Abstract
Glutamate receptors are the most prevalent excitatory neurotransmitter receptors in the vertebrate central nervous system and are important potential drug targets for cognitive enhancement and the treatment of schizophrenia. Allosteric modulators of AMPA receptors promote dimerization by binding to a dimer interface and reducing desensitization and deactivation. The pyrrolidine allosteric modulators, piracetam and aniracetam, were among the first of this class of drugs to be discovered. We have determined the structure of the ligand binding domain of the AMPA receptor subtypes GluA2 and GluA3 with piracetam and a corresponding structure of GluA3 with aniracetam. Both drugs bind to both GluA2 and GluA3 in a very similar manner, suggesting little subunit specificity. However, the binding sites for piracetam and aniracetam differ considerably. Aniracetam binds to a symmetrical site at the center of the dimer interface. Piracetam binds to multiple sites along the dimer interface with low occupation, one of which is a unique binding site for potential allosteric modulators. This new site may be of importance in the design of new allosteric regulators.
Keywords: piracetam, aniracetam, glutamate receptor, AMPA receptor, allosteric modulator, receptor structure
INTRODUCTION
Ionotropic glutamate receptors (iGluRs) are found on most vertebrate central nervous system neurons and are responsible for the majority of excitatory synaptic transmission.1 In addition to important roles in processes such as learning and memory,2–4 these receptors have been associated with disorders such as Parkinson’s and Alzheimer’s diseases, Huntington’s chorea, and neurological disorders including epilepsy and ischemic brain damage.4–6
iGluRs are composed of four subunits arranged around a central ion channel. Three major subtypes have been identified that are characterized by pharmacological properties, sequence, functionality, and biological role into those that are sensitive to: (1) the synthetic agonist N-methyl-D-aspartic acid (NMDA; GluN1, GluN2A-D, GluN3A-B); (2) the synthetic agonist α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA; GluA1-4); and (3) the naturally occurring neurotoxin kainate (GluK1-3). The ligand binding domains of AMPA receptors can exist in two alternatively spliced forms (flip and flop) that differ in the rate of desensitization,7 channel closing rate,8 and sensitivity to allosteric modulators.9
The extracellular agonist-binding domain (S1S2 domain) of ionotropic glutamate receptors consists of two lobes with the agonist-binding pocket located between the two lobes.10 The structures of the S1S2 domain from GluR0,11 GluA2,12–16 GluA3,17 GluA4,18 GluK1,19–21 GluK2,19, 22 GluN1,23 and GluN224 subtypes have been determined in the presence of a variety of agonists, partial agonists, and antagonists. This has provided structural insights into the processes of channel activation and desensitization as well as the structural basis of the differences in the agonist and antagonist selectivity. Recently, the full tetrameric structure of the GluA2 receptor has been determined bound to an antagonist.25 Although the structure was determined in the closed channel form, the availability of this structure provides new insights into the quaternary structure of all iGluRs and will provide a very useful framework for understanding channel activation.
Piracetam was the first nootropic agent discovered26 and was found originally to be effective as a protective agent in hypoxia-induced amnesia.27 Although it affects a number of neuronal proteins, some evidence suggests that it can act at AMPA receptors.28, 29 Aniracetam, another member of the racetam family of drugs, is considerably more potent and acts directly on AMPA receptors.9, 30 Aniracetam and other allosteric modulators of AMPA receptors can block or slow desensitization, slow deactivation, and enhance synaptic plasticity.31, 32 These characteristics have led to clinical trials of several drugs for Alzheimer’s disease and attention deficit disorder,33, 34 but only aniracetam is available for mild dementia in Europe.35 The structure of aniracetam bound to a dimer of the agonist-binding domain of GluA2o has been reported.30 It binds in a symmetrical site at the dimer interface. This is consistent with other allosteric activators, which bind to the dimer interface and slow or prevent the dissociation of the dimer. The dimer forms upon activation of the receptor by agonist36 and is thought to represent the active form of the protein. Preventing dimer dissociation can block or slow desensitization36 as well as having effects on deactivation.30 The binding sites for allosteric activators at the dimer interface is actually a large surface with several subsites, including a central subsite (A) and two pairs of lateral subsites (B/B′ and C/C′; Figure 2).37 A binding site for allosteric activators can consist of one or more of the five subsites. Typically, activators whose binding site includes the A subsite, bind in one copy per dimer. Because of the symmetry of the dimer interface, density corresponding to two overlapping molecules is observed. This is true of aniracetam, CX614, CX546, and PEPA. On the other hand, the benzothiadiazide activators (cyclothiazide, IDRA-21, etc.) bind in two copies per dimer interface at binding sites including the two lateral subsites (B/B′ and C/C′) that are differentially occupied depending upon the structure of the benzothiadiazide.37 This rather large surface with nonoverlapping subsites suggests a means by which activator affinity can be increased, that is, by generating multivalent compounds that can interact with different subsites. The proof of this principle was demonstrated by the dimeric biarylpropylsulfonamide compound synthesized by Kaae and collaborators.38
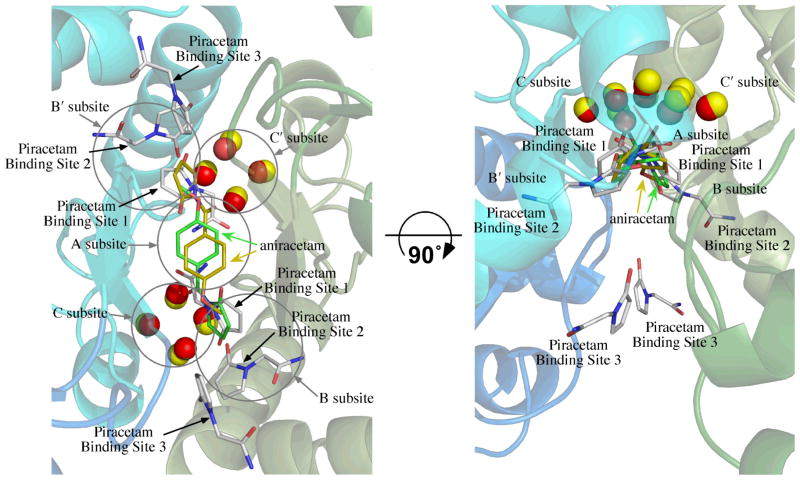
Figure 2.
Overlay of the position of aniracetam bound to GluA2o (fluorowillardiine in the agonist binding site, 2AL5)30 on the structure of piracetam bound to GluA2i (glutamate in the binding site). One molecule of aniracetam is bound to a given dimer interface, but since the interface is symmetrical, two orientations are observed. One is shown with carbons colored yellow and the other with carbons colored green. The water molecules from the aniracetam structure are shown as yellow spheres. The six molecules of piracetam are shown with white carbons and the water molecules as red spheres. The binding sites for piracetam consist of two sets of three distinct sites labeled Binding Sites 1 through 3. Subsites A, B/B′, and C/C′, described previously,37 are shown as circles. The structure is rotated by 90° to illustrate the position of the C/C′ subsites and Piracetam Site 3. Coloring of the backbone is the same as Figure 1.
We report here the three dimensional structures of aniracetam bound to the S1S2 domain of GluA3i and piracetam bound to the S1S2 domains of GluA3i, GluA2i, and GluA2o. In all cases, glutamate was in the agonist-binding site. Aniracetam binds to GluA3i in a manner similar to GluA2o,30 with only small changes of side chain rotameric states. Likewise, piracetam binding did not show any obvious subtype differences, but the density was lower for GluA3. Although it is important to note that subtle differences in outlying regions can affect binding affinity.18 However, piracetam bound in multiple positions along the dimer interface. The most prominent position was at the lateral edge of the dimer interface and represents a unique subsite that has not been reported previously. This new site may be of value for the design of new multivalent allosteric modulators.
EXPERIMENTAL
Materials
Glutamate was obtained from Sigma Aldrich (St. Louis, MO). Piracetam and aniracetam were purchased from Tocris (Ellisville, MO) and were of >99.5% purity as judged by HPLC. The GluA2 S1S2J construct (flop form; denoted GluA2o) was obtained from Eric Gouaux (Vollum Institute).12 The linker region, which replaces the ion channel cassette, consisted of two residues, glycine and threonine. The N754S mutation was prepared to allow binding of flip-specific allosteric modulators.37 This is denoted GluA2i. The construct for the flip form of GluA3 (GluA3i) has been described previously.17
Protein Preparation and Purification
All S1S2 proteins were prepared as described by Furukawa and Gouaux23 with slight modification. Briefly, pET-22b(+) plasmids were transformed in E. coli strain Origami B (DE3) cells and were grown at 37°C to OD600 of 0.9 to 1.0 in LB medium supplemented with the antibiotics (ampicillin and kanamycin). The cultures were cooled to 20°C for 20 min and isopropyl-β-D-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM. Cultures were allowed to grow at 20°C for 20 h. The cells were then pelleted and the S1S2 protein purified using a Ni-NTA column, followed by a sizing column (Superose 12, XK 26/100), and finally an HT-SP-ion exchange-Sepharose column (Amersham Pharmacia). Glutamate (1 mM) was maintained in all buffers throughout purification. After the last column, the protein was concentrated and stored in 20 mM sodium acetate, 1 mM sodium azide, and 10 mM glutamate at pH 5.5.
Crystallization
For crystallization trials, the proteins were concentrated to 0.3 mM using a Centricon 10 centrifugal filter (Millipore, Bedford, MA) in the presence of 10 mM glutamate. The final piracetam and aniracetam concentrations were 5 mM. All crystals were grown at 4°C using the hanging drop technique, and the drops contained a 1:1 (v/v) ratio of protein solution to reservoir solution. The reservoir solution was 16–18% polyethylene glycol (PEG) 8K, 0.1 M sodium cacodylate, and 0.1–0.15 M zinc acetate (pH 6.5).
Crystal structure determination
Data were collected at the Cornell High Energy Synchrotron Source beam line A1 using a Quantum-210 Area Detector Systems charge-coupled device detector. Data sets were indexed and scaled with HKL-2000.39 Structures were solved with molecular replacement and refined using Phenix.40 Coot 0.6 was used for model building.41 The glutamate-bound GluA2 structure, 3DP6, was used as the search model for GluA2, and 3DLN was used for GluA3.17
RESULTS AND DISCUSSION
Binding sites for piracetam on GluA2 S1S2
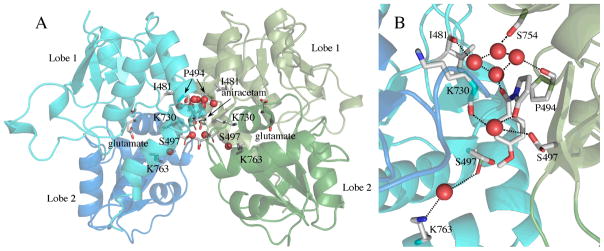
Like other allosteric modulators of AMPA receptors,30, 36–38, 42 piracetam binds to the S1S2 dimer interface of GluA2. Figure 1A shows the position of the dimer in the context of the full-length receptor. The crystal structures of piracetam bound to both the flop form of GluA2 (GluA2o) and the N754S mutation of the flop form, which mimics the flip form (GluAi), were determined. As shown in Figure 1B, piracetam binds in six positions. Because of the symmetry of the dimer, this corresponds to two sets of three distinct binding sites. The differences between GluA2o and GluA2i were limited to several changes in rotameric states of sidechains, so only the structure of GluA2i is shown. In general, the density for piracetam is lower than that of the protein, suggesting either that the modulator has some mobility in the binding site or that only a portion of the binding sites are occupied. The later possibility is consistent with the low affinity of the compound for AMPA receptors.28
Figure 1.
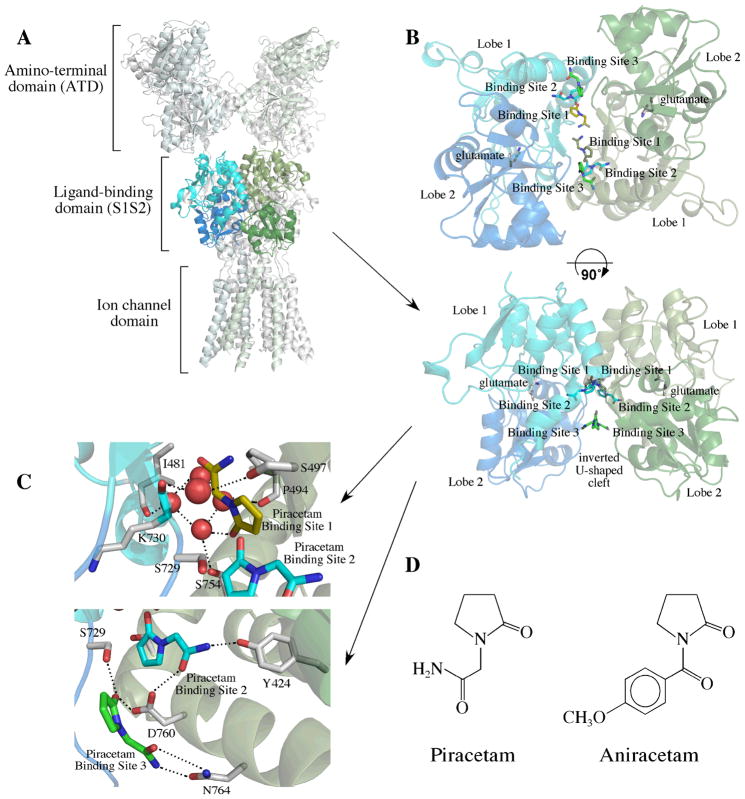
Structure of GluA2 bound to piracetam. (A) Position of the ligand binding domain (S1S2) dimer in the context of the full-length tetrameric receptor. Shown is the crystal structure of GluA2 bound to the antagonist, ZK200775,25 with the ligand binding domain of one monomer colored in shades of blue and the other colored in shades of green. Lobe 1 is the lighter color and Lobe 2 is the darker color. (B) The dimeric form of GluA2i bound to piracetam shown in two orientations. Two symmetrical sets of three binding sites are found at the dimer interface (labeled Binding Sites 1 through 3). Binding Sites 1 and 2 are partially overlapping and would be unlikely to be occupied simultaneously. (C) Details of the binding sites for piracetam showing several water molecules in the binding site and important interactions. (D) Structures of piracetam and aniracetam.
Binding Site 1
For one set of binding sites, the pyrrolidine ring of piracetam partially overlaps the position of the pyrrolidine ring in the structure of aniracetam (a structural analog of piracetam) bound to GluA2 S1S230 (Figure 2). This includes the central subsite (Subsite A) as well as a portion of Subsite B/B′.37 In the cases of aniracetam and CX614,30 both of which bind largely to Subsite A but a portion of Subsite B/B′, the modulators can bind in two symmetrical orientations, but only one at a time. Because piracetam substitutes an amide for the 4-methoxyphenyl group of aniracetam, two copies can potentially bind in this central location. The orientation of the pyrrolidine ring is rotated approximately 180° relative to aniracetam, with the carbonyl pointing into the cleft of the binding surface (Figure 2). Like GluA2 S1S2 bound to aniracetam, four water molecules are buried in the cleft, which form an H-bond network with the carbonyl of the pyrrolidine ring, the backbone carbonyls of P494 and I481, and the sidechain hydroxyl of S754. The one difference between the aniracetam and piracetam structures is that the H-bond to the water is made by the carbonyl oxygen of the pyrrolidine ring for piracetam but by the benzoyl carbonyl for aniracetam. The water molecules fill the C/C′ subsite.
Binding Site 2
A second set of binding sites (Site 2; Figure 1C and Figure 2) overlaps partially the thiazide binding sites in Subsite B/B′.36, 37, 42 This is in contrast to the thiazides which, depending upon the substituent in the 3-position, fill preferentially the C rather than the B/B′ subsite.37 The amide of piracetam interacts with the sidechain hydroxyl of Y424 and the sidechain carboxyl of D760. The proximity of Binding Site 2 to Binding Site 1, suggests that only one of these two binding sites can be occupied at any given time.
Binding Site 3
The third set of binding sites (Site 3; Figure 1) does not correspond to any previously reported allosteric modulator binding sites, represents the strongest density of the three sets of binding sites, and is the most surface exposed of the three. This binding site is within the inverted U-shaped crevice that is formed between the two monomers that make up the dimer. The crevice is an open pocket that faces the membrane and is exposed to solvent (Figures 1A and B). In this case, the amide interacts with the sidechain amide of N764 and the pyrrolidine carbonyl interacts with the sidechain carboxyl of D760 and the sidechain hydroxyl of S729 (Figure 1C).
Binding site on GluA3 S1S2 for piracetam
A structure of GluA3i was obtained in the presence of piracetam. Overall, the density for piracetam was lower than that seen for GluA2i and GluA2o. The densities for Binding Sites 1 and 2 (described above for GluA2) were too low to determine the orientation of piracetam. The density in Binding Site 3 was stronger but not as strong as in GluA2. The orientation within this binding site is identical to that for GluA2o and GluA2i (data not shown).
Binding site on GluA3 S1S2 for aniracetam
The structures of GluA3i S1S2, with glutamate in the agonist-binding site, and bound to aniracetam was solved and shown in Figure 3. The binding site was largely similar to the previous structure of aniracetam bound to GluA2o (fluorowillardiine; Figure 2).30 This is consistent with the fact that the sequence within the binding site differs little between the two subtypes. Minor differences, however, were observed. Small changes in the dihedral angles of aniracetam were observed between GluA2o and GluA3i. As a result, in GluA3i, the sidechain of S497 exists in two distinct rotameric states (Figure 3B). One of these states forms an H-bond with the pyrrolidinone ring of aniracetam. The other (shown with a green carbon in Figure 3B) forms a water-mediated H-bond with the sidechain of K763. In the case of GluA2o, only the water-mediated H- bond with K763 is observed.30 The rotameric state of S729 is also different between GluA2o and GluA3i. In GluA2o, it points into the binding cleft, and in GluA3i, it points out toward the solvent. The GluA3i structure is in the flip form, position 754 is a serine, whereas, it is an asparagine in GluA2o. Although this additional space due to the smaller sidechain results in an additional water molecule in the interface for GluA3i, the effect of the Asn to Ser substitution does not seem to change the binding site significantly, although a small preference for the flop form (N754) has been reported.9 Like piracetam, four water molecules are buried in the binding cleft and H-bond with the carbonyl oxygens of I481 and P494 and the sidechain of S754 (eight water molecules are shown in Figure 3A because both aniracetam orientations are pictured). However, in the case of aniracetam, it is the benzoyl carbonyl that makes the H-bond to the water molecules instead of pyrrolidine carbonyl.
Figure 3.
Structure of GluA3i bound to aniracetam. (A) Dimer of GluA3 S1S2 bound to glutamate and aniracetam. One copy is shown in shades of blue and the other in shades of green. Aniracetam is shown in both orientations (one with carbons in white and the other with carbons in green). The two orientations of the sidechain of S497 are shown with carbons in white in one case and green in the other. Water molecules are shown as red spheres. (B) Details of the aniracetam binding site. Only one orientation of aniracetam, with corresponding contacts is shown. The sidechain of S497 has two orientations, one that forms a water-mediated H-bond with the sidechain of K763 and the other which forms a water mediated H-bonding network with the carbonyl of K730 and aniracetam.
Conclusions based on the structure
Piracetam is a very weak modulator of AMPA receptors, increasing responses (presumably due to a block of desensitization) with a relatively low affinity,28 but having a variety of other effects.43, 44 Whereas the clinical effects of piracetam may not necessarily be mediated by AMPA receptors, the description of its binding sites on AMPA receptors provides insight into possible strategies for designing new, more effective allosteric modulators.
The binding sites for allosteric modulators of AMPA receptors have been determined in a large number of cases using crystallography (aniracetam,30 CX614,30 a dimeric biarylpropylsulfonamide,38 several CTZ derivatives,42 thiazides37). Modulators bind to a rather large surface at the dimer interface, which has been subdivided into a central subsite (Subsite A; Figure 2) and two sets of lateral subsites (Subsites B/B′ and C/C′).37 Aniracetam and CX614 occupy mainly Subsite A30 with some interactions with the B/B′ subsites; whereas, the thiazides can occupy a combination of Subsites B/B′ and C/C′ or Subsite C/C′ alone, but preferentially occupy the C/C′ subsites.37 Cyclothiazide, TCMZ and ALTZ, all with relatively large substituents in the 3-position, span the B/B′ and C/C′ subsites; however, when the size of the subsituent is decreased as in HCTZ, HFMZ, and IDRA-21, the ring is rotated and occupies largely the C/C′ subsites.37 This seems to be due to the fact that the C/C′ subsites are less solvent exposed and more hydrophobic. In contrast, piracetam does not occupy the C/C′ subsites, but rather populates the more superficial B/B′ subsites (Binding Sites 1 and 2). For both aniracetam and piracetam, water fills the C/C′ subsites. The lack of direct interaction of piracetam with N754 (flop) or S754 (flip) would suggest that, unlike cyclothiazide and PEPA, no preference for one of the two alternatively spliced versions of the protein is likely.
One strategy that has been proposed to increase the affinity of allosteric modulators is to produce hybrid compounds that can span several subsites. Both the flop-selective allosteric modulator PEPA (4-[2-(phenylsulphonylamino)ethylthio]-2,6,-difluorophenoxyacetamide)45 and the dimeric biarylpropylsulfonamide compound described by Kaae et al.38 span Subsite A and interact on both sides of the binding surface. The biarylpropylsulfonamide compound can occupy a portion of both the C and C′ subsites with its sulfonamide groups.38 In the case of PEPA, the C (or C′) subsite is fully occupied by the phenyl ring and, on the opposite side of the binding site, the amide of PEPA interacts with N754, conferring flop specificity (Ahmed, Ptak, & Oswald, unpublished data). The structures of GluA2 S1S2 bound to piracetam open a new possibility for drug design. That is, Binding Site 3 (Figures 1C and 2) has not been previously described as a site of interaction of allosteric modulators. One possibility for increasing the affinity of an allosteric modulator would be to covalently link a substituent that could take advantage of the interactions made by piracetam in Binding Site 3.
Aniracetam has some structural similarities to piracetam but is much more specific for AMPA receptors and more potent.46 The binding site is similar in both the GluA2o and the GluA3i structures, and the electron density of the drug in the binding site is higher than piracetam. The similarity of the structures would suggest little AMPA subtype specificity. Aniracetam was a lead compound in developing more potent allosteric modulators of AMPA receptors,47 and many of the more potent modulators (CX614, etc.) bind in a similar manner.30 Replacement of the methyoxybenzoyl group in aniracetam with an acetamide group in piracetam results in a decrease in affinity, but more interestingly the partial population of a several binding sites in the dimer interface, one of which has not been reported for other allosteric activators.
CONCLUSION
Pyrrolidine allosteric modulators bind to both GluA2 and GluA3 in a very similar manner, suggesting little subunit specificity. However, piracetam and aniracetam differ considerably in binding mode. Aniracetam binds very clearly to a symmetrical binding site on the dimer interface. Piracetam, on the other hand, binds to multiple sites with low occupation, one of which is a unique binding site for potential allosteric modulators. The identification of this new site may be of importance in the design of new allosteric regulators.
Table 1.
Structural Statistics
| Structure | piracetam GluA2i | piracetam GluA2o | piracetam GluA3i | aniracetam GluA3i |
|---|---|---|---|---|
| Space Group | P22121 | P22121 | P2221 | P2221 |
| Unit Cell (Å) | a=47.3 b=114.0 c=163.8 | a=47.5 b=114.3 c=163.8 | a=47.4 b=47.6 c=138.0 | a=47.0 b=47.3 c=137.4 |
| X-ray source | CHESS (A1) | CHESS (A1) | CHESS (A1) | CHESS (A1) |
| Wavelength (Å) | 0.977 | 0.977 | 0.977 | 0.977 |
| Resolution (Å) | 50–1.85 (1.88–1.85) | 50–2.1 (2.14–2.10) | 50.0–2.0 (2.03–2.00) | 50.0–1.75 (1.78–1.75) |
| Measured reflections (#) | 449859 | 148547 | 116713 | 183770 |
| Unique reflections (#) | 71963 | 45684 | 19687 | 30553 |
| Data redundancy | 6.0 (2.8) | 3.3 (3.0) | 5.6 (4.6) | 5.8 (3.7) |
| Completeness (%) | 99.0 (88.0) | 89.6 (84.3) | 96.7 (92.0) | 99.2 (93.3) |
| Rsym (%) | 11.0 (53.2) | 8.6 (32.8) | 13.4 (47.8) | 6.8 (29.8) |
| I/σi | 22.6 (2.2) | 16.6 (2.2) | 18.1 (3.6) | 43.1 (4.2) |
| PDB ID | 3LSF | 3LSL | 3LSX | 3LSW |
| Current Model Refinement Statistics | ||||
| Phasing | MR | MR | MR | MR |
| Molecules/AU | 3 | 3 | 1 | 1 |
| Rwork/Rfree (%) | 18.9/23.2 | 20.1/26.3 | 18.0/23.7 | 18.5/22.5 |
| Free R test set size (#/%) | 1918 (2.7) | 1908 (4.4) | 1860 (8.3) | 1908 (5.3) |
| Number of protein atoms | 6036 | 6036 | 2031 | 2044 |
| Number of heteroatoms | 120 | 120 | 50 | 28 |
| Rmsd bond length (Å) | .015 | .013 | .025 | .013 |
| Rmsd bond angles (°) | 1.58 | 1.51 | 2.15 | 1.478 |
to be submitted before publication
Acknowledgments
The authors would like to thank Prof. Eric Gouaux (Vollum Institute) for the GluR2 S1S2J construct and Drs. Gregory Weiland and Christopher Ptak for discussions and significant advice. This work was supported by a grant from the National Institutes of Health (1R21-NS067562 and R01-NS049223). This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
Footnotes
The abbreviations used are: ALTZ, althiazide; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid; iGluR, ionotropic glutamate receptor; CX614, pyrrolidino-1,3-oxazino benzo-1,4-dioxan-10-one; GluA2i, the N754S mutation of GluA2o, which mimics the flip form of GluA2 with respect to binding of allosteric modulators; GluA2o, the flop form of GluA2; GluA3i, the flip form of GluA3; HCTZ, hydrochlorothiazide; HFMZ, hydroflumethiazide; IDRA-21, 7-chloro-3-methyl-3,4-dihydro-2H-benzo[e][1,2,4] thiadiazine 1,1-dioxide; IPTG, isopropyl-β-D-thiogalactoside; NMDA, N-methyl-D-aspartic acid; PEPA, 4-[2-(phenylsulphonylamino)ethylthio]-2,6,-difluorophenoxyacetamide; S1S2, extracellular ligand-binding domains of GluA2 and GluA3; TCMZ, trichlormethiazide.
References
- 1.Dingledine R, Borges K, Bowie D, Traynelis S. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Asztely F, Gustafsson B. Ionotropic glutamate receptors. Their possible role in the expression of hippocampal synaptic plasticity. Mol Neurobiol. 1996;12:1–11. doi: 10.1007/BF02740744. [DOI] [PubMed] [Google Scholar]
- 3.Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- 4.Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci. 2007;30:126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Alt A, Nisenbaum ES, Bleakman D, Witkin JM. A role for AMPA receptors in mood disorders. Biochem Pharmacol. 2006;71:1273–1288. doi: 10.1016/j.bcp.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Soundarapandian MM, Tu WH, Peng PL, Zervos AS, Lu Y. AMPA receptor subunit GluR2 gates injurious signals in ischemic stroke. Mol Neurobiol. 2005;32:145–155. doi: 10.1385/MN:32:2:145. [DOI] [PubMed] [Google Scholar]
- 7.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 8.Pei W, Huang Z, Niu L. GluR3 flip and flop: differences in channel opening kinetics. Biochemistry. 2007;46:2027–2036. doi: 10.1021/bi062213s. [DOI] [PubMed] [Google Scholar]
- 9.Partin KM, Fleck MW, Mayer ML. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci. 1996;16:6634–6647. doi: 10.1523/JNEUROSCI.16-21-06634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oswald RE. Ionotropic glutamate receptor recognition and activation. Advances in Protein Chemistry. 2004;68:313–349. doi: 10.1016/S0065-3233(04)68009-0. [DOI] [PubMed] [Google Scholar]
- 11.Mayer ML, Olson R, Gouaux E. Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol. 2001;311:815–836. doi: 10.1006/jmbi.2001.4884. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA- sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 14.Hogner A, Kastrup J, Jin R, Liljefors T, Mayer M, Egebjerg J, Larsen I, Gouaux E. Structural basis for AMPA receptor activation and ligand selectivity: Crystal structures of five agonist complexes with the GluR2 ligand-binding core. J Mol Biol. 2002;322:93–109. doi: 10.1016/s0022-2836(02)00650-2. [DOI] [PubMed] [Google Scholar]
- 15.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 16.Kasper C, Lunn ML, Liljefors T, Gouaux E, Egebjerg J, Kastrup JS. GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists. FEBS Lett. 2002;531:173–178. doi: 10.1016/s0014-5793(02)03496-8. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed AH, Wang Q, Sondermann H, Oswald RE. Structure of the S1S2 glutamate binding domain of GluR3. Proteins: Structure, Function, and Bioinformatics. 2008;75:628–637. doi: 10.1002/prot.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill A, Birdsey-Benson A, Jones BL, Henderson LP, Madden DR. Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists. Biochemistry. 2008;47:13831–13841. doi: 10.1021/bi8013196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: Molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 20.Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hald H, Naur P, Pickering DS, Sprogoe D, Madsen U, Timmermann DB, Ahring PK, Liljefors T, Schousboe A, Egebjerg J, Gajhede M, Kastrup JS. Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. J Biol Chem. 2007;282:25726–25736. doi: 10.1074/jbc.M700137200. [DOI] [PubMed] [Google Scholar]
- 22.Nanao MH, Green T, Stern-Bach Y, Heinemann SF, Choe S. Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid. Proc Natl Acad Sci U S A. 2005;102:1708–1713. doi: 10.1073/pnas.0409573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 25.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giurgea C, Lefevre D, Lescrenier C, David-Remacle M. Pharmacological protection against hypoxia induced amnesia in rats. Psychopharmacologia. 1971;20:160–168. doi: 10.1007/BF00404369. [DOI] [PubMed] [Google Scholar]
- 27.Giurgea C. The “nootropic” approach to the pharmacology of the integrative activity of the brain. Integrative Psychological and Behavioral Science. 1973;8:108–115. doi: 10.1007/BF03000311. [DOI] [PubMed] [Google Scholar]
- 28.Copani A, Genazzani AA, Aleppo G, Casabona G, Canonico PL, Scapagnini U, Nicoletti F. Nootropic drugs positively modulate alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-sensitive glutamate receptors in neuronal cultures. J Neurochem. 1992;58:1199–1204. doi: 10.1111/j.1471-4159.1992.tb11329.x. [DOI] [PubMed] [Google Scholar]
- 29.O’Neill MJ, Witkin JM. AMPA receptor potentiators: application for depression and Parkinson’s disease. Current drug targets. 2007;8:603–620. doi: 10.2174/138945007780618517. [DOI] [PubMed] [Google Scholar]
- 30.Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaglenova J, Pandiella N, Wijayawardhane N, Vaithianathan T, Birru S, Breese C, Suppiramaniam V, Randal C. Aniracetam reversed learning and memory deficits following prenatal ethanol exposure by modulating functions of synaptic AMPA receptors. Neuropsychopharmacology. 2008;33:1071–1083. doi: 10.1038/sj.npp.1301496. [DOI] [PubMed] [Google Scholar]
- 32.Arai A, Lynch G. Factors regulating the magnitude of long-term potentiation induced by theta pattern stimulation. Brain Res. 1992;598:173–184. doi: 10.1016/0006-8993(92)90181-8. [DOI] [PubMed] [Google Scholar]
- 33.Black MD. Therapeutic potential of positive AMPA modulators and their relationship to AMPA receptor subunits. A review of preclinical data. Psychopharmacology (Berl) 2005;179:154–163. doi: 10.1007/s00213-004-2065-6. [DOI] [PubMed] [Google Scholar]
- 34.Swanson GT. Targeting AMPA and kainate receptors in neurological disease: therapies on the horizon? Neuropsychopharmacology. 2009;34:249–250. doi: 10.1038/npp.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CR, Benfield P. Aniracetam. An overview of its pharmacodynamic and pharmacokinetic properties, and a review of its therapeutic potential in senile cognitive disorders. Drugs Aging. 1994;4:257–273. doi: 10.2165/00002512-199404030-00007. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 37.Ptak CP, Ahmed AH, Oswald RE. Probing the allosteric modulator binding site of GluR2 with thiazide derivatives. Biochemistry. 2009;48:8594–8602. doi: 10.1021/bi901127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaae BH, Harpsoe K, Kastrup JS, Sanz AC, Pickering DS, Metzler B, Clausen RP, Gajhede M, Sauerberg P, Liljefors T, Madsen U. Structural proof of a dimeric positive modulator bridging two identical AMPA receptor-binding sites. Chemistry & biology. 2007;14:1294–1303. doi: 10.1016/j.chembiol.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology, Vol. 276, Macromolecular Crystallography, part A. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Hald H, Ahring PK, Timmermann DB, Liljefors T, Gajhede M, Kastrup JS. Distinct Structural Features of Cyclothiazide are Responsible for Effects on Peak Current Amplitude and Desensitization Kinetics at iGluR2. J Mol Biol. 2009;391:906–917. doi: 10.1016/j.jmb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Winblad B. Piracetam: a review of pharmacological properties and clinical uses. CNS Drug Rev. 2005;11:169–182. doi: 10.1111/j.1527-3458.2005.tb00268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gouliaev AH, Senning A. Piracetam and other structurally related nootropics. Brain Res Brain Res Rev. 1994;19:180–222. doi: 10.1016/0165-0173(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 45.Sekiguchi M, Fleck MW, Mayer ML, Takeo J, Chiba Y, Yamashita S, Wada K. A novel allosteric potentiator of AMPA receptors: 4-[2-(phenylsulfonylamino)ethylthio]-2,6-difluoro-phenoxyacetamide. J Neurosci. 1997;17:5760–5771. doi: 10.1523/JNEUROSCI.17-15-05760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito I, Tanabe S, Kohda A, Sugiyama H. Allosteric potentiation of quisqualate receptors by a nootropic drug aniracetam. J Physiol. 1990;424:533–543. doi: 10.1113/jphysiol.1990.sp018081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang CM, Shi QY, Katchmen A, Lynch G. Modulation of the time course of fast EPSCs and glutamate channel kinetics by aniracetam. Science. 1991;254:288–290. doi: 10.1126/science.254.5029.288. [DOI] [PubMed] [Google Scholar]