Abstract
Comparative whole-genome analyses have demonstrated that horizontal gene transfer (HGT) provides a significant contribution to prokaryotic genome innovation. The evolution of specific prokaryotes is therefore tightly linked to the environment in which they live and the communal pool of genes available within that environment. Here we use the term supergenome to describe the set of all genes that a prokaryotic ‘individual’ can draw on within a particular environmental setting. Conjugative plasmids can be considered particularly successful entities within the communal pool, which have enabled HGT over large taxonomic distances. These plasmids are collections of discrete regions of genes that function as ‘backbone modules’ to undertake different aspects of overall plasmid maintenance and propagation. Conjugative plasmids often carry suites of ‘accessory elements’ that contribute adaptive traits to the hosts and, potentially, other resident prokaryotes within specific environmental niches. Insight into the evolution of plasmid modules therefore contributes to our knowledge of gene dissemination and evolution within prokaryotic communities. This communal pool provides the prokaryotes with an important mechanistic framework for obtaining adaptability and functional diversity that alleviates the need for large genomes of specialized ‘private genes’.
Keywords: supergenome, conjugative plasmids, horizontal gene transfer, communal gene pool, prokaryotic evolution
1. Introduction
Bacteria and Archaea display immense genetic and functional diversity, which has facilitated the shaping of the Earth's biosphere during the past 3.8 billion years (Smets & Barkay 2005). They have evolved an array of efficient genome modification mechanisms that allow adaptation to ever-changing environmental conditions, permitting the colonization of a plethora of ecological niches. Unlike eukaryotes, genetic exchange and proliferation in prokaryotes are independent processes (Levin & Bergstrom 2000). This in turn means that apart from direct mutational changes, variation is not automatically introduced by virtue of self-proliferation (vertical transfer), which corresponds to binary fission in prokaryotes. Their genomes are modified by gene loss and by gene addition through duplication and the acquisition of new genes through horizontal gene transfer (HGT). New genes acquired by HGT are introduced via mobile genetic elements (MGEs) such as plasmids, viruses or transposons or by direct uptake and incorporation of naked DNA by homologous or illegitimate recombination (She et al. 2001; Frost et al. 2005). As a means of ensuring genetic variation in prokaryotes, the recruitment of new genes by HGT could therefore be seen as ‘compensating’ for the lack of sexual recombination, which is why it is often referred to as ‘bacterial sex’ (Narra & Ochman 2006). Hence, Bacteria and Archaea (in this article referred to as prokaryotes) are, most probably because of their unicellularity and absence of nuclei, bound by common molecular mechanisms that clearly distinguish them from eukaryotes with respect to genome evolution.
One of the most exciting outcomes of the recent genomic revolution is the appreciation of the degree to which HGT has aided in the shaping of prokaryotic life on Earth (Jain et al. 2003; Koonin & Wolf 2008). Comparative whole-genome analyses have demonstrated that prokaryotic genomes are exceedingly dynamic and that large amounts of genetic material have continually been added (or lost) through promiscuous genetic exchanges. The analysis of approximately 20 000 genes from the genomes of eight free-living prokaryotes indicated that by circumventing species barriers, HGT has accelerated the introduction of new genes into prokaryotes by a factor of at least 10 000 (Jain et al. 2003). It should therefore be evident that the role of HGT in prokaryotic genome innovation significantly exceeds that of clonal evolution alone.
Most comparative genomic studies conducted in microbial molecular evolution thus far have focused on pathogenic bacteria such as Vibrio cholera, Escherichia coli and Salmonella enterica. These studies have shown that important chromosomally encoded virulence and antibiotic resistance properties have often been acquired by HGT of genomic islands (Dobrindt et al. 2004). It is, however, generally accepted that HGT is important not only in the transfer of pathogenically relevant traits, but also in all aspects of microbial ecology, and thus plays a central role in the evolution and function of complex prokaryotic communities (Sørensen et al. 2005; Pallen & Wren 2007). Evidence documenting the contribution of HGT to prokaryotic evolution has received additional support from metagenomic approaches. For example, it was recently demonstrated that genes encoding for proteorhodopsins, whose abundance is related to their location in the photic zone of the North Pacific Gyre, have been exchanged between Bacteria and Archaea (Frigaard et al. 2006).
It is however, in our opinion, misleading to merely look at the discrete genomes of prokaryotes within these communities in order to infer evolutionary processes. The vast networks of MGEs that facilitate the flow of genes between these prokaryotes and their own interactions and evolutionary developments are a crucial part of the picture.
2. The supergenome
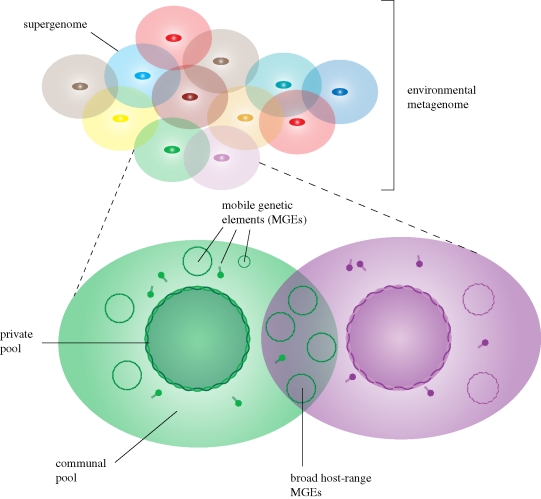
In nature, prokaryotes occupy diverse micro-environmental niches, forming complex communities in which they can readily interact and exchange genes by HGT (Medini et al. 2008). HGT has, not surprisingly, been shown to occur most frequently between organisms that occupy the same environment (Jain et al. 2003). Recent metagenomic surveys have also revealed that, apart from chromosomal sequences, a substantial proportion of environmental samples contain MGE sequences in the form of phages and prophage sequences as well as plasmids (Venter et al. 2004; Tringe et al. 2005). The evolution of specific prokaryotes may therefore be tightly linked to the environment in which they live and to the reservoir of compatible MGEs within that environment. To accommodate a more holistic approach to microbial genomics, we propose the terms: supergenome, which is the total pool of genes readily available to a prokaryotic organism within a particular setting; private pool, which consists of the fixed and ‘idiosyncratic’ genes encoded on the chromosome of the prokaryote; and communal pool which consists of genes encoded on MGEs and that are thus available to all permissive prokaryotes (figure 1). The use of global terms to reflect the dynamic nature of prokaryotic genomes is by no means new. Terms like core genome, which define the genes present in all strains of a prokaryotic species, dispensable genome (or flexible genome), which are genes present in some, but not all, strains of the same species, and pan genome—the sum of the former two—have previously been used (Lawrence & Hendrickson 2005; Medini et al. 2005). These terms were coined in response to the apparently significant differences in gene content that are observed in comparative genomic studies of different strains that were thought to be members of the same species. A weakness with the latter terms, however, is that they focus largely on chromosomal genes and totally ignore the genetic elements that are involved in translocation and dispersal of genes between different species (i.e. MGEs that can traverse the species barrier). Many MGEs can potentially disseminate among all permissive host cells within a given environment at favourable conditions and could therefore not be construed as belonging to any one pan genome. We therefore believe that the supergenome framework is more fitting for describing prokaryotic ‘individuals’ within an environmental setting.
Figure 1.
Overview of the supergenome concept. The supergenome is the total pool of genes readily available to a prokaryotic organism in a particular community setting. It consists of essential genes encoded on the chromosome, termed the private pool, and genes encoded on MGEs (plasmids, transposons, viruses etc.), termed the communal pool.
We also suspect that the evidence of HGT based on genome analysis of pure culture isolates might have underestimated the true impact of HGT on microbial ecology and evolution. This is supported by a recent report that showed a higher degree of genetic recombination, gene loss and acquisition, transposase movement and purifying selection in the genomes within an environmental archaeal population of Ferroplasma acidarmanus, when compared with a pure culture of the same species isolated a few years earlier from the same environment (Allen et al. 2007). Furthermore, the type of retrospective ‘forensic’ analysis employed in most comparative genomic analyses only detects successful recombination events and integration events that were conserved over protracted periods of evolution under various selective regimes (Smets & Barkay 2005). Thus, they probably greatly underestimate the actual flow of genetic information in microbial populations. Genes located on MGEs and encoding traits giving periodic selective advances could, therefore, be recruited and lost by individual cell lines multiple times.
3. A dive into the (deep end of the) communal gene pool
Whether microbial organisms live randomly distributed in bulk environments or in structured biofilm communities, the proximity of potential recipients and donors determines the flow of genetic information and the extent of HGT (and also the preferred mechanism of transfer). Rampant HGT in microbial communities, driven by incessant selective pressures and competition for limiting resources, may over time result in the formation of a distinct communal gene pool that blurs the boundaries of individual prokaryotes (Goldenfeld & Woese 2007). In fact, given the astronomical number of microbial chromosomes that must have existed over the course of evolutionary history, most prokaryotic genes have probably, at one time or another, been transferred by MGEs. What has determined the subsequent success of these transfers has been the ability of genes to be accommodated within the existing genetic frameworks of recipients (within either the private or communal pools) and to increase fitness within the current environmental setting.
The ability of genomic information to flow successfully between prokaryotes is not solely because of their propensity to coexist within highly heterogenic multi-species communities or the separation of their individual genomes by lipid membranes. A highly contributing factor to the extent of HGT is also the strikingly economical manner in which prokaryotic genomes are organized. There is usually very little genomic redundancy in prokaryotes, since a strong ‘deletion bias’ towards small genome sizes exists (Lawrence et al. 2001; Mira et al. 2001; Ochman & Davalos 2006). Furthermore, in prokaryotes, evolution has favoured clustering of cooperating genes, in many cases as ‘selfish’ operons (Lawrence 2003). These two factors, compactness and gene clustering, mean that many conferrable traits can be transferred between discrete replicating elements (chromosomes or plasmids) by the movement of relatively small fragments of DNA. Phylogenetic analyses of complete genomes have also shown that some prokaryotic genes are more likely to be transferred than others (Lawrence & Hendrickson 2003), which, by deduction, would also reflect their propensity to enter the communal pool. Genes involved in transcription, translation and related processes (informational genes) are less likely to have been successfully transferred horizontally than genes involved in amino acid biosynthesis and other housekeeping functions (operational genes), because they are usually members of large molecular complexes, thus ‘the complexity hypothesis’ (Rivera et al. 1998; Jain et al. 1999). Compact operons of operational genes would therefore be more likely to pose an immediate advantage for a wide range of new host cells and would therefore also potentially provide greater selective advantages for members of the communal pool than informational genes.
MGEs are found in all branches of the bacterial tree of life as well as in Archaea and have been detected in terrestrial, marine and clinical environments (Sørensen et al. 2005). Comparative analyses reveal the complexity of MGEs and the presence of a unique set of genes that are clearly distinct from the genes that are typically found on prokaryotic chromosomes. The extent of the genetic diversity within the communal pool and its apparent distinction from the ‘essential’ content presumed for the chromosomal private pool suggest that the communal pool represents a distinct genetic resource. Conversely, it can also been argued that the communal pool of MGEs residing within prokaryotes or engaged in horizontal transfer constitutes a collection of selfish genetic elements whose own evolutionary interests are best served by circumventing, not maintaining, the species boundary, and that the private pool therefore represents a vast resource for MGEs to exploit for their own benefit.
Many factors limit successful evolutionary horizontal gene acquisition. Gene transfer by means of MGEs is first of all limited by restrictions in the host ranges of elements such as viruses and plasmids. Furthermore, various molecular mechanisms counteract uptake and stabilization of foreign DNA molecules as the phylogenetic distances increase (Lawrence & Hendrickson 2003; Thomas & Nielsen 2005). Nonetheless, an intimate relationship critical to the ecology of the hosts seems to have developed between private and communal pools repeatedly. Given that the communal pool in many cases seems tightly linked to its environment, it is also thought to vary considerably from one environment to another.
4. The tools of genetic mobility
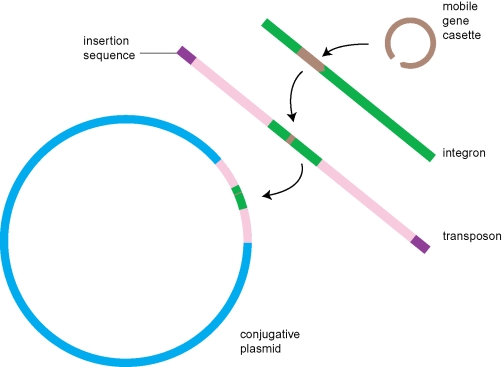
To better appreciate the dynamic nature of genetic rearrangements that happen on the way to, and within, the communal pool, it is necessary to look closer at some of the elements and processes that govern these rearrangements. The physical movement of DNA relies on a number of molecular ‘cut and paste’ mechanisms that are able to manipulate and translocate DNA fragments. Enzymes with these capabilities include recombinases, which facilitate homologous recombination as part of the host-encoded machinery that ensures genome integrity, transposases, which catalyse the movement and insertion (sometimes replicative) of transposons, integrases, which enable insertion of elements such as gene cassettes into integrons by site-specific recombination, and resolvases, which are DNA endonucleases capable of resolving Holiday junctions that arise as a result of genetic recombinations. Many of these enzymes are encoded by an assortment of selfish mobile elements (figure 2) such as insertion sequences, transposons and integrons, which can facilitate gene deletion or capture, and accretion of genetic elements on higher-order mobile elements such as conjugative plasmids (Frost et al. 2005). These genetic rearrangements usually occur within the cytoplasm of a cell and therefore constitute intracellular movement. In describing the communal pool, one can therefore distinguish between: (i) translocative elements, described earlier, that enable the rearrangement, movement and accretion of genes; (ii) operative elements, sets of one or more genes that are moved around without encoding functions related to gene transposition and therefore constitute a type of currency in the form of ‘communal loads’ within the communal gene pool; and (iii) dispersive elements (Baquero 2004), which convey the actual horizontal transfer of communal loads after operative elements have been incorporated via translocative elements. Thus, within the communal gene pool, a clear distinction exists between the permanent members that convey gene transfer (like plasmid backbones) and the operative genes that can be exchanged between replicating elements and might one day become integrated into the private pool of a prokaryote. Dispersive elements such as conjugative plasmids could therefore be said to constitute permanent ‘vessels of the communal pool’, which maintain the fluidity of the supergenome.
Figure 2.
The modular and hierarchical composition of MGEs. Gene cassettes are inserted into integrons by integrase-mediated site-specific recombination. Integrons may be inserted into composite transposons (mobile gene islands flanked by transposase-encoding insertion sequences), which in turn may be inserted into a dispersive element like a conjugative plasmid. The plasmid thus becomes a vessel for the transportation of genetic information within the communal gene pool.
Most studied prokaryotes can accommodate several extra-chromosomal elements (sometimes in excess of 20 plasmids, constituting as much as a quarter of the total amount of genetic information in that cell (Casjens et al. 2000; Frost et al. 2005)). Thus, within the cytoplasm of prokaryotes, there often exist several independent replicons, but only the chromosome is confined to the cell. Genetic mobility should therefore not merely be interpreted in terms of transportation of genes across the cell barriers of prokaryotes, but rather as a perpetual flow between discrete reproductive units. It is therefore not unusual to observe genetic rearrangements between plasmids, or plasmid fusions (co-integration) resulting from homologous recombinations or similar mechanisms.
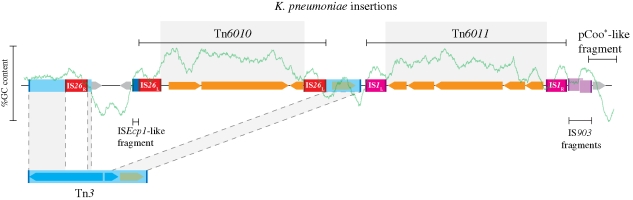
The mobilization of genes from chromosomes to dispersive elements, mediated by insertion sequences, is a well-documented segue into the communal gene pool. The conjugative plasmid pOLA52 from E. coli provides a useful illustration of this. This plasmid contains a 5 kb fragment that confers the formation of type III fimbriae that significantly enhance the ability to form biofilms with other Enterobacteriaceae that also harbour the plasmid (Burmølle et al. 2008; Norman et al. 2008). The fragment is thought to have been produced by the insertion of two IS1 elements, resulting in the capture of an entire operon from the chromosome of a Klebsiella pneumoniae (thus forming a composite transposon). In a similar manner, another 5 kb fragment on the same plasmid, which encodes an efflux pump capable of conferring resistance towards a great variety of antimicrobial substances (Hansen et al. 2007), was also captured from K. pneumoniae (figure 3).
Figure 3.
Graphical representation of the mosaic structure of the ‘genetic adaptive load’ region of the IncX1 plasmid pOLA52. Two composite transposons (Tn6010 and Tn6011) have been inserted into the plasmid, which confer biofilm formation and multi-drug resistance. Both composite transposons carry foreign chromosomal regions, which can also be identified by a markedly higher average in GC composition (green curve), showing more than 99% homology to K. pneumoniae MGH 78578. The Tn6010 transposon has inserted itself within a region that has disrupted a resident Tn3 transposon without disturbing an ampicillin-resistance conferring gene (bla). Adapted from Norman et al. (2008).
The intracellular location of an operative element not only affects the potential dissemination of that element, but also its rate of evolution. The movement of a gene onto a discrete replicating element that exists in a higher copy number, for example, from a chromosome onto a plasmid, in effect represents a gene duplication equal to the copy number of the plasmid. Having a gene located on a plasmid therefore allows for a lot of ‘tinkering’, and the communal pool would therefore seem like an ideal location for gene evolution. Curiously, not all operative genes occurring on MGEs like conjugative plasmids can be traced back to a chromosomal origin, which raises the question whether there are communal genes that have never successfully penetrated the private pool.
5. Mechanisms of, and barriers to, horizontal gene transfer
The ability to move genes between cells is a prerequisite for membership to the communal pool. The dispersive elements of the communal pool can be transferred between prokaryotes by transformation, transduction or conjugation. Transformation is the uptake of DNA into cells from the surrounding environment and relies on the presence of plasmid or chromosomal DNA fragments that are often released as a result of cell death or active excretion. This mechanism therefore, at least in theory, requires only a viable recipient, and usually a mechanism for the active uptake of extracellular DNA (competence) (Chen & Dubnau 2004). Integration of DNA transported via transformation into the recipient relies on DNA repair enzymes (recombinases) of the host and can occur via either homologous recombination, which, as the name implies, requires a great degree of homology between the donor and the recipient DNA, or the so-called illegitimate recombination (Hülter & Wackernagel 2008).
Transduction is the transportation of DNA through bacteriophages. This mechanism requires that a phage replicates within the donor organism and, in the process of DNA packaging, occasionally incorporates DNA fragments from the host into the phage capsid. Phages are then released into the environment in which they can inject their DNA into a new host.
Finally, many dispersive elements are able to be transferred by conjugation, a multi-step process that requires cell contact, as described below.
The specific requirements of each of the three transfer mechanisms suggest that they occur with different probabilities within various habitats. In natural environments, naked DNA becomes especially vulnerable towards degradation from nucleases (DNases) or heavy metals, although in sands and clay it is known to adsorb to colloid particles that greatly increase stability (Davison 1999). The packaging of DNA into protein capsules provides a substantially better protection from the surrounding environment than during transformation and also ensures a much more efficient means of entering the host cell. Most known phages, however, have very specific host ranges, and transduction is therefore, like transformation, thought to provide for mainly intraspecies gene transfer similar to sexual recombination in eukaryotes. Like transduction, conjugation also keeps DNA separated from the destructive forces of the extracellular environment by protective walls at all times. Given the immense diversity of prokaryotes, the requirement for compatible cell contact would seem to be a greatly limiting step for HGT via conjugation. However, considerable evidence has accumulated over the years for conjugation between prokaryotes over large taxonomic distances (Musovic et al. 2006). Some studies have even demonstrated trans-kingdom conjugation of plasmids from prokaryotes to eukaryotes. Conjugation would, therefore, seem like a safe way to transport large DNA fragments with high fidelity, although only between physically proximate donors and recipients. It is therefore believed to be the driving factor in making the communal gene pool communal. Some environments are believed to be more conductive for conjugation than others. Microbial biofilms with high density of highly active bacterial cells are well-known examples of such hotspots of HGT (Sørensen et al. 2005). Hence, both the dynamics and the size of the communal gene pool are restricted by environmental considerations. The evolution of prokaryotes, therefore, depends on the environment, not only for driving selection like eukaryotic organisms, but also for a source of genetic innovation. Hence, the size of the supergenome of a prokaryote will depend on the composition of the relevant microbial community.
MGEs have traditionally been divided into plasmids, phages or transposons. Out of these three, conjugative plasmids are by far the best characterized and most widely studied. The increasing numbers of available MGE sequences in GenBank, however, have shown that several elements are chimeric and resemble more than one of these groups. Elements such as integrative conjugational elements, transposable prophages, integrative plasmids and mobile integrons have therefore contributed to muddling the picture considerably. An emerging view is that MGEs should instead be considered as mosaics of functional blocks, or modules, of genes (Osborn & Böltner 2002; Toussaint & Merlin 2002). Indeed, this perspective is the basis for the ACLAME database (Leplae et al. 2004). The conjugative plasmid could therefore be considered a particularly successful collaboration between a number of genetic elements that in conjunction have enabled HGT to occur over large taxonomic distances. We believe that conjugative plasmids represent one of the most interesting constituents of the communal gene pool and focus on them in the remainder of this article.
6. The world of conjugative plasmids
The term ‘plasmid’ was coined in 1952 by the late Joshua Lederberg (Lederberg 1952) who was also the first to describe the process of bacterial conjugation, long before any plasmid structure had been seen. Early studies of the conjugation archetype, the F-prime ‘sex factor’ (now simply called the F plasmid), were also a major contributor to the development of recombinant DNA technology and the establishment of plasmid biology as a separate field (Cohen 1993; Helinski 2000). The study of conjugative plasmids was also stimulated by the explosive emergence of antibiotic resistance that followed shortly after our entrance into the era of antibiotic treatment for bacterial infections. For many years, conjugative plasmids were therefore associated mainly with the overuse of antimicrobial agents and, later on, increasing anthropogenic pollution with heavy metals or petroleum derivatives. Thus, the majority of conjugative plasmids were studied because they, in one way or another, directly affected the well being of humans. By now, however, more than a thousand plasmids from all three domains of life and almost every conceivable environmental niche, even those with little or no exposure to human meddling, have been described. Furthermore, ongoing microbial genome sequencing projects, as well as metagenomic surveys, reveal new plasmid sequences almost on a daily basis. The picture that has emerged from this vast amount of data seems to be that the role of conjugative plasmids could be equally important for bacterial evolution in pristine environments.
(a). What are conjugative plasmids?
The most common structure of plasmids is that of extra-chromosomal covalently closed circular (and supercoiled) units of DNA, although several examples of linear plasmids also exist. In fact, the most important defining feature of plasmids is their separateness from the host chromosome and hence their ability to replicate autonomously. Integrative conjugational elements, such as the conjugative transposon Tn916 from Enterococcus faecalis or the ICEBs1 element from Bacillus subtilis, can only maintain themselves stably by integrating into the chromosome and should therefore not be considered plasmids, even though they employ a form of replication during conjugation (Burrus et al. 2002).
The second, and perhaps most well-known, property of conjugative plasmids is their ability to horizontally transmit genes by conjugation, which also makes them a vital part of any communal pool. The ability to transmit horizontally confers a number of selective advantages to conjugative plasmids, in that it provides autonomy from host replication and also offers an abundance of alternative hosts within heterogeneous populations. The ability to spread over a diverse set of hosts thus evades the danger of becoming extinct with one particular cell population if environmental conditions were to suddenly change unfavourably. When one considers these advantages, it is therefore not surprising that many diverse mechanisms of horizontal plasmid transfer have evolved independently (Zechner et al. 2000).
Another identifying feature of plasmids has typically been that from the viewpoint of the host strain: they are dispensable. However, the discovery of plasmids larger than 1 Mb, the so-called megaplasmids, containing putatively host-essential genes, has also contributed in making the distinction between ‘chromosome’ and ‘plasmid’ somewhat harder. In one case, the 1.18 Mb extra-chromosomal element of Brucella melitensis 16 M was defined as a second chromosome (DelVecchio et al. 2002), while the larger (1.68 Mb) extra-chromosomal element pSymB in Sinorhizobium meliloti was defined as a plasmid (Galibert et al. 2001). Such definitions can therefore sometimes seem to be a matter of semantics, pertaining to the use of the words ‘expendable’ or ‘essential’, rather than science.
(b). Organization of conjugative plasmids
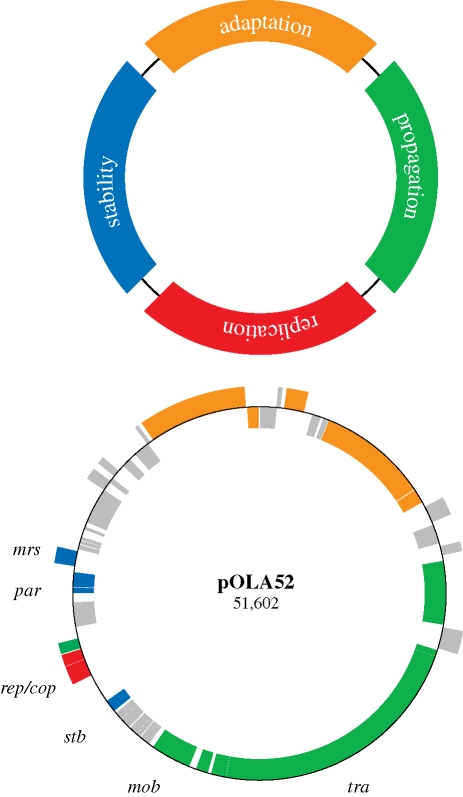
Plasmids have long been considered to be modular in nature since they often contain discrete regions of genes, clustered together in functional groups, that are responsible for various aspects of plasmid maintenance and propagation (Thomas, 2000; de la Cueva-Méndez & Pimentel 2007). Plasmids are therefore said to consist of a ‘backbone’ of plasmid-selfish modules (figure 4), often combined with a block of ‘accessory elements’ containing mosaics of translocative elements and operative elements that encode beneficial traits for the host cell and, potentially, other resident prokaryotes, within specific environmental niches (Eberhard 1990). Even though not technically discrete modules, such regions could therefore be referred to as ‘adaptation modules’.
Figure 4.
An overview of the organization of an archetypical conjugative plasmid comprised four gene modules: stability (blue), replication (red), propagation (green) and adaptation (orange) compared with the organization of the annotated IncX1 plasmid pOLA52 isolated from E. coli. Genes encoding unknown functions or functions not directly related to the four modules are indicated with grey.
Backbone modules usually consist of a relatively modest number of compactly arranged genes, often accompanied by cis-elements related to their function (origins of replication or transfer, centromere sites for plasmid segregation, etc.). The simplest of plasmids is only around 1–2 Kb in size and in most cases do not even encode any proteins. These plasmids only meet the one requirement of persistence: the ability to replicate. In order to be maintained, replication modules (rep) must ensure that replication proceeds in accordance with the host cell growth cycle without allowing copy numbers to reach a level that imposes an unreasonable metabolic burden on the host. Conversely, plasmid copy numbers must not fall below a level that leads to plasmid-free (cured) segregants or allows other resident plasmids a competitive advantage (Paulsson 2002). Plasmid replication modules are therefore subjected to both intercellular and intracellular, often opposing, selection pressures that have driven the evolution of stringent replication control mechanisms (Thomas 2000). In many cases, rep modules are therefore often coupled with regions (cop) that ensure the maintenance of stable copy numbers. This usually means that if copy numbers drop below the desired level, plasmids replicate more than once a cell cycle, while plasmid replication stops almost completely if copy numbers are too high.
Even though the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome), which currently lists some 1600 plasmid genomes (as of January 2009), shows that plasmids can be as small as 0.85 Kb (pRKU1 from Thermotoga petrophila), conjugative plasmids are usually relatively large, owing to the number of additional genes required for the conjugation machinery (mob, tra), and the subsequent requirement for plasmid stabilization that comes with large plasmids (discussed subsequently). The smallest known conjugative plasmid from proteobacteria is currently R338, which is approximately 34 kb in size. Smaller plasmids, which do not possess conjugation machineries, often rely on mobilization or conduction (piggybacking on a transmissible plasmid by co-integration) for horizontal transfer.
While smaller plasmids can be found in excess of hundreds of copies, conjugative plasmids are typically found in low copy numbers (<10 copies/cell), which reflects the selective advantage of minimizing the metabolic burden on the host (Paulsson 2002). It has not been clearly determined whether plasmid copy numbers reflect adaptation of the replication mechanisms to increasing plasmid size, or whether certain replication mechanisms have been better able to accommodate a constant influx of genetic elements resulting from mobile element insertions, co-integration and homologous recombination. Ultimately, the upper limit for plasmid size may also depend on the host strain carrying the plasmid and the heterogeny of plasmid carriage within that cell (Sherley et al. 2003). A recent survey of metadata from about a 1000 available plasmid sequences (Slater et al. 2008) also revealed a connection between host environment and plasmid size.
Nevertheless, low copy numbers provide many challenges for conjugative plasmids that require the presence of additional backbone modules, and these also increase plasmid size. Modules that ensure stable vertical transmission and segregational fidelity (par, mrs) are often found in close proximity to the rep and cop modules on conjugative plasmids, with the exception of tra modules.
These plasmids usually also carry modules dedicated to plasmid addiction (stb), which ensures killing of plasmid-free segregants. Further refinement can be acquired by the addition of central control regions, similar to those observed in the IncP family of plasmids, that coordinate the expression of individual backbone modules, which probably also greatly minimizes the burden imposed upon the host cell.
Even though conjugative plasmid backbones often maintain a great degree of conservation, as well as synteny, within discrete incompatibility groups, this is not always the case. Some studies of isolates from different genera or different parts of the world (Norman et al. 2008; Sevastsyanovich et al. 2008) indicate that backbone genes have different phylogenetic origins for different modules, and sometimes even for individual module genes (Fernandez-Lopez et al. 2006). This suggests that plasmid innovation can occur, from time to time, by interacting with other plasmid backbones, which leads to insertion or replacement of backbone modules or to allelic replacements of individual genes. The addition of central control regions, however, probably helps in preserving a more fixed set of backbone modules, as has been observed with the IncP plasmids (Thomas 2000; Bahl et al. 2007c). Even though plasmids display lower levels of genetic mosaicism than most bacteriophages (Hatfull et al. 2008), nevertheless, functional plasmid modules are still susceptible to genetic recombination events or to allelic replacements, provided that the overall functionality of the plasmid is not compromised. This also provides important clues as to how plasmids evolve.
The modular or genetic mosaicism observed in plasmid backbones, although relatively low, makes it difficult to infer an accurate evolutionary history of any one particular plasmid group, or to find robust methods of plasmid classification beyond the traditional division into incompatibility groups (e.g. IncF, IncP, IncN, IncX, etc.) (Couturier et al. 1988).
Phylogenetic congruence, or lack thereof, between backbone genes and their host cells can also help in providing clues as to the degree of coevolution that plasmid modules have undergone within certain taxonomic groups of hosts, for example, within the Enterobacteriaceae (Norman et al. 2008).
(i). Modules affecting plasmid replication and copy control
A large number of different mechanisms have evolved for the vertical transmission of circular plasmids (Giraldo 2003), but most follow either of two schemes: one is the theta mechanism, in which replication initiates by melting the double-stranded DNA at the oriV, thus allowing for assembly of the replisome. Replication may then proceed either uni- or bi-directionally (leading to the formation of a structure resembling the greek letter θ). In the other mechanism, rolling circle replication (RCR) is initiated from a 3′-OH primer, generated by nicking one strand of the plasmid, and then proceeds by strand displacement (del Solar et al. 1998).
Previously, it was thought that θ-replicating plasmids proliferated within Gram-negative bacteria, while RCR plasmids came from Gram-positive bacteria and archaea (incidentally, the RCR mechanism is also mimicked by the DNA-transfer mechanisms of many plasmids of Gram-negative origin). At present, both mechanisms have been observed in plasmids and phages from both Gram-positive and Gram-negative bacteria, while only RCR plasmids have been found in archaea (Giraldo 2003). Future studies should reveal if this has been a consequence of sampling bias.
Plasmid copy number is almost universally controlled at the level of replication initiation by employing plasmid-encoded trans-activators or inhibitors. These are usually in the form of Rep replicases (del Solar et al. 1998). This also greatly limits the degree of homology that two plasmids can have, since similar, and thus incompatible, replication modules will begin interfering with each other's copy number control mechanisms. Hence, plasmid incompatibility can potentially restrict the access to part of the communal gene pool when similar plasmids are present. Plasmids such as the IncF, IncI, IncX or IncN group are, for example, rarely found outside the Enterobacteriaceae, and are therefore referred to as narrow host-range plasmids, while others, like the intensely studied IncP plasmids, are able to disseminate over a surprisingly wide variety of bacteria (Musovic et al. 2006). Host range is therefore first and foremost determined by the plasmid replication mechanism, since a requisite of plasmid dissemination is an ability to replicate within every host through which it passes.
(ii). Modules affecting plasmid stability
Apart from maintaining stable copy numbers with replication, plasmids must cope with enzymatic degradation, damaging insertions of foreign DNA and homologous recombination that leads to the formation of plasmid multimers. If a plasmid fails to be included in a daughter cell, that lineage becomes plasmid-free or ‘cured’. The metabolic burden imposed on the host by a plasmid means that cured lineages will be favoured in the absence of a genetic requirement that is met by the plasmid. If a strain is cured, the communal load of the lost plasmid will therefore in effect become extinct over time, unless the selective pressure is restored (hampering the plasmid-free cells), or the communal load has been successfully transmitted onto the host chromosome or another plasmid (Bergström et al. 2000). Segregational fidelity is therefore highly influential on the perseverance of plasmids in bacterial populations and the continued existence of communal genes. Several plasmid mechanisms that promote plasmid stability have been identified.
Multimer resolution. Plasmid multimer formation potentially prevents proper segregation into daughter cells (Summers et al. 1993). Many smaller plasmids have therefore acquired site-specific recombinase systems, also called multimer resolution systems (mrs), that prevent the destabilizing effects that arise from the formation of plasmid multimers. The most well studied of these is the Xer–cer system of the ColE1 plasmid, which possesses a recombination site (cer) where multimers are resolved by the host-encoded XerCD complex of E. coli as well as other host-encoded proteins (Summers 1998).
Larger conjugative plasmids usually also contain resolvase genes for multimer resolution, an example is the parCBA system of the IncP-1 plasmid RK2. Resident transposons on larger plasmids are also able to contribute to stability by encoding resolvases that enable multimer resolution, sometimes with greater efficiency than the plasmid systems (Tolmasky et al. 2000).
Active partitioning. Plasmids maintained at high copy numbers can normally rely on adequate distribution simply by chance, because of random diffusion. Low copy number conjugative plasmids, however, have to rely on active mechanisms, usually a nucleoprotein complex (the segrosome), which ensures that plasmids are actively moved into position prior to cell division in a manner analogous to eukaryotic mitotic division. This mechanism is, in the majority of observed cases, encoded on par loci consisting of two genes, often given the generic parA and parB denomination, that encode trans-acting proteins and a centromere-like cis-region sometimes named the parS site (Tolmasky et al. 2000; Hayes & Barilla 2006). In most cases, one gene (parA) encodes an ATPase, which provides the energy required for the intracellular movement and distribution of plasmids, which binds to a DNA-binding protein (encoded by parB) that, in turn, binds to the plasmid centromere region. Cross-talk between partitioning mechanisms has also been known to lead to plasmid incompatibility and should therefore also drive the evolution of these systems towards diversity (Bouet et al. 2007).
Plasmid addiction. Another effective strategy in maintaining plasmid stability is to simply get rid of potential intercellular competition (i.e. cured lineages) if it arises, by recruiting the so-called post-segregational killing (PSK) or addiction systems. Usually one gene product of these systems performs an action that leads to growth–arrest or killing of the host cell, while the other encodes a much less stable product that counteracts these effects. This means that homeostasis is only maintained as long as both genes, and by extension, the replicating elements that carry them, are present. Some examples are the ccdAB toxin–antitoxin (TA) system of the F plasmid, or the similar, but unrelated, parDE system of the IncP plasmid RK2, where toxin arrests DNA replication by inhibiting the host cell gyrase, unless bound by the cognate antitoxin protein. Another example is the hok/sok system of the plasmid R1 from E. coli, where antisense RNA prevents translation of toxin mRNA that would otherwise translate into a host cell killing (Hok) protein product.
TA systems display the properties of true selfish genetic elements, in that they impose a metabolic burden on the host and ensure their own presence by killing cells that have eliminated them. Their pervasiveness on plasmids and MGEs, however, is a testament to the vertical stability they offer any replicating elements that carry them. A multitude of theories as to the true purpose of TA systems have been proposed, especially to explain the physiological role of chromosomal TA systems (Gerdes et al. 2005; Magnuson 2007; Saavedra De Bast et al. 2008). One particularly interesting theory is that chromosomal TA systems act as anti-addiction mechanisms that prevent killing in the event of plasmid loss (Saavedra De Bast et al. 2008). Chromosomal TA systems would thus drive the evolution of plasmid-encoded TA systems so that the toxins were no longer recognized by the anti-addiction modules.
Restriction–modification (RM) systems have been less studied with respect to plasmid stability, but have nevertheless also been found on plasmids. RM systems can be regarded as selfish mobile elements such as transposons, but with a strong stabilizing effect on the DNA molecule in which it inserts (Kobayashi 2001).
(iii). Modules affecting plasmid propagation
Plasmid propagation by conjugation can be seen as the successful coupling of two, otherwise unrelated, functions: mating pair formation (Mpf) and RCR (relaxosome formation) (Llosa et al. 2002). The initial requirement for conjugation is Mpf, formation of a complex where the donor and recipient connect physically. In most reported cases, this proceeds through synthesis of a type IV secretion system (T4SS—discussed subsequently), which generates filaments (conjugation pili) that extend from the donor cell to reach suitable recipient cells within its proximity and subsequently retract to facilitate cell contact (Christie et al. 2005; Clarke et al. 2008). Pilus morphology (thick flexible, thin flexible or rigid), which previously served as a way of distinguishing plasmid incompatibility groups, also determines whether conjugation proceeds optimally in liquid media (planktonic cells) or on solid surfaces (in biofilms) (Bradley 1980; Bradley et al. 1980). Pilus synthesis can therefore be a central determinant in the extent of gene dissemination within the communal pool because it determines which cells a donor can physically connect with (i.e. the conjugation host range) and the preferred medium of transfer.
The second step in the conjugation process (DNA processing and transfer) involves relaxase-mediated nicking of the plasmid at the plasmid origin of transfer (oriT) and formation of the relaxosome, a nucleoprotein that contains single-stranded plasmid DNA, the relaxase and a number of other proteins (some host encoded). A cognate coupling protein usually helps the relaxosome to dock with T4SS, where it is subsequently transported into the recipient cell. Finally, RCR ensures that a second strand is synthesized in both the donor and the recipient.
On conjugative plasmids, both functions are encoded on the plasmid backbone, sometimes in two separate modules, but mobilizable plasmids only carry the genetic information necessary for relaxosome formation and DNA processing, and therefore do not provide the burden of pilus synthesis. Mpf could therefore be said to be an altruistic process (from the conjugative plasmids point of view), in that it benefits the plasmid that encodes it and the mobilizable plasmids that are capable of being transferred with it. Furthermore, the relatively recent implication of conjugative pili in establishing bacterial biofilms (Ghigo 2001) lends further support to the view that Mpf modules are altruistic entities. Bacterial biofilms are known to protect against many harmful substances (like antimicrobials) and bacteriophage attacks and also provide a rich matrix for HGT. Biofilm formation might even represent the default state of most bacteria (Jefferson 2004). Mpf, therefore, not only benefits the carrying plasmid, but also mobilizable plasmids as well as the whole community of prokaryotes that can participate in biofilm formation (Burmølle et al. 2007).
Finally, conjugation can also prevent plasmid loss by reinfecting plasmid-free cells. This also, potentially, eliminates ‘cheating’ prokaryotes that benefit from the expression of plasmid-encoded traits such as excreted antimicrobial- or xenophobic-degrading enzymes. Conjugation therefore also contributes to plasmid stability since, theoretically, it rids the population of intercellular competition as long as transfer rates are higher than cell division rates. Reinfection has recently been demonstrated, through mathematical modelling, to greatly enhance the stability of plasmids (Lili et al. 2007), which has also been supported by experiments performed in high-density bacterial mats as well as on rat endothelial cells (Bahl et al. 2007a,b). The competitive advantages offered by a conjugation machinery alone would therefore further explain the great evolutionary success of conjugative plasmids. Stability through reinfection might also account for the existence of cryptic plasmids, although perhaps too many plasmids have been deemed cryptic owing to limited knowledge of the functions of the encoded proteins.
Type IV secretion systems. Many T4SSs that enable the translocation of macromolecules over the cell envelope have been described. T4SSs are normally divided into those that facilitate conjugation of DNA and those that deliver transfer effector proteins or toxins into infected host cells (Cascales & Christie 2003). Uptake of naked DNA has also been known to occur through T4SS mechanisms, as is the case with the gut prokaryote Helicobacter pylori. What makes plasmid-encoded T4SSs unique is that their gene products physically participate in their own gene dissemination.
Assembly of a T4SS often requires expression of 10 or more genes that are usually encoded on a single operon. These systems almost universally retain synteny, grouping genes into submodules whose gene products act within the cytoplasm, the periplasm or on the outer membrane (Cao & Saier 2001; Christie et al. 2005). This is probably a testament to the degree of organization required for the functionality of a machinery of such complexity. Curiously, phylogenies inferred from the T4SS genes have shown evidence of gene shuffling within T4SS submodules and that T4SSs, in some cases, have switched function more than once over the course of evolutionary history (Cao & Saier 2001; Frank et al. 2005; Nystedt et al. 2008). The contrast between the rather strict genetic organization and the dynamic nature of T4SS functionality would therefore seem to make them attractive subjects for the study of adaptive evolution of apparently ‘irreducibly complex’ genetic systems.
Most often, the expression of T4SS modules involved in conjugation is repressed (Bradley et al. 1980; Ghigo 2001), probably because the synthesis of conjugative pili represents a burden on the host cell, either from the energy expenditure, or because plasmid-specific phages can attack cells by attaching to conjugative pili. Expression of conjugative pili can normally be triggered by external cues such as a change in temperature (Alonso et al. 2005), but also by encountering plasmid-free cells, which subsequently results in the epidemic spread of the plasmid through whole plasmid-free populations. This phenomenon is known as transitory de-repression (Lundquist & Levin 1986). Plasmid transfer rates will therefore not only depend on varying physical conditions in the environment, but also on the existing distribution of the plasmid through the population. The extent of gene flow within a given communal gene pool would therefore seem to depend greatly on the levels of expression and the distribution of T4SS genes.
Surface and entry exclusion (protection against invasion). Entry exclusion is the property by which some plasmids physically prevent the entry of similar plasmids by T4SS-mediated conjugation. This is usually achieved either by interfering with the attachment of the conjugation pilus to the recipient cell surface (surface exclusion) or by preventing the transfer of DNA between existing mating pairs (entry exclusion). The latter is presumably achieved by preventing the binding of inner membrane components of T4SS (Zechner et al. 2000). Surface or entry exclusion seems like a particularly favourable stability strategy since it excludes closely related plasmids that could interfere with plasmid segregation or replication control and could potentially out-compete established plasmids. It is therefore a safeguard against incompatibility. Furthermore, entry exclusion can benefit the host cell by avoiding DNA transfer that could potentially pose a further metabolic burden. This mechanism may also increase the efficiency of dispersal of a plasmid in a population of bacteria without overall energy being wasted on conjugation between cells already containing plasmids (Zechner et al. 2000).
In most cases, the genes encoding entry exclusion are embedded within T4SS operons and are therefore parts of the Mpf modules. Although T4SSs are involved in a broad variety of functions, entry exclusion genes seem universally linked to the transportation of DNA. Thus, although phylogenetically related T4SSs that transport proteins exist, these do not seem to encode any genes that resemble entry exclusion genes.
Post-conjugation establishment. Entry of a plasmid into a new host (transconjugant) often results in derepression of several backbone genes, because of the absence of control proteins within the new cell, which helps in establishing the plasmid after conjugation. Low concentrations of copy-control molecules, for example, cause a dramatic rise in the initiation of plasmid replication, thereby ensure a quick recovery of copy numbers within the transconjugant. Some conjugative plasmids also carry antirestriction mechanisms that protect them from host-encoded restriction endonucleases, or mechanisms that minimize the triggering of the host SOS response (Althorpe et al. 1999). In many ways, these modules therefore represent stability mechanisms for horizontal dissemination, analogous to vertical stability mechanisms such as plasmid segregation or PSK.
If a plasmid were slow to establish itself within a new host, this might also force the integration of operative elements into the new chromosome, either by homologous recombination or by transposition events. Conjugation into less permissible hosts could therefore in effect abolish any HGT-mediated ‘evolutionary leaps’ achieved by transferring operative elements from one genome to another.
(iv). Modules affecting plasmid host adaptation
Most analysed plasmids carry a number of operative elements that enable host prokaryotes to succeed within specific environmental niches. In essence, they represent the most vital part of the communal pool. These elements can include virulence factors that enable colonization of eukaryotic cells, protection against antimicrobial or heavy metal substances or the ability to metabolize certain carbon sources. In most cases, accessory elements of foreign origin can be recognized on a plasmid by significant differences in the GC content from the plasmid backbone (e.g. figure 3).
The adaptive regions of a plasmid do not usually interfere with the normal order of backbone modules. Normally, adaptation modules are highly mosaic in structure and contain multiple insertion of IS sequences, transposons or integrons of different origins. This is probably due to the fact that the compact arrangement of plasmid essential genes within backbone modules makes them highly vulnerable to insertion of translocative elements, which could compromise the functionality of essential genes. The presence of insertion sequences within the adaptive regions of a plasmid backbone usually also provides multiple sites where additional insertions can happen without interfering with overall plasmid functionality.
(c). Are plasmids parasites or endosymbionts?
Over the years, plasmids have been regarded in a manner of different ways: (i) as parasitic entities (Lundquist & Levin 1986; Kado 1998) that prey on the resources of their host (which are themselves preyed upon by lesser mobile elements such as transposons, integrons or insertion sequences), (ii) facultative symbionts that provide their prokaryotic hosts with means of obtaining increased fitness at a trade-off of the resources required for their own existence (Eberhard 1990; Slater et al. 2008), or (iii) a collection of selfish functional genetic modules that coexist as discrete semi-autonomous pieces of DNA within a vast continuum of cellular life (Toussaint & Merlin 2002).
As the previous sections have described, some plasmid modules can be perceived as conferring selfish traits upon the host (PSK), while others confer altruism by promoting HGT and biofilm formation (Mpf modules). The life of these MGEs, almost universally, occurs within the cytoplasm of prokaryotic cells, so one could therefore add another layer of complexity by regarding the intracellular space of prokaryotes as tiny ecological niches where plasmids thrive in competition with other plasmids, for resources and the protection of the host cell membrane, as long as the total amount of extra-chromosomal DNA can be accommodated. At this level, plasmids are therefore driven by their selfish interests or the combined interests of their backbone modules. This competition, combined with the incompatibility of certain vital backbone functions, has therefore not only driven the evolution of conjugative plasmids, but has also helped to determine the adaptive gene contents of communal pools.
What the ‘true nature’ of conjugative plasmids is, therefore, is an open question, but it could to a large extent depend on the composition of prokaryotic communities and other environmental factors that shape the backbones of plasmids. Regardless, the analogy of conjugative plasmids as vessels of the communal pool that acts to transport adaptive traits between individual prokaryotes still rings true.
There is little doubt that conjugative plasmids can impose a metabolic burden upon their host and that carriage of traits that aid in local adaptation is, at least, a way of making transient plasmid carriage more advantageous (Eberhard 1990). Intuitively, it would seem that quickly integrating communal genes into the host chromosome would spare host cells the inconvenience of plasmid maintenance. Several investigations into the parasitic nature of conjugative plasmids, using various mathematical models, have speculated on this (Bergström et al. 2000; Levin & Bergstrom 2000; Lili et al. 2007). Rather surprisingly, the most recent study provided a strong indication that conjugative plasmids can be maintained in homogeneous prokaryotic populations by oscillations between plasmid-carrying and plasmid-free cells alone, regardless of the presence of adaptive traits (i.e. cryptic plasmids). As mentioned earlier, this has also been supported by some experimental evidence (Bahl et al. 2007a,b).
A direct correlation between high transfer rates and stable maintenance of conjugative plasmids has some rather striking implications for the communal pool hypothesis. This suggests that within communities where horizontal transfer rates are high, plasmid conjugation could perpetuate the formation of large communal gene pools, thereby allowing locally beneficial traits to be shared among prokaryotic populations restricted only by the host ranges of resident plasmids. Hence, a large communal pool might lead to an overall decrease in prokaryote genome sizes, assuming that only strictly host essential genes would be required within the private pools. Conversely, if transfer rates were to drop below those needed to sufficiently sustain conjugative plasmids, this might drive the communal pool back into the private pools (and chromosomes) of individual prokaryotes, perhaps resulting in an overall increase in the genome size. In the future, it will be interesting to experimentally test this hypothesis. Some plasmids, such as the E. coli F plasmid, posses specific mechanisms for integrating into the host chromosome and replicate in conjunction with it. In retrospect, this might represent a useful survival strategy for periods of limited horizontal transfer, as well as a method for acquiring new adaptive traits that may be useful when high transfer rates are restored. On rare occasions, such integrations can also turn the bacterial chromosome into one large conjugative unit (known as chromosome mobilization).
7. How do we study the communal gene pool?
Since the evolution of a prokaryote depends on the surrounding environment as a source for genetic innovation, the supergenome will depend upon the composition of the microbial community of which it is a part. Detailed understanding of the structure and composition of natural bacterial communities is therefore a requisite for understanding the dynamics of bacterial evolution. A case in point is clinical microbiology, where researchers have traditionally focused on only studying the biology of pathogenic microbes in monocultures, or in conjunction with their hosts. To investigate the supergenome of such a system, a much more holistic approach is required. In practical terms, it would entail the inclusion of the entire human microbiome as a subject or, perhaps more manageably, a focused study of the communal gene pool within the human microbiome. But how do we study the communal gene pool in highly complex natural microbial communities?
As described earlier, much work has been done in understanding how plasmids can replicate, regulate copy numbers, are maintained in the bacterial cell, pick up accessory elements or are transferred to other related or unrelated cultural bacteria. Much is now known about the diversity of plasmids that roam within well-studied genera such as E. coli and Pseudomonas, or the many adaptive traits found on plasmids that have relevance for humans, such as resistance to antibiotics and metals and degradative pathways. But since these plasmids are being investigated while they are under extreme accelerated selective forces, they are hardly representative of the natural diversity of plasmids. Bacteria associated with humans, animals or sewage systems are living in conjugational hotspots where they are in close contact with potential donors and recipients and multiple plasmids often reside in the same cell. IS elements and other homologies between plasmids would surely recombine more frequently in these strains than in strains where only one plasmid resides.
This suggests that to understand the role of the communal gene pool in bacterial evolution in general, one should focus on studying MGEs in natural environments. Do they carry accessory genes or are they merely streamlined molecular parasites? If they carry extra genetic loads, are these randomly collected genes or are they genes that encode beneficial traits for coping with life in the environment?
A few studies have focused on the exploration of environmental MGEs. These studies require that one must isolate plasmids without imposing predetermined phenotypes upon them. Recent studies suggest how this might be possible. For example, a 45 kb cryptic plasmid belonging to a new family was isolated based only on its ability to mobilize a resistance plasmid, a general property shared by most conjugative plasmids. This plasmid, pIPO2T, is widespread in rhizosphere populations and contains a suite of ORFs with unknown functions in addition to genes predicted to be involved in plasmid replication, maintenance and conjugative transfer (Tauch et al. 2002). In other studies, an ingenious transposon capture method was used to isolate non-specific plasmid DNA from the human gut flora, thereby recovering several small cryptic plasmids (Jones & Marchesi 2007), and a PCR- based method was used to amplify and sequence environmental ORFs related to class 1 integrons (Stokes et al. 2001). In the well known ‘Sargasso paper’ (Venter et al. 2004), at least 10 putative plasmids ranging from <10 to >100 kb were sequenced and assembled without any selective demands imposed. These plasmids were shown to encode homologues of the usual plasmid maintenance and transfer genes and putative genes for UV light, arsenate, mercury, copper and cadmium resistance in addition to ORFs of unknown function. In a comparative metagenomic study, an overrepresentation of plasmid-related sequences in soil metagenomes was found when compared with the Sargasso sequences (Tringe et al. 2005). Hence, in these and in future metagenomic datasets, there are likely to be many unbiased plasmid sequences to mine.
With the recent invasion of second-generation sequencing such as the 454-pyrosequencing technology, more ambitious studies can now be undertaken for exploring environmental DNA, both at the F single plasmid scale and at the a metagenomic scale, for a fraction of the cost and effort previously required. In most environments, approaches designed to analyse the non-communal gene pool are advantageous, because of the enormous number of these sequences present. Plasmid-specific pyrosequencing approaches are also being developed (Szczepanowski et al. 2008). In this approach, a selective pressure towards isolating resistance plasmid DNA is used, but no specific organismal pressure is added. Even more direct and non-selective metabolomic approaches will be needed to accomplish the unveiling of the communal gene pool efficiently.
To date, the ecology and fate of plasmids in the environment are poorly understood. However, the fast intra- and inter-cellular exchange of genetic material involving MGEs allows us to study both the dissemination (the epidemiology) and the maintenance of genes associated with plasmids in complex communities such as biofilms or soil microcosms. New culture-independent fluorometric detection techniques allow us to estimate the extent and host range of plasmid transfer, even for strains in which the plasmids cannot be stably maintained (Sørensen et al. 2005). These techniques, combined with powerful high-throughput sequencing, are moving plasmid biology to a new environmental era where our insight into bacterial evolution is bound to expand dramatically.
Footnotes
One contribution of 11 to a Theme Issue ‘The network of life: genome beginnings and evolution’.
References
- Allen E. E., Tyson G. W., Whitaker R. J., Detter J. C., Richardson P. M., Banfield J. F.2007Genome dynamics in a natural archaeal population. Proc. Natl Acad. Sci. 104, 1883–1888 (doi:10.1073/pnas.0604851104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso G., Baptista K., Ngo T., Taylor D. E.2005Transcriptional organization of the temperature-sensitive transfer system from the IncHI1 plasmid R27. Microbiology 151, 3563–3573 (doi:10.1099/mic.0.28256-0) [DOI] [PubMed] [Google Scholar]
- Althorpe N. J., Chilley P. M., Thomas A. T., Brammar W. J., Wilkins B. M.1999Transient transcriptional activation of the Incl1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol. Microbiol. 31, 133–142 (doi:10.1046/j.1365-2958.1999.01153.x) [DOI] [PubMed] [Google Scholar]
- Bahl M. I., Hansen L. H., Licht T. R., Sørensen S. J.2007aConjugative transfer facilitates stable maintenance of IncP-1 plasmid pKJK5 in Escherichia coli cells colonizing the gastrointestinal tract of the germfree rat. Appl. Environ. Microbiol. 73, 341–343 (doi:10.1128/AEM.01971-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl M. I., Hansen L. H., Sørensen S. J.2007bImpact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol. Lett. 266, 250–256 (doi:10.1111/j.1574-6968.2006.00536.x) [DOI] [PubMed] [Google Scholar]
- Bahl M. I., Hansen L. H., Goesmann A., Sørensen S. R. J.2007cThe multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established [alpha], [beta] and [delta] sub-groups. Plasmid 58, 31–43 (doi:10.1016/j.plasmid.2006.11.007) [DOI] [PubMed] [Google Scholar]
- Baquero F.2004Opinion—From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2, 510–518 (doi:10.1038/nrmicro909) [DOI] [PubMed] [Google Scholar]
- Bergström C. T., Lipsitch M., Levin B. R.2000Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J. Y., Nordstrom K., Lane D.2007Plasmid partition and incompatibility—the focus shifts. Mol. Microbiol. 65, 1405–1414 (doi:10.1111/j.1365-2958.2007.05882.x) [DOI] [PubMed] [Google Scholar]
- Bradley D. E.1980Morphological and serological relationships of conjugative pili. Plasmid 4, 155–169 (doi:10.1016/0147-619x(80)90005-0) [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Taylor D. E., Cohen D. R.1980Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 143, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmølle M., Hansen L. H., Sørensen S. J.2007Establishment and early succession of a multispecies biofilm composed of soil bacteria. Microb. Ecol. 54, 352–362 (doi:10.1007/S00248-007.9222-5) [DOI] [PubMed] [Google Scholar]
- Burmølle M., Bahl M. I., Jensen L. B., Sørensen S. J., Hansen L. H.2008Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154, 187–195 (doi:10.1099/mic.0.2007/010454-0) [DOI] [PubMed] [Google Scholar]
- Burrus V., Pavlovic G., Decaris B., Guedon G.2002Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46, 601–610 (doi:10.1046/j.1365-2958.2002.03191.x) [DOI] [PubMed] [Google Scholar]
- Cao T. B., Saier M. H., Jr2001Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147, 3201–3214 [DOI] [PubMed] [Google Scholar]
- Cascales E., Christie P. J.2003The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149 (doi:10.1038/nrmicro753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S., et al. 2000A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35, 490–516 (doi:10.1046/j.1365-2958.2000.01698.x) [DOI] [PubMed] [Google Scholar]
- Chen I., Dubnau D.2004DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 (doi:10.1038/nrmicro844) [DOI] [PubMed] [Google Scholar]
- Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E.2005Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451 (doi:10.1146/annurev.micro.58.030603.123630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Maddera L., Harris R. L., Silverman P. M.2008F-pili dynamics by live-cell imaging. Proc. Nat. Acad. Sci. 105, 17 978–17 981 (doi:10.1073/pnas.0806786105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N.1993Bacterial plasmids: their extraordinary contribution to molecular genetics. Gene 135, 67–76 (doi:10.1016/0378-1119(93)90050-D) [DOI] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K.1988Identification and classification of bacterial plasmids. Microbiol. Mol. Biol. Rev. 52, 375–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J.1999Genetic exchange between bacteria in the environment. Plasmid 42, 73–91 (doi:10.1006/plas.1999.1421) [DOI] [PubMed] [Google Scholar]
- de la Cueva-Méndez G., Pimentel B.2007Gene and cell survival: lessons from prokaryotic plasmid R1. EMBO Rep. 8, 458–464 (doi:10.1038/sj.embor.7400957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M., Az-Orejas R.1998Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelVecchio V. G., et al. 2002The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Nat. Acad. Sci. USA 99, 443–448 (doi:10.1073/pnas.221575398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J.2004Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424 (doi:10.1038/nrmicro884) [DOI] [PubMed] [Google Scholar]
- Eberhard W. G.1990Evolution in bacterial plasmids and levels of selection. Q. Rev. Biol. 65, 3–22 (doi:10.1086/416582) [DOI] [PubMed] [Google Scholar]
- Fernandez-Lopez R., Garcillan-Barcia M. P., Revilla C., Lazaro M., Vielva L., de la C. F.2006Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30, 942–966 (doi:10.1111/j.1574-6976.2006.00042.x) [DOI] [PubMed] [Google Scholar]
- Frank A. C., Alsmark C. M., Thollesson M., Andersson S. G. E.2005Functional divergence and horizontal transfer of type IV secretion systems, Mol. Biol. Evol. 22, 1325–1336 (doi:10.1093/molbev/msi124) [DOI] [PubMed] [Google Scholar]
- Frigaard N. U., Martinez A., Mincer T. J., DeLong E. F.2006Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439, 847–850 (doi:10.1038/nature04435) [DOI] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A.2005Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (doi:10.1038/nrmicro/235) [DOI] [PubMed] [Google Scholar]
- Galibert F., et al. 2001The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668–672 (doi:10.1126/science.1060966) [DOI] [PubMed] [Google Scholar]
- Gerdes K., Christensen S. K., Lobner-Olesen A.2005Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 (doi:10.1038/nrmicro1147) [DOI] [PubMed] [Google Scholar]
- Ghigo J. M.2001Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445 (doi:10.1038/35086581) [DOI] [PubMed] [Google Scholar]
- Giraldo R.2003Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 26, 533–554 (doi:10.1111/j.1574-6976.2003.tb00629.x) [DOI] [PubMed] [Google Scholar]
- Goldenfeld N., Woese C.2007Biology's next revolution, Nature 445, 369 (doi:10.1038/445369a) [DOI] [PubMed] [Google Scholar]
- Hansen L. H., Jensen L. B., Sørensen H. I., Sørensen S. J.2007Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60, 145–147 (doi:10.1093/jac/dkm167) [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Cresawn S. G., Hendrix R. W.2008Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 159, 332–339 (doi:10.1016/j.resmic.2008.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Barilla D.2006The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4, 133–143 (doi:10.1038/nrmicro1342) [DOI] [PubMed] [Google Scholar]
- Helinski D. R.2000Introduction to plasmids: a selective view of their history. In Plasmid biology (eds Funnell B. E., Phillips G. J.) pp. 1–21 Washington, DC: ASM Press [Google Scholar]
- Hülter N., Wackernagel W.2008Double illegitimate recombination events integrate DNA segments through two different mechanisms during natural transformation of Acinetobacter baylyi. Mol. Microbiol. 67, 984–995 (doi:10.1111/j.1365-2958.2007.06096.x) [DOI] [PubMed] [Google Scholar]
- Jain R., Rivera M. C., Lake J. A.1999Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Nat. Acad. Sci. USA 96, 3801–3806 (doi:10.1073/pnas.96.7.3801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Rivera M. C., Moore J. E., Lake J. A.2003Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20, 1598–1602 (doi:10.1093/molbev/msg154) [DOI] [PubMed] [Google Scholar]
- Jefferson K. K.2004What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173 [DOI] [PubMed] [Google Scholar]
- Jones B. V., Marchesi J. R.2007Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat. Methods 4, 55–61 (doi:10.1038/nmeth964) [DOI] [PubMed] [Google Scholar]
- Kado C. I.1998Origin and evolution of plasmids. Antonie van Leeuwenhoek 73, 117–126 (doi:10.1023/A:1000652513822) [DOI] [PubMed] [Google Scholar]
- Kobayashi I.2001Behavior of restriction–modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29, 3742–3756 (doi:10.1093/nar/29.18.3742) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I.2008Genomics of Bacteria and Archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36, 6688–6719 (doi:10.1093/nar/gkn668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G.2003Gene organization: selection, selfishness, and serendipity. Ann. Rev. Microbiol. 57, 419 (doi:10.1146/annurev.micro.57.030502.090816) [DOI] [PubMed] [Google Scholar]
- Lawrence J. G., Hendrickson H.2003Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50, 739–749 (doi:10.1046/j.1365-2958.2003.03778.x) [DOI] [PubMed] [Google Scholar]
- Lawrence J. G., Hendrickson H.2005Genome evolution in bacteria: order beneath chaos. Curr. Opin. Microbiol. 8, 572–578 (doi:10.1016/j.mib.2005.08.005) [DOI] [PubMed] [Google Scholar]
- Lawrence J. G., Hendrix R. W., Casjens S.2001Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9, 535–540 (doi:10.1016/S0966-842x(01)02198-9) [DOI] [PubMed] [Google Scholar]
- Lederberg J.1952Cell genetics and hereditary symbiosis. Physiol. Rev. 32, 403–430 [DOI] [PubMed] [Google Scholar]
- Leplae R., Hebrant A., Wodak S. J., Toussaint A.2004ACLAME: a classification of mobile genetic elements. Nucleic Acids Res. 32Suppl.1, D45–D49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R., Bergstrom C. T.2000Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Nat. Acad. Sci. USA 97, 6981–6985 (doi:10.1073/pnas.97.13.6981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lili L. N., Britton N. F., Feil E. J.2007The persistence of parasitic plasmids. Genetics 177, 399–405 (doi:10.1534/genetics.107.077420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M., Gomis-Ruth F. X., Coll M., de la Cruz F. F.2002Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8 (doi:10.1046/j-1365-2958.2002.03014.x) [DOI] [PubMed] [Google Scholar]
- Lundquist P. D., Levin B. R.1986Transitory derepression and the maintenance of conjugative plasmids. Genetics 113, 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R. D.2007Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189, 6089–6092 (doi:10.1128/JB.00958-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R.2005The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594 (doi:10.1016/j.gde.2005.09.006) [DOI] [PubMed] [Google Scholar]
- Medini D., Serruto D., Parkhill J., Relman D. A., Donati C., Moxon R., Falkow S., Rappuoli R.2008Microbiology in the post-genomic era, Nat. Rev. Microbiol. 6, 419–430 (doi:10.1038/nrmicro01901) [DOI] [PubMed] [Google Scholar]
- Mira A., Ochman H., Moran N. A.2001Deletional bias and the evolution of bacterial genomes. Trends Genet. 17, 589–596 (doi:10.1016/S0168-9525(01)02447-7) [DOI] [PubMed] [Google Scholar]
- Musovic S., Oregaard G., Kroer N., Sorensen S. J.2006Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among Gram-positive and gram-negative bacteria indigenous to the barley rhizosphere. Appl. Environ. Microbiol. 72, 6687–6692 (doi:10.1128/AEM.00013-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narra H. P., Ochman H.2006Of what use is sex to bacteria? (Abstract). Current Biology 16, R705–R710 [DOI] [PubMed] [Google Scholar]
- Norman A., Hansen L. H., She Q., Sørensen S. J.2008Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60, 59–74 (doi:10.1016/j.plasmid.2008.03.003) [DOI] [PubMed] [Google Scholar]
- Nystedt B., Frank A. C., Thollesson M., Andersson S. G. E., 2008Diversifying selection and concerted evolution of a type IV secretion system in Bartonella. Mol. Biol. Evol. 25, 287–300 (doi:10.1093/molbev/msm252) [DOI] [PubMed] [Google Scholar]
- Ochman H., Davalos L. M.2006The nature and dynamics of bacterial genomes. Science 311, 1730–1733 (doi:10.1126/science.1119966) [DOI] [PubMed] [Google Scholar]
- Osborn M. A., Böltner D.2002When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48, 202–212 (doi:10.1016/S0147-619x(02)00117-8) [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Wren B. W.2007Bacterial pathogenomics. Nature 449, 835–842(doi:10.1038/nature06248) [DOI] [PubMed] [Google Scholar]
- Paulsson J.2002Multileveled selection on plasmid replication. Genetics 161, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. C., Jain R., Moore J. E., Lake J. A.1998Genomic evidence for two functionally distinct gene classes. Proc. Nat. Acad. Sci. USA 95, 6239–6244 (doi:10.1073/pnas.95.11.6239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra De Bast M., Mine N., Van Melderen L.2008Chromosomal toxin–antitoxin systems may act as antiaddiction modules. J. Bacteriol. 190, 4603–4609 (doi:10.1128/JB.00357-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevastsyanovich Y. R., et al. 2008Diversity of IncP-9 plasmids of Pseudomonas. Microbiology 154, 2929–2941 (doi:10.1099/mic.0.2008/017939.0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q., Peng X., Zillig W., Garrett R. A.2001Genome evolution: gene capture in archaeal chromosomes. Nature 409, 478 (doi:10.1038/35054138) [DOI] [PubMed] [Google Scholar]
- Sherley M., Gordon D. M., Collignon P. J.2003Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid 49, 79–85 (doi:10.1016/S0147-619x(02)00107-5) [DOI] [PubMed] [Google Scholar]
- Slater F. R., Bailey M. J., Tett A. J., Turner S. L.2008Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 66, 3–13 (doi:10.1111/j.1574.6941.2008.00505.x) [DOI] [PubMed] [Google Scholar]
- Smets B. F., Barkay T.2005Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Rev. Microbiol. 3, 675–678 (doi:10.1038/nrmicro1253) [DOI] [PubMed] [Google Scholar]
- Sørensen S. J., Bailey M., Hansen L. H., Kroer N., Wuertz S.2005Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710 (doi:10.1038/nrmicro1232) [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Holmes A. J., Nield B. S., Holley M. P., Nevalainen K. M. H., Mabbutt B. C., Gillings M. R.2001Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67, 5240–5246 (doi:10.1128/AEM.67.11.5240-5246.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D.1998Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29, 1137–1145 (doi:10.1046/j.1365-2958.1998.01012.x) [DOI] [PubMed] [Google Scholar]
- Summers D. K., Beton C. W., Withers H. L.1993Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol. Microbiol. 8, 1031–1038 (doi:10.1111/j.1365-2958.1993.tb01648.x) [DOI] [PubMed] [Google Scholar]
- Szczepanowski R., Bekel T., Goesmann A., Krause L., Krömeke H., Kaiser O., Eichler W., Pühler A., Schlüter A.2008Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analyzed by the 454-pyrosequencing technology. J. Biotechnol. 136, 54–64 (doi:10.1016/j.jbiotec.2008.03.020) [DOI] [PubMed] [Google Scholar]
- Tauch A., et al. 2002The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology 148, 1637–1653 [DOI] [PubMed] [Google Scholar]
- Thomas C. M.2000Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491 (doi:10.1046/j.1365-2958.2000.02006.x) [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Nielsen K. M.2005Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (doi:10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- Tolmasky M. E., Colloms S., Blakely G., Sherratt D. J.2000Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146, 581–589 [DOI] [PubMed] [Google Scholar]
- Toussaint A., Merlin C.2002Mobile elements as a combination of functional modules. Plasmid 47, 26–35 (doi:10.1006/plas.2001.1552) [DOI] [PubMed] [Google Scholar]
- Tringe S. G., et al. 2005Comparative metagenomics of microbial communities. Science 308, 554–557 (doi:10.1126/science.1107851) [DOI] [PubMed] [Google Scholar]
- Venter J. C., et al. 2004Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (doi:10.1126/science.1093857) [DOI] [PubMed] [Google Scholar]
- Zechner E. L., et al. 2000Conjugative-DNA processes. In The horizontal gene pool (ed. Thomas C. M.), pp. 87–174 Amsterdam, The Netherlands: Harwood Academic Publishers [Google Scholar]