Abstract
Mitochondrial biogenesis occurs under basal conditions and is an adaptive response initiated by cells to maintain energetic demands and metabolic homeostasis after injuries targeting mitochondrial function. Identifying pharmacological agents that stimulate mitochondrial biogenesis is a critical step in the development of new therapeutics for the treatment of these injuries and to test the hypothesis that these agents will expedite recovery of cell and organ function after acute organ injuries. In this study, we examined the effects of N-[2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl]quinoxaline-2-carboxamide (SRT1720) on mitochondrial biogenesis and function in primary cultures of renal proximal tubule cells (RPTCs). We also tested the ability of this compound to restore mitochondrial functions after oxidant-induced RPTC injury. SRT1720 (3–10 μM) induced mitochondrial biogenesis in RPTCs within 24 h as determined by elevations in mitochondrial DNA copy number, increased expression of the mitochondrial proteins NADH dehydrogenase 1β subcomplex subunit 8 (NDUFB8) and ATP synthase β, and elevated mitochondrial respiration rates and ATP levels. Induction of mitochondrial biogenesis depended on mammalian sirtuin 1 (SIRT1) deacetylase activity, correlated with deacetylated nuclear peroxisome proliferator-activated receptor coactivator (PGC)-1α, and occurred in the absence of AMP-dependent kinase (AMPK) activation. Finally, SRT1720 treatment accelerated recovery of mitochondrial functions after acute oxidant injury. This study demonstrates that SRT1720 can induce mitochondrial biogenesis through SIRT1 activity and deacetylated PGC-1α, but not AMPK, in RPTCs within 24 h after oxidant injury. The results support further study of mitochondrial biogenesis as a repair process and a pharmacological target in acute organ injuries and disorders plagued by mitochondrial impairment.
Mitochondrial dysfunction is a primary pathological consequence of ischemic or toxic insults. In ischemic acute kidney injury (AKI), de-energization of the mitochondria and persistent energy depletion may hinder critical energy-dependent repair mechanisms and lead to irreversible cell injury, limiting restoration of organ function (Weinberg et al., 2000; Feldkamp et al., 2005). As such, there is therapeutic potential for agents that promote mitochondrial function to treat injuries characterized by mitochondrial impairment.
Mitochondrial biogenesis occurs under basal conditions and is an adaptive response initiated by cells to maintain energy demands or heat expenditure after injury, cold exposure, or caloric restriction (Puigserver et al., 1998; Yin et al., 2008). A primary regulator of mitochondrial biogenesis is the nuclear transcriptional coactivator peroxisome proliferator-activated receptor coactivator (PGC)-1α. Through induction of uncoupling proteins-2, nuclear respiratory factors 1/2, and as a coactivator of the promoter region of mitochondrial transcription factor A and the mitochondrial transcription specificity factors B1 and B2, PGC-1α has significant influence on mitochondrial function (Puigserver et al., 1998; Wu et al., 1999; Puigserver and Spiegelman, 2003; Gleyzer et al., 2005; Lin et al., 2005).
PGC-1α is highly expressed in metabolic tissues, and its expression and activity are regulated by a network of receptors, including the nuclear hormone receptors thyroid hormone and peroxisome proliferator-activated receptor γ (Puigserver et al., 1998; Wrutniak-Cabello et al., 2001); signaling pathways, such as the mitogen-activated protein kinase and Ca2+/calmodulin-dependent kinase pathways (Barger et al., 2001; Wu et al., 2002); and posttranslational phosphorylation, methylation, and acetylation modifications (Barger et al., 2001; Puigserver et al., 2001; Teyssier et al., 2005; Coste et al., 2008). In addition, PGC-1α transcription is regulated by the activity of signaling molecules and transcription factors such as protein kinase B, forkhead transcription factor, and myocyte enhancer factor-2 (Czubryt et al., 2003; Daitoku et al., 2003).
Under energetic crises, primary responders for maintaining energy and nutrient homeostasis are AMP-activated kinase (AMPK) and SIRT1. Rather than exclusive mechanisms of adaptation, recent evidence points to concurrent regulation and convergent mechanisms induced by AMPK and SIRT1 in response to changes in cellular energy levels and redox states, with a primary target of both pathways converging on PGC-1α (Cantó et al., 2009). AMPK monitors cellular energy levels, inducing ATP synthesis and inhibiting ATP expenditure when ATP levels are low (Hardie et al., 2003), by regulating expression of mitochondrial and metabolic genes via direct phosphorylation of PGC-1α (Jäger et al., 2007). SIRT1 is a nuclear protein that is also activated in response to energy depletion and promotes induction of genes that regulate metabolic adaptation to low energy levels. As a member of a conserved family of NAD+-dependent deacetylase enzymes known as the sirtuins, SIRT1 monitors cellular energy levels and becomes active in response to elevated NAD+/NADH ratios (Landry et al., 2000). SIRT1 catalyzes the deacetylation and activation of PGC-1α in both in vitro and in vivo systems (Nemoto et al., 2005; Rodgers et al., 2005), which may contribute to a protective role in metabolic regulation and resistance to oxidative stress (Howitz et al., 2003; Bordone and Guarente, 2005).
Several small molecules have been reported, such as resveratrol and isoflavone-derived compounds (Howitz et al., 2003; Rasbach and Schnellmann, 2008), to induce mitochondrial biogenesis in renal proximal tubule cells (RPTCs). SRT1720 was reported to be a SIRT1 activator, and exposure of this compound led to deacetylation of SIRT1 target proteins in both cells and animals (Milne et al., 2007; Feige et al., 2008). In genetic and diet-induced obese and diabetic rodents, 4 to 10 weeks of SRT1720 treatment improves insulin sensitivity and reduces plasma glucose levels while enhancing skeletal muscle mitochondrial activity (Milne et al., 2007). In addition, C2C12 cells treated with SRT1720 express elevated citrate synthase activity and ATP levels, suggesting induction of mitochondrial biogenesis (Smith et al., 2009). However, the acute effects of this compound in primary cultures of RPTCs cells, which better mimic the metabolic properties of cells in vivo compared with glycolytic cell lines, on mitochondrial biogenesis have not been explored. Furthermore, the effects of this compound in targeted injury models with mitochondrial impairment also have not been characterized.
Mitochondrial dysfunction contributes to oxidant-induced renal cell injury (Nowak et al., 1998), and PGC-1α plays a predominant role in the recovery of mitochondrial function after the initial injury (Rasbach and Schnellmann, 2007b). Overexpression of PGC-1α accelerates recovery of mitochondrial and cellular functions after oxidant injury (Rasbach and Schnellmann, 2007a), but because of the toxicity limitations in using adenovirus in vivo, there is a need for pharmacological agents that stimulate mitochondrial biogenesis to treat injuries characterized by mitochondrial impairment. In this study, we examined the mechanism of SRT1720 induced mitochondrial biogenesis and function in renal epithelial cells and tested the hypothesis that stimulation of mitochondrial function accelerates recovery from an acute cellular and mitochondrial injury.
Materials and Methods
Reagents.
PGC-1α (H300), NDUFB8, ATP synthase β, and GAPDH antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), Invitrogen (Carlsbad, CA), Abcam Inc. (Cambridge, MA), and Fitzgerald (Concord, MA), respectively. Acetylated lysine, phosphorylated AMPK (Thr172), and AMPK antibodies were obtained from Cell Signaling Technology Inc. (Danvers, MA). Sirtinol and nicotinamide (NAM) were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals were obtained from Sigma-Aldrich.
SRT1720 Synthesis and SIRT1 Activation.
SRT1720 was synthesized according to a procedure described previously (Milne et al., 2007) and was confirmed by NMR and mass spectrometry. The final product was purified by high-performance liquid chromatography. SIRT1 deacetylase activity was measured using a fluorescence-based SIRT1 activity kit (BIOMOL Research Laboratories, Plymouth Meeting, PA) according to the manufacturer's protocol as described previously (Rasbach and Schnellmann, 2008).
Isolation and Culture of Renal Proximal Tubules.
Female New Zealand White rabbits (∼2 kg) were purchased from Myrtle's Rabbitry (Thompson Station, TN). Renal proximal tubules were isolated by using an iron oxide uptake method described previously (Rasbach and Schnellmann, 2007b). Cells were cultured on 35-mm dishes in a medium consisting of 1:1 Dulbecco's modified Eagle's medium/Ham's F-12 (lacking glucose, phenol red, and sodium pyruvate), and supplemented with HEPES (15 mM), glutamine (2.5 mM), pyridoxine HCl (1 μM), sodium bicarbonate (15 mM), and lactate (6 mM). Hydrocortisone (50 nM), selenium (5 ng/ml), human transferrin (5 μg/ml), bovine insulin (10 nM), and l-ascorbic acid-2-phosphate (50 μM) were added daily to fresh culture medium. Experiments with RPTCs were conducted on the sixth day after plating when the cells had reached a confluent monolayer. Treatments were administered for 24 h unless otherwise noted. For M tert-butyl hydroperoxide (TBHP) injury experiments, cells were exposed to 400 μM TBHP for 45 min, at which time TBHP media were replaced with fresh media.
Preparation of Cell Lysates for Immunoblot Analysis.
Twenty-four hours after treatment, RPTCs were harvested in radioimmunoprecipitation assay lysis buffer consisting of 25 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS. Lysates were sonicated, and total protein was measured by BCA. Immunoblot analysis was performed as described previously (Rasbach and Schnellmann, 2007b).
Immunoprecipitation.
Cells were harvested from pooled culture dishes in a homogenization buffer consisting of 50 mM Tris-HCl, 1 mM β-mercaptoethanol, 1 mM EDTA, and 0.32 M sucrose. Cells were disrupted by sonication, and nuclei were collected by centrifugation at 900g for 10 min. After centrifugation, the nuclear pellet was resuspended in a nuclear lysis buffer consisting of 10 mM Tris, 500 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM sodium pyrophosphate, 1 mM NaVO4, 1 mM NaF, and protease inhibitors. Immunoprecipitations were carried out according to a protocol by Roche Diagnostics (Indianapolis, IN). Nuclear protein lysate (500 μg) and PGC-1α (5 μg) antibody were used for experiments. Immunoprecipitates were analyzed by immunoblotting using antibodies against acetylated lysine residues and PGC-1α. Supernatants collected from immunoprecipitations were analyzed for histone H3 expression as a control for initial nuclear protein input.
Quantitative Real-Time PCR.
Total RNA was isolated from cells with TRIzol reagent (Invitrogen). cDNA was synthesized from 5 μg of RNA template using a SuperScript II Reverse Transcriptase kit (Invitrogen). PCRs were carried out by using 2.5 μl of cDNA template combined with Brilliant II SYBR Green master mix (Stratagene, La Jolla, CA) at a final concentration of 1× and primers (Integrated DNA Technologies, Inc., Coralville, IA) at a concentration of 400 nM. Sequences of primers used for real-time PCRs were: PGC-1α (FW: 5′-AGG AAA TCC GAG CTG AGC TGA ACA-3′, REV: 5′-GCA AGA AGG CGA CAC ATC GAA CAA-3′) and GAPDH (FW: 5′-GAG CTG AAC GGG AAA CTC AC-3′, REV: 5′-CAC TGT TGA AGT CGC AGG AG-3′).
Mitochondrial DNA Content.
Real-time PCR was used to determine relative quantities of mitochondrial DNA content in SRT1720-treated cells and control cells. After a 24-h treatment, total genomic DNA was extracted by using a DNeasy Blood and Tissue kit (QIAGEN, Valencia, CA). DNA was quantified by measuring A260 values, and 50 ng of total DNA was used for PCRs. Primers specific to the mitochondrial-encoded ND6 gene (FW: ACT GCG ATG GCA ACT GAG GAG TAT, REV: ACC ATA ACT ATA CAA CGC CGC CAC) were used to assess mitochondrial DNA copy numbers. Primers designed against the nuclear-encoded Pou5f1 gene (FW: 5′-GGC CTA TGT CTT TTC CTC TGG-3′, REV: 5′-TCC AGG TTC TCT CTC CCT AGC-3′) were used for normalization.
Oxygen Consumption and ATP Levels.
Basal and FCCP-uncoupled oxygen consumption (QO2) and ATP levels were measured 24 h after treatment with SRT1720 and/or TBHP. QO2 was measured by using a Clark oxygen electrode as described previously (Nowak et al., 1998). ATP content was measured by using an ATP bioluminescent assay kit (BIOMOL Research Laboratories) as described previously and normalized to cellular protein (Rasbach and Schnellmann, 2007b).
Statistical Analysis.
Data are presented as means ± S.E.M. and were subjected to one-way analysis of variance. Multiple means were compared post hoc by using Student-Newman-Keuls test and were considered statistically different when p < 0.05. RPTCs isolated from a single rabbit represented an individual experiment (n = 1) and were repeated until an n of at least six was obtained.
Results
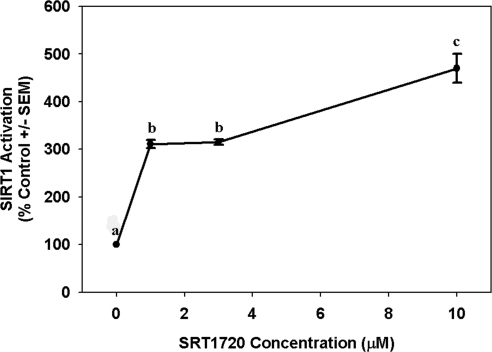
SRT1720 was reported to activate SIRT1 (Milne et al., 2007; Feige et al., 2008), and because SIRT1 activation can increase PGC-1α activity and mitochondrial functioning, we conducted a series of experiments to determine whether SRT1720 mediates mitochondrial biogenesis in primary cultures of RPTCs and, if so, by what mechanism. To verify SRT1720 potency, a fluorescence-based SIRT1 activity assay kit measuring deacetylation of a peptide target was used to examine SIRT1 deacetylase activity when exposed to SRT1720. SRT1720 increased SIRT1 activity in a concentration-dependent manner with a 3-fold increase in SIRT1 at 1 and 3 μM SRT1720 and a 5-fold increase at 10 μM (Fig. 1).
Fig. 1.
SRT1720 enhances SIRT1 deacetylase activity. Recombinant SIRT1 enzyme was incubated with acetylated peptide, NAD+, and various concentrations of SRT1720. Increasing concentrations of SRT1720 corresponded with increased SIRT1 activity, which was measured indirectly from a fluorescent signal produced that was relative to levels of deacetylated product. Data are presented as mean percentage of control ± S.E.M. Different superscripts indicate data are significantly different from each other (p < 0.05).
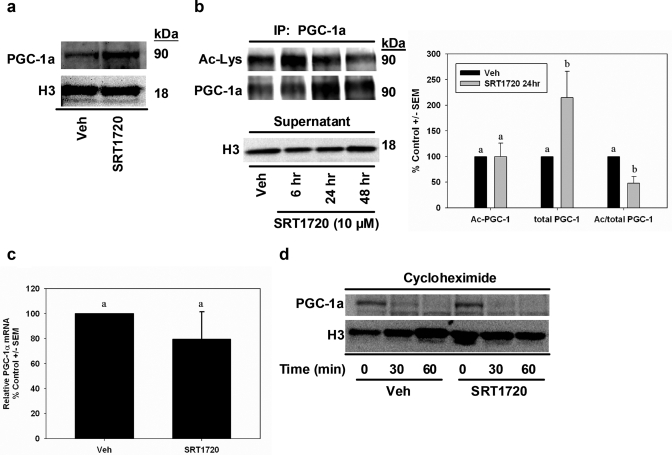
Because SIRT1 can modulate PGC-1α expression and/or activity by deacetylation (Nemoto et al., 2005; Rodgers et al., 2005), we assessed the expression and acetylation state of PGC-1α in RPTCs exposed to SRT1720 or vehicle for 24 h. Immunoblot analysis of nuclear lysates revealed elevated PGC-1α expression (Fig. 2a). To further examine nuclear PGC-1α content and acetylation state, PGC-1α protein was immunoprecipitated from nuclear lysates and subjected to immunoblot analysis with antibodies to acetylated lysine residues and PGC-1α (Fig. 2b). Time course analysis of acetylated PGC-1α consistently revealed reduced acetylation with 48-h SRT1720 treatment, with no differences at 24 h. Total PGC-1α levels in the immunoprecipitate were elevated at 24 and 48 h. The ratio of acetylated to total PGC-1α was decreased approximately 50% in SRT1720 cells at 24 h, indicating more active PGC-1α in the nucleus with SRT1720 treatment. We confirmed equal loading by measuring histone H3 in the supernatant from the immunoprecipitation experiments by immunoblot analysis (Fig. 2b).
Fig. 2.
Nuclear deacetylated PGC-1α expression is elevated in SRT1720-treated cells. a, nuclear lysates were fractionated from vehicle- and SRT1720-treated cells, and PGC-1 expression was assessed by immunoblot analysis. Histone H3 expression verified equal protein input within sample groups. b, acetylation state of PGC-1α was examined in nuclear lysates by immunoprecipitating PGC-1α followed by immunoblot analysis of acetylated lysine residues and PGC-1α. Because SRT1720 lysates contained more PGC-1α, supernatants from immunoprecipitations were subjected to histone H3 immunoblot analysis for input control. c, PGC-1α mRNA expression in SRT1720 and vehicle cells was determined by real-time PCR using primers designed to measure PGC-1α transcripts and GAPDH as internal control. d, PGC-1α degradation was examined in SRT1720 cells by extracting nuclear protein at 0, 30, and 60 min after cycloheximide exposure and immunoblotting for PGC-1α expression. Histone H3 expression verified protein input. Data are presented as mean percentage of control ± S.E.M. Different superscripts indicate data are significantly different from each other (p < 0.05).
Because active PGC-1α promotes transcription of the PGC-1 gene by an autoregulatory feedback loop (Czubryt et al., 2003), we examined transcript levels of PGC-1α by real-time PCR but found no differences between vehicle and SRT1720-treated cells (Fig. 2c). Because modifications to PGC-1α may regulate degradation of the protein, we tested whether the increased expression of PGC-1α was caused by decreased proteasomal degradation. We have characterized previously the degradation of PGC-1α in RPTCs and showed that it has a short half-life (37 min) (Rasbach et al., 2008). RPTCs were treated with vehicle or SRT1720 for 24 h, and then protein translation was inhibited with cycloheximide (100 μM) and samples taken 30 and 60 min later. Nuclear lysates were probed for PGC-1α expression by immunoblot analysis. No changes in PGC-1α degradation were observed in SRT1720 and vehicle-treated RPTCs (Fig. 2d). Taken together, these data provide evidence that SRT1720 treatment induced accumulation of deacetylated nuclear PGC-1α in RPTCs that was not the result of either elevated PGC-1α transcription at 24-h treatment or decreased proteasomal degradation.
Mitochondrial biogenesis was determined by assessing mitochondrial DNA copy number, expression of mitochondrial proteins, and mitochondrial function after 24 h of SRT1720 treatment. Relative mitochondrial DNA copy number was determined by using quantitative real-time PCR to examine the ratio of a select mitochondrial-encoded gene over nuclear DNA in SRT1720 and vehicle-treated cells (Fig. 3a). There was a 3.5-fold increase in mitochondrial-encoded NADH dehydrogenase subunit 6 (ND6) DNA in SRT1720 cells compared with controls. Nuclear-encoded Pou5f1 was used for normalization.
Fig. 3.
SRT1720 induces mitochondrial biogenesis in RPTCs within 24 h. a, mitochondrial DNA copy numbers were assessed by real-time PCR. DNA isolated from RPTCs treated with vehicle or 10 μM SRT1720 was analyzed by real-time PCR for relative quantities of the mitochondrial gene ND6 and the nuclear gene Pou5f1. b, mitochondrial proteins ATP synthase β and NDUFB8 were measured by immunoblot analysis in cells treated with 1, 3, or 10 μM SRT1720. GAPDH immunoblots were performed to verify equal protein input. c, mitochondrial function was assessed in vehicle and SRT1720-treated cells. Basal and FCCP-uncoupled respiration and ATP levels were measured in RPTCs treated with 1, 3, or 10 μM SRT1720. Total protein was measured by BCA and used for normalization of data. Data are presented as mean percentage of control ± S.E.M. Different superscripts indicate data are significantly different from each other (p < 0.05).
Then, the effect of SRT1720 treatment on mitochondrial protein levels was explored. SRT1720 (10 μM) elevated ATP synthase β, a nuclear-encoded protein within the F1 subunit of the ATP synthase, 1.5-fold over controls (Fig. 3b). NDUFB8, a nuclear-encoded complex I subunit, also was elevated approximately 1.5-fold over control by SRT1720.
Mitochondrial function was determined by measuring cellular respiration and ATP levels in RPTCs. Compared with controls, basal respiration was elevated approximately 1.5-fold with 3 or 10 μM treatments at 24 h (Fig. 3c). Uncoupled respiration was elevated approximately 1.5-fold at the same concentrations. Finally, ATP levels were also elevated (1.8-fold) over vehicle controls (Fig. 3c). Taken together, the elevations in mitochondrial DNA, proteins, and functional capacity provide strong evidence that mitochondrial biogenesis occurs in RPTCs with SRT1720 treatment.
To verify that the RPTC mitochondrial biogenesis produced by SRT1720 depends on SIRT1 activation, pharmacological inhibitors were used to block SIRT1 activity before SRT1720 exposure, and then mitochondrial DNA content and function were analyzed. SRT1720 treatment elevated mitochondrial DNA content compared with vehicle-treated cells, whereas cells exposed to SRT1720 in the presence of the SIRT1 inhibitor NAM (100 μM) did not show any changes in mitochondrial DNA (Fig. 4a). In addition, RPTCs exposed to SRT1720 for 24 h demonstrated elevations in ATP levels compared with vehicle cells (Fig. 4b). Pretreatment of RPTCs with the synthetic SIRT1 inhibitor sirtinol (100 μM) or NAM prevented the SRT1720-mediated increased ATP levels at 24 h. The data from these experiments verify that SIRT1 is required for SRT1720-induced mitochondrial biogenesis.
Fig. 4.
SRT1720-induced mitochondrial biogenesis is SIRT1-dependent. a, RPTCs were treated with 10 μM SRT1720 alone or in the presence of the SIRT1 inhibitor NAM (100 μM) for 1 h before SRT1720 addition. Mitochondrial DNA levels were analyzed by real-time PCR for the mitochondrial gene ND6. The nuclear-encoded gene Pou5f1 was used for normalization. b, RPTCs were treated with SRT1720 alone or in combination with the SIRT1 inhibitors nicotinamide or sirtinol. ATP levels were measured 24 h after SRT1720 addition. Total protein was measured by BCA and used for normalization. Data are presented as mean percentage of control ± S.E.M. Different superscripts indicate data are significantly different from each other (p < 0.05).
AMPK, a primary energy regulator, monitors AMP/ATP levels and activates energy-producing mechanisms when this ratio is elevated (Hardie et al., 2003). AMPK regulates energy supply by directly phosphorylating modulators of metabolic pathways, including PGC-1α (Jäger et al., 2007). Indeed, PGC-1α has at least two sites available for AMPK-mediated phosphorylation, and activators of SIRT1, such as resveratrol, can also induce activation of AMPK (Zang et al., 2006). To determine whether SRT1720 also induces AMPK activation, RPTCs were treated for 1 and 24 h with SRT1720 or vehicle and activation of AMPK was detected by immunoblotting for phosphorylated AMPK (Thr172). AICAR and metformin were used concurrently as positive controls for AMPK activation. At both 1 and 24 h, there was no effect on pAMPK levels by SRT1720, whereas a significant induction was observed with metformin treatment at both time points (Fig. 5). Contrary to previous reports in other systems (Zang et al., 2006), we did not observe any changes in pAMPK with AICAR treatment. Total AMPK levels did not change with any treatment. These data provide evidence that SRT1720 acts through SIRT1 activation and not concurrent activation of AMPK.
Fig. 5.
SRT1720 does not activate AMPK. RPTCs treated with 10 μM SRT1720 or vehicle for 1 or 24 h were subjected to immunoblot analysis using antibodies to detect phosphorylated AMPK (Thr172), total AMPK, and GAPDH. The known AMPK activators AICAR (500 μM) and metformin (1 mM) were used as positive controls for pAMPK antibody. GAPDH expression was analyzed for load control. Different superscripts indicate data are significantly different from each other (p < 0.05).
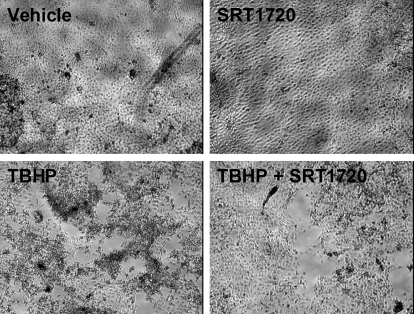
Because PGC-1α and mitochondrial biogenesis have a pivotal role in the recovery of RPTCs from oxidant-induced mitochondrial dysfunction (Rasbach and Schnellmann, 2007a,b), we tested the hypothesis that pharmacological activation of mitochondrial biogenesis after injury would expedite recovery of mitochondrial functions in RPTCs. RPTCs were incubated with 400 μM TBHP to induce oxidant injury. At 6 h after injury, RPTCs were treated with SRT1720 to stimulate mitochondrial biogenesis. At 24 h, mitochondrial function and cell morphology of injured RPTCs treated with SRT1720 or vehicle were examined. Uncoupled respiration and ATP levels were approximately 60% of control in TBHP-injured RPTCs at 24 h (Fig. 6). In contrast, injured cells treated with SRT1720 demonstrated partial recovery of uncoupled respiration and full recovery of ATP levels 24 h after injury (Fig. 6). Correlating with partial recovery of mitochondrial functions, recovery of RPTC morphology was observed in injured cells treated with SRT1720. Six hours after TBHP exposure, the injury was characterized by a loss of approximately 50% of cells as visualized by denuded areas of the dish as cells had sloughed off the plate surface, and a generalized shrinkage and rounding of adherent cells. RPTCs treated with SRT1720 for 24 h after injury reverted to a preinjury state characterized by reorganization and migration of surviving cells returning to a confluent monolayer and dome formation indicative of polarized RPTCs (Fig. 7). This recovery was not as apparent in vehicle-treated injured cells. The data from these experiments indicate that SIRT1 activation can reverse oxidant-induced mitochondrial dysfunction, and recovery of mitochondrial numbers and function may aid in recovery of RPTC morphology after acute injury.
Fig. 6.
Mitochondrial function is rescued in SRT1720-treated cells after oxidant injury. RPTCs injured with the oxidant TBHP were treated with 10 μM SRT1720 or an equal volume of dimethyl sulfoxide 6 h after injury and ATP levels and FCCP-uncoupled respiration were measured 24 h after injury. Total protein was measured by BCA for data normalization. Data are presented as mean percentage of control ± S.E.M. Different superscripts indicate data are significantly different from each other (p < 0.05).
Fig. 7.
RPTC morphology is partially recovered in cells treated with SRT1720 after TBHP toxicity. RPTCs exposed to 400 μM TBHP were treated with 10 μM SRT1720 or dimethyl sulfoxide 6 h after injury and then examined by light microscopy (10× magnification) for changes in cell morphology at 24 h after injury.
Discussion
Mitochondrial dysfunction is a common mechanism in the etiology of organ injuries and diseases characterized by metabolic insufficiency. Mitochondrial health is essential for cell and organ function because of their role in ATP production, fatty acid and lipid metabolism, signaling pathways, and apoptosis. Despite potential for treating disorders characterized by mitochondrial impairment, very few therapies target the mitochondria to promote its function. In this study, we demonstrated that pharmacologically induced mitochondrial biogenesis enhanced mitochondrial function in RPTCs and restored function after an acute injury.
SRT1720 stimulated mitochondrial biogenesis in RPTCs within 24 h of exposure. Elevated levels of mitochondrial DNA, proteins, and function were observed with 10 μM treatment. The findings agree with results we have published previously linking isoflavone-induced mitochondrial biogenesis with SIRT1 activation (Rasbach and Schnellmann, 2008), and others who have demonstrated mitochondrial biogenesis with resveratrol in other cell types (Lagouge et al., 2006; Csiszar et al., 2009). The pharmacological advantage of SRT1720 over isoflavones is that SRT1720 produces mitochondrial biogenesis within 24 h, a key requirement if targeting acute organ injury.
Although SRT1720 was reported previously as an SIRT1 activator, its mechanism of mitochondrial biogenesis in a cellular system is incomplete. Previous studies examining SRT1720-induced mitochondrial biogenesis have based their interpretations primarily on indirect mitochondrial measurements, such as respiration and ATP levels and electron transport chain activity, predominantly in skeletal muscle cell lines (Feige et al., 2008; Smith et al., 2009). In this study, we sought to explore SRT1720-induced mitochondrial biogenesis in primary kidney cell cultures, which better mimic the metabolic properties of renal cells in vivo, not only by examining alterations in functional output but also by examining direct measurements of mitochondrial protein and DNA expression. When primary RPTC cultures were incubated with SRT1720, mitochondrial proteins NDUFB8 and ATP synthase β, and mitochondrial DNA copy numbers were elevated compared with vehicle-treated cells (Fig. 3), indicating mitochondrial biogenesis occurred within 24 h. Furthermore, we confirmed that elevations in mitochondrial components corresponded with increased mitochondrial output by examining cellular respiration and ATP production. Finally, we verified that the observed effects of SRT1720 depended on SIRT1 activity by using pharmacological inhibitors of SIRT1 (Fig. 4), similar to what has been shown previously in other cell types using SIRT1 short hairpin RNA (Feige et al., 2008).
SIRT1 activation results in deacetylation of target proteins, and several substrates have been identified, including PGC-1α. SRT1720 elevated expression of deacetylated nuclear PGC-1α at 24 h in RPTCs (Fig. 2, a and b). The elevated expression was neither the result of increased PGC-1 transcription, because we did not observe any changes in PGC-1α mRNA expression (Fig. 2c), nor an increased resistance to proteasomal-targeted degradation (Fig. 2d). When SRT1720-treated cells were exposed to the inhibitor of protein translation cycloheximide, nuclear PGC-1α degraded at the same rate as vehicle cells, indicating the protein is still susceptible to proteasomal degradation (Sano et al., 2007; Rasbach et al., 2008). Taken together, these data indicate that SRT1720 did not induce PGC-1α transcription or increase stability of the protein. However, it is possible that the increased expression of nuclear PGC-1α may have been the result of an earlier transcriptional event that was missed by examining the 24-h time point or may be the result of increased nuclear sequestration.
We did not observe any activation of AMPK with SRT1720 as examined by immunoblotting for Thr172-phosphorylated AMPK or in total AMPK (Fig. 5), which is consistent with previous reports that this compound exerts its effects in an AMPK-independent mechanism (Feige et al., 2008). It is interesting to note that metformin induced a robust phosphorylation of AMPK within 1 h of treatment that was maintained for at least 24 h. In contrast, AICAR, which has been shown previously to induce phosphorylation of AMPK in other cell types (Zang et al., 2006), did not have any effect on pAMPK expression after 1- or 24-h treatment in RPTCs. We have not explored the reason for the differential effects of AICAR in RPTCs and other cell types.
PGC-1α is an emerging therapeutic target for mitochondrial abnormalities because of its regulatory role in controlling metabolic processes and mitochondrial activities and biogenesis within the cell. Enhancing PGC-1α expression or activity has proven effective in reversing the phenotypic consequences of mitochondrial impairment. Mitochondrial myopathies can be rescued through transgenic expression of PGC-1α or the pharmacologic peroxisome proliferator-activated receptor pan-agonist bezafibrate, both of which induce mitochondrial biogenesis, enhance respiratory capacity, conserve ATP levels, and prolong life span (Wenz et al., 2008). Pharmacological stimulation or adenoviral up-regulation of PGC-1α rescued mitochondrial function and bioenergetics and restored insulin signaling in insulin-resistant skeletal muscle cells (Pagel-Langenickel et al., 2008). Finally, the benefits of exercise and caloric restriction to rescue or protect against metabolic deficiencies have been linked to enhanced PGC-1α activity (Koves et al., 2005; Boily et al., 2008).
Recent evidence indicates that induction of PGC-1α and mitochondrial biogenesis is a critical adaptive response to maintain energy levels and metabolic demands required during recovery from certain acute injuries to cells and organs (Rasbach and Schnellmann, 2007b; Wang et al., 2008; Yin et al., 2008). In response to partial hepatectomy, CCAAT/enhancer-binding protein β transcriptionally induces PGC-1α to maintain metabolic homeostasis and energy demands of the regenerating liver (Wang et al., 2008). In response to oxidant-induced mitochondrial dysfunction in RPTCs, induction of PGC-1α and mitochondrial biogenesis is an adaptive repair mechanism initiated by the cell, which can be stimulated by PGC-1α overexpression (Rasbach and Schnellmann, 2007a,b). Here, we show that pharmacologically induced mitochondrial biogenesis also rescues mitochondrial functions after oxidant-induced injury. Within 24 h, SRT1720 reversed mitochondrial dysfunction and ATP depletion resulting from TBHP toxicity (Fig. 6).
Although the majority of studies investigating PGC-1α-mediated mitochondrial regulation through AMPK or SIRT1 are focusing on its role in chronic or age-related metabolic deficiencies (Guarente, 2007; Milne and Denu, 2008), this pathway offers a unique target for the treatment of acute organ injuries that are also plagued by mitochondrial impairment. As observed in this study and previous reports (Rasbach and Schnellmann, 2007a,b), mitochondrial biogenesis has a pivotal role in recovery of critical mitochondrial functions in oxidant-injured renal cells. Acute organ injuries, such as ischemic AKI, are characterized by de-energization of the mitochondria and loss of mitochondrial proteins and depletion of cellular energy stores (Weinberg et al., 2000; Bonventre and Weinberg, 2003; Feldkamp et al., 2005), which could exacerbate cell death and organ failure or limit energy-dependent repair processes if mitochondrial function is not restored. These studies provide evidentiary basis to study the involvement of mitochondrial repair processes in the recovery from organ injuries such as AKI and highlight the therapeutic potential of pharmacological inducers of mitochondrial biogenesis to rescue mitochondrial function in injuries and disorders plagued by mitochondrial impairment.
This study was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM084147] and the National Institute of Environmental Health Sciences Training Program in Environmental Stress Signaling [Grant T32-ES012878].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.161992.
- SRT1720
- N-[2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl]quinoxaline-2-carboxamide
- AKI
- acute kidney injury
- PGC
- peroxisome proliferator-activated receptor coactivator
- AMPK
- AMP-activated kinase
- SIRT1
- mammalian sirtuin 1
- NAM
- nicotinamide
- RPTC
- renal proximal tubule cell
- NDUFB8
- NADH dehydrogenase 1β subcomplex subunit 8
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TBHP
- tert-butyl hydroperoxide
- BCA
- bicinchoninic acid
- PCR
- polymerase chain reaction
- FW
- forward
- REV
- reverse
- QO2
- oxygen consumption
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- ND6
- NADH-ubiquinone oxidoreductase chain 6
- AICAR
- aminoimidazole carboxamide ribonucleotide
- p
- phosphorylated.
References
- Barger PM, Browning AC, Garner AN, Kelly DP. (2001) p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem 276:44495–44501 [DOI] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, et al. (2008) SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One 3:e1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre JV, Weinberg JM. (2003) Recent advances in the pathophysiology of ischemic acute renal failure. J Am Soc Nephrol 14:2199–2210 [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. (2005) Calorie restriction, SIRT1, and metabolism: understanding longevity. Nat Rev Mol Cell Biol 6:298–305 [DOI] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. (2008) The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1α. Proc Natl Acad Sci USA 105:17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar A, Labinskyy N, Pinto JT, Ballabh P, Zhang H, Losonczy G, Pearson K, de Cabo R, Pacher P, Zhang C, et al. (2009) Resveratrol induces mitochondrial biogenesis in endothelial cells. Am J Physiol Heart Circ Physiol 297:H13–H20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czubryt MP, McAnally J, Fishman GI, Olson EN. (2003) Regulation of peroxisome proliferator-activated receptor γ coactivator 1 α (PGC-1α) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci USA 100:1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. (2003) Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52:642–649 [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. (2008) Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8:347–358 [DOI] [PubMed] [Google Scholar]
- Feldkamp T, Kribben A, Weinberg JM. (2005) Assessment of mitochondrial membrane potential in proximal tubules after hypoxia-reoxygenation. Am J Physiol Renal Physiol 288:F1092–F1102 [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC. (2005) Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol 25:1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72:483–488 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Scott JW, Pan DA, Hudson ER. (2003) Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546:113–120 [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191–196 [DOI] [PubMed] [Google Scholar]
- Jäger S, Handschin C, St-Pierre J, Spiegelman BM. (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci USA 104:12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. (2005) Peroxisome proliferator-activated receptor-γ coactivator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 280:33588–33598 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. (2006) Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA 97:5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1:361–370 [DOI] [PubMed] [Google Scholar]
- Milne JC, Denu JM. (2008) The Sirtuin family: therapeutic targets to treat diseases of aging. Curr Opin Chem Biol 12:11–17 [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. (2005) SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem 280:16456–16460 [DOI] [PubMed] [Google Scholar]
- Nowak G, Aleo MD, Morgan JA, Schnellmann RG. (1998) Recovery of cellular functions following oxidant injury. Am J Physiol Renal Physiol 274:F509–F515 [DOI] [PubMed] [Google Scholar]
- Pagel-Langenickel I, Bao J, Joseph JJ, Schwartz DR, Mantell BS, Xu X, Raghavachari N, Sack MN. (2008) PGC-1α integrates insulin signaling, mitochondrial regulation, and bioenergetic function in skeletal muscle. J Biol Chem 283:22464–22472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. (2001) Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell 8:971–982 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. (2003) Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr Rev 24:78–90 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829–839 [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Green PT, Schnellmann RG. (2008) Oxidants and Ca2+ induce PGC-1alpha degradation through calpain. Arch Biochem Biophys 478:130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG. (2007a) PGC-1α overexpression promotes recovery from mitochondrial dysfunction and cell injury. Biochem Biophys Res Commun 355:734–739 [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG. (2007b) Signaling of mitochondrial biogenesis following oxidant injury. J Biol Chem 282:2355–2362 [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG. (2008) Isoflavones promote mitochondrial biogenesis. J Pharmacol Exp Ther 325:536–543 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- Sano M, Tokudome S, Shimizu N, Yoshikawa N, Ogawa C, Shirakawa K, Endo J, Katayama T, Yuasa S, Ieda M, et al. (2007) Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor γ coactivator 1α. J Biol Chem 282:25970–25980 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Kenney RD, Gagne DJ, Frushour BP, Ladd W, Galonek HL, Israelian K, Song J, Razvadauskaite G, Lynch AV, et al. (2009) Small molecule activators of SIRT1 replicate signaling pathways triggered by calorie restriction in vivo. BMC Syst Biol 3:31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. (2005) Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev 19:1466–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peiris TH, Mowery A, Le Lay J, Gao Y, Greenbaum LE. (2008) CCAAT/enhancer binding protein-β is a transcriptional regulator of peroxisome-proliferator-activated receptor-γ coactivator-1α in the regenerating liver. Mol Endocrinol 22:1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JM, Venkatachalam MA, Roeser NF, Nissim I. (2000) Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci USA 97:2826–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Diaz F, Spiegelman BM, Moraes CT. (2008) Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab 8:249–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wrutniak-Cabello C, Casas F, Cabello G. (2001) Thyroid hormone action in mitochondria. J Mol Endocrinol 26:67–77 [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. (2002) Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296:349–352 [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124 [DOI] [PubMed] [Google Scholar]
- Yin W, Signore AP, Iwai M, Cao G, Gao Y, Chen J. (2008) Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke 39:3057–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang M, Xu S, Maitland-Toolan KA, Zuccollo A, Hou X, Jiang B, Wierzbicki M, Verbeuren TJ, Cohen RA. (2006) Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes 55:2180–2191 [DOI] [PubMed] [Google Scholar]