Schlag PM, Jockusch BM, Scherneck S. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J Exp Med. 2000;191(10):1675–1686. doi: 10.1084/jem.191.10.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Pore N, Cerniglia GJ, Mick R, Georgescu MM, Bernhard EJ, Hahn SM, Gupta AK, Maity A. Phosphatase and tensin homologue deficiency in glioblastoma confers resistance to radiation and temozolomide that is reversed by the protease inhibitor nelfinavir. Cancer Res. 2007;67(9):4467–4473. doi: 10.1158/0008-5472.CAN-06-3398. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- Kaldis P. Another piece of the p27Kip1 puzzle. Cell. 2007;128(2):241–244. doi: 10.1016/j.cell.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6(12):1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Agami R. Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle. 2007;6(22):2742–2749. doi: 10.4161/cc.6.22.4900. [DOI] [PubMed] [Google Scholar]
- Lee JG, Kay EP. Involvement of two distinct ubiquitin E3 ligase systems for p27 degradation in corneal endothelial cells. Invest Ophthalmol Vis Sci. 2008;49(1):189–196. doi: 10.1167/iovs.07-0855. [DOI] [PubMed] [Google Scholar]
- Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2(4):339–345. [PubMed] [Google Scholar]
- Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, Franssen E, Slingerland JM. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med. 2002;8(10):1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- Migita T, Oda Y, Masuda K, Hirata A, Kuwano M, Naito S, Tsuneyoshi M. Inverse relationship between E-cadherin and p27Kip1 expression in renal cell carcinoma. Int J Oncol. 2008;33(1):41–47. [PubMed] [Google Scholar]
- Motti ML, Califano D, Baldassarre G, Celetti A, Merolla F, Forzati F, Napolitano M, Tavernise B, Fusco A, Viglietto G. Reduced E-cadherin expression contributes to the loss of p27kip1-mediated mechanism of contact inhibition in thyroid anaplastic carcinomas. Carcinogenesis. 2005;26(6):1021–1034. doi: 10.1093/carcin/bgi050. [DOI] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8(1):9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12(24):2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79(4):551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9(10):1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- St Croix B, Sheehan C, Rak JW, Florenes VA, Slingerland JM, Kerbel RS. E-Cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1) J Cell Biol. 1998;142(2):557–571. doi: 10.1083/jcb.142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste MC, Nalbant P, Adamson ED, Hahn KM. Vinculin controls PTEN protein level by maintaining the interaction of the adherens junction protein beta-catenin with the scaffolding protein MAGI-2. J Biol Chem. 2005;280(7):5676–5681. doi: 10.1074/jbc.M405561200. [DOI] [PubMed] [Google Scholar]
- Uegaki K, Kanamori Y, Kigawa J, Kawaguchi W, Kaneko R, Naniwa J, Takahashi M, Shimada M, Oishi T, Itamochi H, Terakawa N. PTEN is involved in the signal transduction pathway of contact inhibition in endometrial cells. Cell Tissue Res. 2006;323(3):523–528. doi: 10.1007/s00441-005-0082-3. [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Luscher B. Post-translational regulation of the tumor suppressor p27(KIP1) Cell Mol Life Sci. 2008;65(20):3255–3264. doi: 10.1007/s00018-008-8296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14(8):461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A. 2001;98(7):3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenmayer N, Jandrig B, Rothkegel M, Schluter K, Arnold W, Haensch W, Scherneck S, Jockusch BM. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15(4):1600–1608. doi: 10.1091/mbc.E03-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Zhang W, Yang Y, Liang YL, Wang LY, Jin JW, Cai XM, Zha XL. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics. 2006;6(22):6095–6106. doi: 10.1002/pmic.200500321. [DOI] [PubMed] [Google Scholar]
- Yan X, Fraser M, Qiu Q, Tsang BK. Over-expression of PTEN sensitizes human ovarian cancer cells to cisplatin-induced apoptosis in a p53-dependent manner. Gynecol Oncol. 2006;102(2):348–355. doi: 10.1016/j.ygyno.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Zou L, Hazan R, Roy P. Profilin-1 overexpression restores adherens junctions in MDA-MB-231 breast cancer cells in R-cadherin-dependent manner. Cell Motil Cytoskeleton. 2009;66(12):1048–1056. doi: 10.1002/cm.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer. 2007;97(10):1361–1371. doi: 10.1038/sj.bjc.6604038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig 1: Immunostaining of phospho-S10-p27 of GFP and GFP-Pfn1 expressers (bar – 30 μm).
Supplemental Fig 2: SYBR-green fluorescence tracings of GAPDH and p27 RT-PCR products in real-time qRT-PCR experiments.