Abstract
Whole-genome sequences from the choanoflagellate Monosiga brevicollis, the placozoan Trichoplax adhaerens and the cnidarian Nematostella vectensis have confirmed results from comparative evolutionary developmental studies that much of the developmental toolkit once thought to be characteristic of bilaterians appeared much earlier in the evolution of animals. The diversity of transcription factors and signalling pathway genes in animals with a limited number of cell types and a restricted developmental repertoire is puzzling, particularly in light of claims that such highly conserved elements among bilaterians provide evidence of a morphologically complex protostome–deuterostome ancestor. Here, I explore the early origination of elements of what became the bilaterian toolkit, and suggest that placozoans and cnidarians represent a depauperate residue of a once more diverse assemblage of early animals, some of which may be represented in the Ediacaran fauna (c. 585–542 Myr ago).
Keywords: Cambrian explosion, evo-devo, cnidaria, Ediacaran, genetic toolkit
1. Introduction
A variety of transcription factors, signalling pathway genes and other regulatory elements once thought to be characteristic of bilaterians, and associated with their diverse cell types and more complex morphologies, have now been documented among more ancient clades that lack diverse cell types and complex morphogenesis. This raises questions about the current function of these elements (Srivastava et al. 2008) as well as their ancestral role during the early evolution of animals. The precursors of the bilaterian developmental toolkit have been documented in non-bilaterian animals and choanoflagellates by comparative studies and confirmed by whole-genome sequences of the choanoflagellate Monosiga brevicollis (the closest known relative of metazoans), the placozoan Trichoplax adhaerens and the cnidarian Nematostella vectensis. The whole genome of the sponge Amphimedon queenlandica is expected shortly. Table 1 shows the genome size, number of cell types and various features of developmental complexity in these groups, and includes Drosophila melanogaster for comparison.
Table 1.
Genome size and the number of inferred genes are from the whole-genome studies cited in the text; that of Amphimedon is from Fahey et al. (2008). The number of cell types is from Valentine et al. (1994), except for Trichoplax, which is from Srivastava et al. (2008); and the number of miRNAs comes from Sempere et al. (2006), Gimson et al. (2008) and Wheeler et al. (2009). Note that according to Wheeler et al. (2009), the seven miRNAs in demosponges are not homologous to any eumetazoan miRNAs. The 10 transcription factors families are the homeodomian, forkhead, p53, Myc, Sox/TCF, ETS, HOX, NHR, POU and T-box, with the data for Monosiga from King et al. (2008), and other taxa from Larroux et al. (2008); true HOX class genes are missing from Monosiga and Amphimedon. The number of basic helix-loop-helix (bHLH) genes from Simionato et al. (2007) except for Trichoplax which is from Srivastava et al. (2008).
| Monosiga brevicollis | Amphimedon queenslandica | Trichoplax adhaerens | Nematostella vectensis | Drosophila melanogaster | |
|---|---|---|---|---|---|
| genome size (Mb) | 41.6 | 167 | 98 | 450 | 180 |
| no. of genes | 9100 | ? | 11 514 | 18 000 | 14 601 |
| no. of cell types | 1 | 12 | 4 | 20 | 50 |
| no. of miRNA | 0 | 7 | 7 | 3 | 49 |
| no. of metazoan transcription factors/families | ?/5 | 57/?6 | 35/9 | min. 87/10 | min. 87/10 |
| no. of bHLH genes | 0 | 16 | 27 | 68 | 59 |
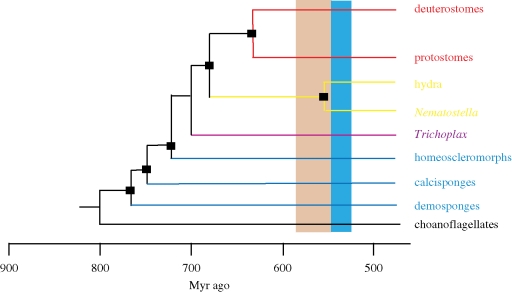
This analysis requires a robust metazoan phylogeny (figure 1). Of note are the paraphyletic sponges, with demosponges-, calcisponges- and the homeoscleromorphs-independent clades (Sperling et al. 2007), and Trichoplax positioned between sponges and cnidarians (Srivastava et al. 2008). The acoel flatworms are not included below the protostome–deuterostome (P/D) divergence, following Philippe et al. (2007). The phylogenetic affinities of the acoels have long been contentious, with most studies supporting their placement as basal bilaterians (Ruiz-Trillo et al. 1999; Baguana & Riutort 2004; Baguna et al. 2008; Hejnol & Martindale 2008a). By contrast, Philippe et al. (2007) employed a new model to overcome the long-branch attraction problems that have previously plagued phylogenetic positioning of this clade, suggesting that acoels are deuterostomes, and are secondarily primitive. I view the placement of acoels as basal bilaterians as the more likely, but it is not critical to the following discussion. Indeed, recent developmental studies of acoels (Hejnol & Martindale 2008a,b) strengthen the arguments here. The tree in figure 1 is calibrated with the molecular clock results of Peterson et al. (2008), with metazoans originating during the Cryogenian and bilaterians during the Ediacaran.
Figure 1.
A phylogenetic tree for metazoans scaled against the molecular clock dates of Peterson et al. (2008). The ages of nodes are well calibrated by the molecular clock results of Peterson et al. and are shown as black boxes.
2. The protostome–deuterostome toolkit
The similarity of a variety of complex morphogenetic pathways across protostomes and deuterostomes (based initially on studies of Drosophila, Cenorhabditis and Mus, but recently expanding to a more phylogenetically diverse array of animals) led to the conclusion that their last common ancestor not only possessed these developmental systems, but the complex morphologies that they produce in modern organisms (Slack et al. 1993; De Robertis & Sasai 1996; Ohno 1996; Valentine et al. 1999; Carroll et al. 2001; Erwin & Davidson 2002; Erwin 2006; De Robertis 2008; Raff 2008).
The complexity of this toolkit varies as new studies and broader comparative studies are reported, but minimally includes the following: anterior/posterior (A/P) patterning with seven or eight HOX genes (de Rosa et al. 1999; Balavoine et al. 2002), and the associated microRNA responsible for inhibiting translation of HOX mRNAs (de Robertis 2008); the HOX genes part of a larger super cluster of at least eight other ANTP-class genes including the ParaHox and NK cluster genes (Butts et al. 2008); dorsal/ventral (D/V) patterning controlled by the sog/chordin dpp/BMP2/4 system (Arendt & Nubler-Jung 1994; De Robertis & Sasai 1996); anterior patterning via ems/Emx and otd/Otx and a tripartite brain (Arendt & Nubler-Jung 1999; Reichert & Simeone 2001; Arendt et al. 2008) with posterior patterning via evenskipped/evx and caudal/cdx; segmentation through engrailed and Delta–Notch (Holland et al. 1997; Balavoine & Adoutte 2003; Stollenwerk et al. 2003; Tautz 2004); eye formation controlled by a dense network of genes, including Pax6 and ey (Quiring et al. 1994; Halder et al. 1995; Gehring 2004), but see Arendt et al. (2004) and Fernald (2000); endoderm formation and a regionalized through gut via GATA transcription factors, brachyury and goosecoid (Arendt et al. 2001); heart formation via Nkx2.5/tinman (Harvey 1996; Bodmer & Venkatesh 1998; Olson 2006); and distal-less involvement in appendage formation (Panganiban et al. 1997; Panganiban & Rubenstein 2002; Pueyo & Couso 2005). The pattern of acquisition of microRNAs tracks increasing morphological complexity and provides important information on the evolution of developmental control (Sempere et al. 2006; Gimson et al. 2008; Wheeler et al. 2009).
The support for a complex P/D ancestor is not as strong as many proponents suggest. While A/P and D/V patterning seem inescapably part of the P/D ancestor, the assumption that all conserved elements were necessarily involved in the same morphogenetic roles as today may not be valid. The ongoing debate over the extent of nervous system centralization in the P/D ancestor illustrates the problem. In both flies and vertebrates, the boundaries of the tripartite brain reflect patterning by Otd/Otx, Pax 2/5/8 and the anteriormost Hox genes, leading to the widespread view that these divisions were present in the P/D ancestor (Arendt & Nubler-Jung 1999; Reichert & Simeone 2001; Ghysen 2003; Hirth et al. 2003; Lichtneckert & Reichert 2005; Denes et al. 2007; Mizutani & Bier 2008). Yet, the hemichordate Saccoglossus kowalevskii lacks a centralized brain, possessing a diffuse nerve net, albeit with some degree of anterior neuronal concentration. The ectodermal patterning of these three genes (and 19 others) is similar to that in animals with a tripartite brain (Lowe et al. 2003). Lowe et al. concluded that ancestral deuterostomes, and probably the P/D ancestor, similarly had a diffuse nerve net rather than a centralized brain (a conclusion that anatomists had previously reached based on comparative morphology). More recent studies, particularly comparisons between the annelid Platyneris and Saccoglassus, have revealed a more complex situation. Arendt et al. (2008) illustrate the conservation of the mediolateral patterning of the neurectoderm, as well as a conservation of neuron types, suggesting a ‘conservation of molecular architectures’ (p. 1527). But can we infer ancestral morphologies from these conserved architectures? Arendt et al. do not consider the issue within a phylogenetic context, which favours a diffuse nerve net, as Lowe originally argued. Lowe's continuing studies of Saccoglossus nerve system development confirm that the networks of regulatory interaction have been conserved, but he cautions that this information provides little insight into ancestral morphology (Lowe 2008). It is clear, however, that inferring ancestral morphologies from conserved genes, even from conserved networks of regulatory interactions, is not straightforward. Homology of a regulatory pattern does not demonstrate homologous morphologic structures.
Davidson and I (Erwin & Davidson 2002) made the more general argument that many of these conserved processes were responsible for cell specification and regional patterning rather than initiation of complex morphogenetic development. We suggested that pattern formation developmental control was subsequently intercalated into these simpler networks to produce specific patterning systems within different clades. From this perspective, the P/D ancestor would have been much simpler than suggested by others, composed of a suite of regionally patterned specialized cell types, but not necessarily with the more complex structures observed today. One way of addressing this issue is to trace these highly conserved systems back into animal clades that arose earlier in evolution, but that lack complex morphology.
3. Genomic precursors
There are many cnidarian model systems, but the development of the sea anemone N. vectensis has received considerable attention and the entire genome sequence was recently released (Ryan et al. 2007b). Other cnidarians have also been studied, including the coral Acropora and Hydra, a hydrozoan. Anthozoans split from hydrozoans perhaps 548 Ma (figure 1), and the development of the medusoid life-history stage in hydrozoans was accompanied by the origin of a number of cell-type-specific genes (Hwang et al. 2007). Thus, anthozoans such as Nematostella may be more informative about the last common ancestor of cnidarians + higher bilaterians. Developmental studies of Nematostella have revealed unexpected complexity in its developmental toolkit. Orthologues of many genes previously considered characteristic of higher bilaterians arose before the origin of cnidarians. It is now clear that different lineages of higher bilaterians lost many genes present in ancestral metazoans, even as the diversity of particular gene families expanded through gene duplication.
Bilaterians have six major signalling pathways: Wnt, TGF-β, Notch, hedgehog, Jak/STAT and RT; all are present in cnidarians (Technau et al. 2005; Matus et al. 2008), as are a diversity of transcription factors (Putnam et al. 2007). All 12 of the known Wnt subfamilies occur in cnidarians, with overlapping expression along the oral–aboral axis, involvement in cell-type specification and gastrulation (Kusserow et al. 2005). Fifty-six homeodomain families are present in Nematostella (Ryan et al. 2007a) and representatives of four different Pax gene classes (Piatigorsky & Kozmik 2004; Matus et al. 2007). More remarkably for an animal with only two tissue layers, at least seven genes associated with the formation of mesoderm in bilaterians are expressed in the developing endoderm of Nematostella (Martindale et al. 2004). Thus, germ-layer specification was already present in the ancestral forms. The conservation of these elements in cnidarians does not necessarily mean that they had the same morphogenetic role as in more complex bilaterians, rather, new or more enhanced roles may have developed later (Ball et al. 2004). The staggered expression of Hox genes along the body axis and the asymmetric expression of dpp (a TGF-β family gene), presumably reflecting a D/V axis (Finnerty et al. 2004), have been associated with bilateral symmetry. Comparison of synteny patterns led Hui et al. (2008) to propose that Nematostella has both a Hox and a Parahox cluster, indicating their duplication before the divergence of cnidarians. A suite of TGF-β genes and antagonists as well as homeodomain proteins are expressed asymmetrically along the A/P axis, and are interpreted by Matus et al. (2006) as the precursors of a bilaterally organized central nervous system.
As emphasized by Baguna et al. (2008), this distribution of characters challenges traditional views of metazoan evolution, in particular, by separating the origin of a D/V axis from the origin of mesoderm. Taken as read, it suggests that the last common ancestor of cnidarians and higher bilaterians had A/P and possibly D/V polarity, muscle cells of mesodermal affinities and a simple eye. But perhaps the most critical issues are proposals that triploblasty originated before the cnidarian divergence, and thus that the diploblastic condition of Hydra and some other cnidarians is a derived condition (Boero et al. 1998; Martindale et al. 2004; Seipel & Schmid 2005, 2006; Boero et al. 2007). Evidence in favour of this position includes the histologically identical striated muscles in the entocodon of hydromedusae, the expression patterns of ‘mesodermal’ genes in cnidarians, cited above and the coelom-like hydromedusan subumbrellar structure. If these conclusions are correct, then the last common ancestor of cnidarians plus higher bilaterians possessed the toolkit for bilaterality and triploblasty, and possessed at least some elements of mesodermal muscle development. By contrast, Ball et al. (2007) critically reexamined the evidence for axial patterning and specifically the homology of the cnidarian oral–aboral axis with the eumetazoan A/P axis. As with mesoderm formation, Ball et al. (2007) make the provocative suggestion that the diversity of cnidarian body plans may represent an independent derivation of axial patterning and other regional patterning systems from those of bilaterians. For interpreting the fossils of the Ediacaran, the important point is that cnidarians possess the developmental tools for axial patterning and the production of triploblastic body plans, whether or not body axes and mesoderm of higher bilaterians actually are homologous to cnidarian structures.
Placazoans are 1–2 mm, disc-shaped animals with two epithelial layers and four cell types, but they lack specialized nerve, sensory or muscle cells. Their phylogenetic affinities have been controversial, but appear to have been resolved by the recent sequencing of the whole genome of T. adhaerens (Srivastava et al. 2008; see commentary by Miller & Ball 2008). Both Bayesian and maximum likelihood analyses strongly support placazoans as the sister group to cnidarians + bilaterians (figure 1), and reject previous suggestions that Trichoplax is either a derived cnidarian or a bilaterian. The repertoire of transcription factors and signalling pathways is remarkable: 35 homeobox TFs, including ANTP, PRD, POU and SIX-class genes that are associated with regional patterning in bilaterians; a variety of factors associated with cell-type specification; the components of a Wnt/β catenin signalling pathway, responsible for axial patterning in bilaterians, cnidarians (Lee et al. 2007) and demosponges (Adamska et al. 2007) and the TGF-β pathway, active in D/V patterning in bilaterians, and evidently in cnidarians (Matus et al. 2006) and demosponge larvae (Adamska et al. 2007) as well. The hedgehog pathway is evidently absent and the Notch and JAK/STAT pathways are incomplete, although these were evidently lost as they have been identified in sponges. Despite their lack of a nervous system, placozoans respond to stimuli and have genes associated with synapse formation, for neurotransmitters, and photoreception (Srivastava et al. 2008). Given their developmental complexity, Srivastava et al. (2008) and Miller & Ball (2008) raise the issue that there may well be as yet unidentified body plans generated by Trichoplax that may make use of this complexity. This is an interesting possibility to explore, but here I am concerned with the attributes of the ancestral node rather than their current expression.
Sponges seem to be developmentally unsophisticated organisms, lacking true muscle or nerve cells, tissues or regionalization of discrete cell lineages, and more akin to a colony of cells than true animals. However, developmental studies of the demosponge A. queenlandica have revealed a suite of developmental attributes including cell specification, patterning of distinct cell layers and morphogenetic gradients. These attributes must have arisen between the split from choanoflagellates and the origin of metazoa. Moreover, molecular phylogenetics has now shown sponges are not a single clade (Phylum Porifera), but three different clades (figure 1). Many details of the sponge developmental toolkit have been identified (Fahey et al. 2008; reviewed in Muller et al. 2004). The Wnt/catenin and TGF-β pathways provide axial and D/V patterning (Adamska et al. 2007). Other transcription factors have been identified, including a variety of ANTP (HOX), Pax, POU, T-box, Sox, Mef2, PRD and LIM class genes; some are expressed in specific tissues and thus establish regionalized domains of gene expression (Muller et al. 2004; Larroux et al. 2006, 2008). Complicating the issue is that demosponges have almost certainly lost a number of transcription factors, including NK and ANTP genes (Peterson & Sperling 2007) There are, however, important differences between demosponges and eumetazoans: there are relatively few genes within each class of transcription factors; the expansion of most of classes occurred after the origin of the demosponges (Kusserow et al. 2005; Simionato et al. 2007). Moreover, based on a comparison of the intergenic regions within the NK homeobox cluster of Amphimedon, Fahey et al. (2008) propose that demosponges possess more limited regulatory machinery than eumetazoans. Thus, the last common ancestor of all metazoa was able to specify multiple cell types, establish body axes and array different cell types along these axes and produce multicellular structures (Larroux et al. 2006), but evidently lacked the regulatory complexity and depth of transcription factors and microRNAs required to produce complex gene regulatory networks (GRNs), and thus more complex morphological structures.
Choanoflagellates are the nearest sister group to the metazoa, a relationship long recognized by their similarity to sponge choanocytes. Recent comparative genomic studies and the recent sequencing of the genome of M. brevicollis have identified a remarkable developmental diversity of cell adhesion, extra cellular matrix, signal transduction and cellular differentiation elements, including 78 protein domains shared by choanoflagellates and metazoans but no other groups (King et al. 2003, 2008; King 2004). The wnt and TGF-β signalling pathways are absent, and there is only a single gene representing what develops into the JAK/STAT pathway. Some metazoan transcription factor families such as p53, Sox/TCF and Myc are present, but the HOX, ETS, POU and T-box families are all missing (King et al. 2008). Those families that are present are less diverse than in metazoa. For example, five immunoglobulin domains were identified in M. brevicollis versus the 150–1500 that may exist in vertebrates. More importantly, many of these protein domains do not have the characteristic architectures of metazoan adhesion proteins.
This comparative framework reveals a progressive increase in the regulatory complexity from choanoflagellates to sponges, placozoans, cnidarians and finally, complex bilaterians. The four primary signalling pathways appeared relatively early, but there was an approximately threefold increase in homeobox and bHLH genes between sponges and cnidarians, although with a different amount of gene duplication in different lineages (Simionato et al. 2007; Larroux et al. 2008). Cell-type specification, regional pattern formation and some degree of axis formation are evident in sponges, and all three have increased substantially by the appearance of cnidarians, although some of these tools may have been acquired independently in sponges and cnidarians. The presence of many ‘bilaterian’ developmental tools in morphologically simpler organisms reinforces the view that the original role of these genes and regulatory networks was in the formation of specialized cell types in specific regions of the body, not necessarily in producing complex multicellular structures (Erwin & Davidson 2002). Evidence from acoels further substantiates this view (Hejnol & Martindale 2008a). The increasing diversity of microRNAs provides further evidence of increasing regulatory control (Sempere et al. 2006; Gimson et al. 2008; Wheeler et al. 2009). Gene loss is correlated with the increasing morphologic complexity of body plans, with deuterostomes retaining more of the ancestral gene families than ecdysozoans, although it may be premature to claim, as De Robertis (2008) does, that such losses play a fundamental role.
4. The ediacaran fauna in light of development
Rocks from the Ediacaran Period (635–542 Ma) immediately preceding the explosion of animals diversity in the Early Cambrian, contain a diversity of centimetre to metre-long fronds, discs and more complex forms, some of them superficially similar to modern animal groups. Although some members of this assemblage of fossils, known as the Ediacaran biota (575–542 Ma), display apparent bilateral symmetry, none has evidence of appendages, eyes, a mouth (with one exception) or other characteristics of the higher bilaterians. This puzzling array of features has led to persistent controversy over what sorts of organisms these fossils represent. Paleontologists have allied particular fossils with the arthropods, annelids and other bilaterian clades, while others believe they represent early animals, but not bilaterians. Others have suggested they are not even animals, but rather lichen, fungi or even prokaryotes (for recent reviews see Gehling et al. 2005; Narbonne 2005; Fedonkin et al. 2007a; Xiao & Laflemme 2009).
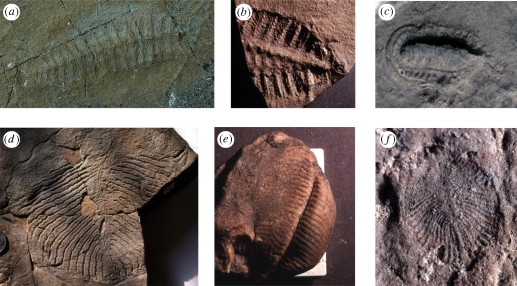
Within the Ediacaran fauna, several different morphological and constructional clusters can be distinguished, although the extent to which they represent monophyletic clades is unclear. One group, known as the rangeomorphs from the genus Rangea, includes a variety of fronds and brush-like forms, composed of alternately arranged frodlets with a fractal structure (figure 2a,b). The erniettomorphs include Ernietta (figure 2e) and Pteridinium, and have been interpreted as having quilted tubes, much like an air mattress, alternately arrayed along a midline. They appear to have been constructed of a reasonably tough and tear-resistant material. Dickinsonia (figure 2f) is similar to the erniettamorphs but has a higher degree of bilaterial symmetry, A/P differentiation and evidence of muscular tissue. It is, in some ways, an intermediate to a more heterogeneous assemblage of roughly bilaterally symmetrical forms, some of which appear to have segmentation, such as Spriginna and Yorgia (figure 2d). There are also a host of other forms that are more difficult to categorize, including a variety of discs that probably represent the holdfast of fronds, and forms with other symmetries. The one probable bilaterian in the Ediacaran assemblage is Kimberella, which apparently had a rasping mouthpart structure similar to the radula of a gastropod (figure 2c).
Figure 2.
Ediacaran fossils mentioned in the text. (a) Fracofusus from Mistaken Point, Newfoundland, 14 cm. (b) Rangea from southern Namibia; 5 cm. (c) Kimberella from White Sea, Russia; anterior to right, with the anterior elongation, 9 cm. (d) Yorgia from the White Sea, Russia; anterior (?) up, about 18 cm. (e) Ernietta from southern Namibia, approximately 15 cm. (f) Dickinsonia from Ediacaran Hills, south Australia, 6 cm.
My concern here is assessing the variety of developmental strategies. Many Ediacaran organisms exhibit strong axial growth, as seen in a variety of frondose forms, including the rangeamorphs, in fronds such as Charnia and Charnodiscus, in the erniettomorphs and in Dickinsonia and its allies. Anterior–posterior differentiation is evident in Dickinsonia, and expands to include anterior specialization in Kimberella, Marywadea, Parvancorina, Sprigginna and Yorgia. Dorsal–ventral differentiation and alternating growth along a midline have been described from all of the above taxa, as well as the erniettomorphs such as Ernietta. A more complicated issue is that of segmentation. Segmentation is distinct from serial repetition, and the Delta–Notch signalling system is involved in defining segmental boundaries in vertebrates and spiders (where Notch patterns the ectoderm, rather than the mesoderm, as in vertebrates) (Stollenwerk et al. 2003; Tautz 2004; Pueyo et al. 2008; reviewed in Erwin 2006). While the Notch signalling pathway is present in cnidarians, the Delta–Notch cascade appears to be a later innovation. Thus, if true segmentation could be shown in Ediacaran organisms, it would be evidence of developmental sophistication beyond that of cnidarians. The rangeomorphs exhibit a fractal rather than a segmental growth pattern, and in the suite of forms that appear most ‘segmented’, there is rarely evidence of internal repetition of structures. Clear evidence of internal structures occurs in only a few forms. With the exception of the apparent anterior proboscis of Kimberella (Fedonkin et al. 2007b), it is not obvious that any Ediacaran organisms possess developmental attributes that require their placement within the higher bilaterians. The presence of Kimberella in rocks dated to 555 Ma, suggesting that the P/D split must have occurred by this time. This does not necessitate that other Ediacaran organisms may have been complex bilaterians, although it is certainly possible. However, even the association of Kimberella with bilaterians could be spurious if cnidarians actually possessed the developmental tools for triploblasty, as discussed earlier.
Paleontologists who assigned many of these taxa to higher bilaterian clades (annelids, arthropods, etc) often did so because of apparent similarities in overall form, and from the presumption that cnidarians lacked the developmental processes to produce such morphologies. Our new understanding of the developmental tool kit of sponges, placozoans and cnidarians now raises the possibility that elements of the Ediacaran fauna may belong to these clades, or represent now-extinct clades positioned between the sponges and the P/D split.
5. Discussion
We have witnessed the boom and bust of several plausible metrics relating organismic complexity to various features of the genome, including genome size and gene content (Carroll et al. 2001; Copley 2008). Comparative developmental studies and whole-genome sequencing of more basal metazoans, as well as a choanoflagellate, have now demonstrated the difficulty in unambiguously identifying the role of even highly conserved genes. As Hejnol & Martindale (2008a) note, the critical question is how and when the various genes were assembled into the various developmental GRNs responsible for cell-type specification and regional patterning. Comparative developmental studies and whole-genome sequencing of early metazoans have demonstrated that cell-type specification, axial differentiation and regional patterning occur within sponges and cnidarians. Even when the phylogenetic patterning of these networks is known, however, problems of establishing homology between the developmental networks and the resulting morphology are likely to remain.
The very ancient divergence between the two cnidarian classes Anthozoa (including Nematostella) and Hydrozoa (including Hydra) revealed by molecular data raises the possibility that current cnidarian diversity may represent a depauperate sample of the original phylogenetic and morphologic diversity of the clade. Although some Ediacaran organisms evidently had more sophisticated morphogenesis than extant cnidarians, it appears that the clade possessed the tools to build such morphologies. A/P and D/V differentiation, anterior patterning, muscular contraction and response to sensory stimulation are all within the scope of cnidarian-grade organisms. Placozoans are the basal-most eumetazoans, yet possesses a number of key elements of the bilaterian toolkit, further emphasizing the developmental potential of the earliest nodes on the metazoan tree. Although Dickinsonia and some other Ediacaran organisms could have been members of the protostome or deuterostome clades, their developmental complexity, and our growing knowledge of the developmental complexity of basal metazoans, now make it possible, perhaps even likely, that most Ediacarans belonged to clades below the P/D split.
Acknowledgements
D.H.E. acknowledges support from the NASA National Astrobiology Institute, the National Museum of Natural History and the Santa Fe Institute and appreciates comments on an earlier draft from Kevin Peterson and an anonymous reviewer.
Footnotes
One contribution of 11 to a Theme Issue ‘The network of life: genome beginnings and evolution’.
References
- Adamska M., Degnan S. M., Green K. M., Adamski M., Craigie A., Larroux C., Degnan B. M.2007Wnt and TGF-beta expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE 2, e1031 (doi:10.1371/journal.pone.0001031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D., Nubler-Jung K.1994Inversion of dorso-ventral axis. Nature 371, 26 (doi:10.1038/371026a0) [DOI] [PubMed] [Google Scholar]
- Arendt D., Nubler-Jung K.1999Comparison of early nerve cord development in insects and vertebrates. Development 126, 2309–2325 [DOI] [PubMed] [Google Scholar]
- Arendt D., Technau U., Wittbrodt J.2001Evolution of the bilateria larval foregut. Nature 409, 81–85 (doi:10.1038/35051075) [DOI] [PubMed] [Google Scholar]
- Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A., Wittbrodt J.2004Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 (doi:10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- Arendt D., Denes A. S., Jekely G., Tessmar-Raible K.2008The evolution of nervous system centralization. Phil. Trans. R. Soc. B 363, 1523–1528 (doi:10.1098/rstb.2007.2242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguana J., Riutort M.2004The dawn of bilaterian animals: the case of acoelomorph flatworms. BioEssays 26, 1046–1057 [DOI] [PubMed] [Google Scholar]
- Baguna J., Martinez P., Paps J., Riutort M.2008Back in time: a new systematic proposal for the bilateria. Phil. Trans. R. Soc. B 363, 1481–1491 (doi:10.1098/rstb.2007.2238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balavoine G., Adoutte A.2003The segmented Urbilateria: a testable scenario. Int. Comp. Biol. 43, 137–147 (doi:10.1093/icb/43.1.137) [DOI] [PubMed] [Google Scholar]
- Balavoine G., de Rosa R., Adoutte A.2002Hox clusters and bilaterian phylogeny. Mol. Phyl. Evol. 24, 366–373 (doi:10.1016/S1055-7903(02)00237-3) [DOI] [PubMed] [Google Scholar]
- Ball E. E., Hayward D. C., Saint R., Miller D. J.2004A simple plan—cnidarians and the origins of developmental mechanisms. Nat. Rev. Genet. 5, 567–577 (doi:10.1038/nrg1402) [DOI] [PubMed] [Google Scholar]
- Ball E. E., De Jong D. M., Schierwater B., Shinzato C., Hayward D. C., Miller D. J.2007Implications of cnidarian gene expression patterns for the origins of bilateralty—is the glass half full or half empty? Int. Comp. Biol. 47, 701–711 (doi:10.1093/icb/icm028) [DOI] [PubMed] [Google Scholar]
- Bodmer R., Venkatesh T. V.1998Heart development in Drosophila and vertebrates: conservation of molecular mechanisms. Dev. Genet. 22, 181–186 (doi:10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2) [DOI] [PubMed] [Google Scholar]
- Boero F., Gravili C., Pagliara P., Piraino S., Bouillon J., Schmid V.1998The cnidarian premises of metazoan evolution: from triploblasty, to coelom formation, to metamery. Ital. J. Zool. 65, 5–9 (doi:10.1080/11250009809386722) [Google Scholar]
- Boero F., Schierwater B., Piraino S.2007Cnidarian milestones in metazoan evolution. Int. Comp. Biol. 47, 693–700 (doi:10.1093/icb/icm041) [DOI] [PubMed] [Google Scholar]
- Butts T., Holland P. W. H., Ferrier D. E. K.2008The urbilaterian super-hox cluster. Trends Genet. 24, 259–262 (doi:10.1016/j.tig.2007.09.006) [DOI] [PubMed] [Google Scholar]
- Carroll S., Grenier J., Weatherbee S.2001From DNA to diversity Malden, MA: Blackwell Scientific [Google Scholar]
- Copley R. R.2008The animal in the genome: comparative genomics and evolution. Phil. Trans. R. Soc. B 363, 1453–1461 (doi:10.1098/rstb.2007.2235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis E. M.2008Evo-devo: variations on ancestral themes. Cell 132, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis E. M., Sasai Y.1996A common plan for dorsoventral patterning in bilateria. Nature 380, 37–40 (doi:10.1038/380037a0) [DOI] [PubMed] [Google Scholar]
- de Rosa R., Grenier J. K., Andreeva T., Cook C. E., Adoutte A., Akam M., Carroll S. B., Balavoine G.1999Hox genes in brachiopods and priapulids and protostome evolution. Nature 399, 772–776 (doi:10.1038/21631) [DOI] [PubMed] [Google Scholar]
- Denes A. S., Jekely G., Steinmetz P. R., Raible F., Snyman H., Prud’homme B., Ferrier D. E., Balavoine G., Arendt D.2007Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129, 277–288 (doi:10.1016/j.cell.2007.02.040) [DOI] [PubMed] [Google Scholar]
- Erwin D. H.2006The developmental origins of animal body plans. In Neoproerozoic geobiology and paleobiology (eds Xiao S. H., Kaufman A. J.), pp. 157–197 Dordrecht, The Netherlands: Kluwer Press [Google Scholar]
- Erwin D. H., Davidson E. H.2002The last common bilaterian ancestor. Development 129, 3021–3032 [DOI] [PubMed] [Google Scholar]
- Fahey B., Larroux C., Woodcroft B. J., Degnan B. M.2008Does the high gene density in the sponge NK homeobox gene cluster reflect limited regulatory capacity? Biol. Bull. 214, 205–217 [DOI] [PubMed] [Google Scholar]
- Fedonkin M. A., Gehling J. G., Grey K., Narbonne G. M., Vickers-Rich P.2007aThe rise of animals Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- Fedonkin M. A., Simonetta A., Ivantsov A. Y.2007bNew data on Kimberella, the Vendian mollusc-like organism (White Sea region, Russia): paleontological and evolutionary implications. In The rise and fall of the Ediacaran biota (eds Vickers-Rich P., Komarower P.), pp. 157–179 London, UK: Geological Society [Google Scholar]
- Fernald R. D.2000Evolution of eyes. Curr. Opin. Neurobiol. 10, 444–450 (doi:10.1016/S0959-4388(00)00114-8) [DOI] [PubMed] [Google Scholar]
- Finnerty J. R., Pang K., Burton P., Paulson D., Martindale M. Q.2004Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304, 1335–1337 (doi:10.1126/science.1091946) [DOI] [PubMed] [Google Scholar]
- Gehling J. G., Droser M. L., Jensen S. R., Runnegar B. N.2005Ediacara organisms: relating form to function. In Form and function: fossils and development (ed. Briggs D. E. G.), pp. 43–66 New Haven, CT: Peabody Museum of Natural History, Yale University [Google Scholar]
- Gehring W. J.2004Historical perspective on the development and evolution of eyes and photoreceptors. Int. J. Dev. Biol 48, 707–717 (doi:10.1387/ijdb.041900wg) [DOI] [PubMed] [Google Scholar]
- Ghysen A.2003The origin and evolution of the nervous system. Int. J. Dev. Biol. 47, 555–562 [PubMed] [Google Scholar]
- Gimson A., Srivastava M., Fahey B., Woodcroft B. J., Chiang H. R., King N., Begnan B. M., Rokhsar D. S., Bartel D. P.2008Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197 (doi:10.1038/nature07415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Gehring W. J.1995Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788–1792 (doi:10.1126/science.7892602) [DOI] [PubMed] [Google Scholar]
- Harvey R. P.1996NK-2 homeobox genes and heart development. Dev. Biol. 178, 203–216 (doi:10.1006/dbio.1996.0212) [DOI] [PubMed] [Google Scholar]
- Hejnol A., Martindale M. Q.2008aAcoel development supports a simple planula-like urbilaterian. Phil. Trans. R. Soc. B 363, 1493–1501 (doi:10.1098/rstb.2007.2239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A., Martindale M. Q.2008bAcoel development indicates the independent evolution of the bilaterian mouth and anus. Nature 546, 382–386 (doi:10.1038/nature07309) [DOI] [PubMed] [Google Scholar]
- Hirth F., Kammermeier L., Frei E., Walldorf U., Noll M., Reichert H.2003An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development 130, 2365–2373 (doi:10.1242/dev.00438) [DOI] [PubMed] [Google Scholar]
- Holland L. Z., Kene M., Williams N. A., Holland N. D.1997Sequence and embryonic expression of the amphioxis engrailed gene (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development 124, 1723–1732 [DOI] [PubMed] [Google Scholar]
- Hui J. H. L., Holland P. W. H., Ferrier D. E. K.2008Do cnidarians have a ParaHox cluster: analysis of synteny around a Nematostella homeobox gene cluster. Evol. Dev. 10, 725–730 [DOI] [PubMed] [Google Scholar]
- Hwang J. H., Ohyanagi H., Hayakawa S., Osato N., Nichimiya-Fujisawa C., Ikeo K., David C. N., Fujisawa T., Gohobori T.2007The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of Hydra. Proc. Natl Acad. Sci. USA 104, 14 735–14 740 (doi:10.1073/pnas.0703331104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N.2004The unicellular ancestry of animal development. Dev. Cell 7, 313–325 (doi:10.1016/j.devcel.2004.08.010) [DOI] [PubMed] [Google Scholar]
- King N., Hittinger C. T., Carroll S. B.2003Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301, 361–363 (doi:10.1126/science.1083853) [DOI] [PubMed] [Google Scholar]
- King N., et al. 2008The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788 (doi:10.1038/nature06617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A., et al. 2005Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160 (doi:10.1038/nature03158) [DOI] [PubMed] [Google Scholar]
- Larroux C., Fahey B., Liubicich D., Hinman V., Guathier M., Gongora M., Green K., Worheide G., Leys S. P., Degnan B. M.2006Developmental expression of transcription factor genes in a demosponge: insights into the origins of metazoan multicellularity. Evol. Dev. 8, 150–173 (doi:10.1111/j.1525-142X.2006.00086.x) [DOI] [PubMed] [Google Scholar]
- Larroux C., Luke G. N., Koopman P., Rokhsar D., Shimeld S. M., Degnan B. M.2008Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996 (doi:10.1093/molbev/msn047) [DOI] [PubMed] [Google Scholar]
- Lee P. N., Kumburegama S., Marlow H. Q., Martindale M. Q., Wikramanayake A. H.2007Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis is mediated by Dishevelled. Dev. Biol. 310, 169–187 (doi:10.1016/j.ydbio.2007.05.040) [DOI] [PubMed] [Google Scholar]
- Lichtneckert R., Reichert H.2005Insights into the urbilaterian brain: conserved genetic patterning mechanisms in insect and vertebrate brain development. Heredity 94, 1–13 [DOI] [PubMed] [Google Scholar]
- Lowe C. J.2008Molecular genetic insights into deuterostome evolution from the direct-developing hemichordate Saccoglossus kowalevskii. Phil. Trans. R. Soc. B 363, 1569–1578 (doi:10.1098/rstb.2007.2247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C. J., Wu M., Salic A., Evans L., Lander E., Strant-Thomann N., Gruber C. E., Gerhart J., Kirschner M.2003Anteroposterior patterning in hemichordates and the origin of the chordate nervous system. Cell 113, 853–865 (doi:10.1016/S0092-8674(03)00469-0) [DOI] [PubMed] [Google Scholar]
- Martindale M. Q., Pang K., Finnerty J. R.2004Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 131, 2463–2474 (doi:10.1242/dev.01119) [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Pang K., Marlow H., Dunn C. W., Thomsen G. H., Martindale M. Q.2006Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc. Natl Acad. Sci. USA 103, 11 195–11 200 (doi:10.1073/pnas.0601257103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D. Q., Pang K., Daly M., Martindale M. Q.2007Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol. Dev. 9, 25–38 [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Magie C. R., Pang K., Martindale M. Q., Thomsen G. H.2008The hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan hedgehog pathway evolution. Dev. Biol. 313, 501–518 (doi:10.1016/j.ydbio.2007.09.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. J., Ball E. E.2008Animal evolution: Trichoplax, trees and taxonomic turmoil. Curr. Biol. 18, R1003–R1005 (doi:10.1016/j.cub.2008.09.016) [DOI] [PubMed] [Google Scholar]
- Mizutani C. M., Bier E.2008EvoD/Vo: the origins of BMP signalling in the neurectoderm. Nat. Rev. Genet. 9, 663–677 (doi:10.1038/nrg2417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. E., Wiens M., Adell T., Gamulin V., Schroder H. C., Muller I. M.2004Bauplan of urmetazoa: basis for genetic complexity of metazoa. Int. Rev. Cytol. 235, 53–92 (doi:10.1016/S0074-7696(04)35002-3) [DOI] [PubMed] [Google Scholar]
- Narbonne G. M.2005The Ediacara biota: neoproterozoic origin of animals and their ecosystems. Annu. Rev. Earth Planet. Sci. 33, 421–442 (doi:10.1146/annurev.earth.33.092203.122519) [Google Scholar]
- Ohno S.1996The notion of the Cambrian pananimalia genome. Proc. Natl Acad. Sci. USA 93, 8475–8478 (doi:10.1073/pnas.93.16.8475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. N.2006Gene regulatory networks in the evolution and development of the heart. Science 312, 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G., Rubenstein J. L. R.2002Developmental functions of the Distal-less/Dlx homeobox genes. Development 129, 4371–4386 [DOI] [PubMed] [Google Scholar]
- Panganiban G. E. F., et al. 1997The origin and evolution of animal appendages. Proc. Natl Acad. Sci. USA 94, 5162–5166 (doi:10.1073/pnas.94.10.5162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. J., Sperling E. A.2007Poriferan ANTP genes: primitively simple or secondarily reduced? Evol. Dev. 9, 405–409 [DOI] [PubMed] [Google Scholar]
- Peterson K. J., Cotton J. A., Gehling J. G., Pisani D.2008The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Phil. Trans. R. Soc. B 363, 1435–1443 (doi:10.1098/rstb.2007.2233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., Brinkmann H., Martinez P., Riutort M., Baguna J.2007Acoel flatworms are not platyhelminthes: evidence from phylogenomics. PLoS ONE 2, e717 (doi:10.1371/journal.pone.0000717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J., Kozmik Z.2004Cubozoan jellyfish: an Evo/Devo model for eyes and other sensory systems. Int. J. Dev. Biol. 48, 719–729 (doi:10.1387/ijdb.041851jp) [DOI] [PubMed] [Google Scholar]
- Pueyo J. I., Couso J. P.2005Parallels between the proximal-distal development of vertebrate and arthropod appendages: homology without an ancestor? Curr. Opin. Genet. Dev. 15, 439–446 (doi:10.1016/j.gde.2005.06.007) [DOI] [PubMed] [Google Scholar]
- Pueyo J. I., Lanfear R., Couso J. P.2008Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl Acad. Sci. USA 105, 16 614–16 619 (doi:10.1073/pnas.0804093105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H., et al. 2007Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (doi:10.1126/science.1139158) [DOI] [PubMed] [Google Scholar]
- Quiring R., Walldorf U., Kloter U., Gehring W. J.1994Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science 265, 785–789 (doi:10.1126/science.7914031) [DOI] [PubMed] [Google Scholar]
- Raff R. A.2008Origins of the other metazoan body plans: the evolution of larval forms. Phil. Trans. R. Soc. B 363, 1473–1479 (doi:10.1098/rstb.2007.2237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H., Simeone A.2001Developmental genetic evidence for a monophyletic origin of the bilaterian brain. Phil. Trans. R. Soc. Lond. B 356, 1533–1544 (doi:10.1098/rstb.2001.0972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I., Riutort M., Littlewood D. T. J., Herniou E. A., Baguna J.1999Acoel flatworms: earliest extant bilaterian metazoans, not members of Platyhelminthes. Science 283, 1919–1923 (doi:10.1126/science.283.5409.1919) [DOI] [PubMed] [Google Scholar]
- Ryan J. F., Burton P. M., Mazza M. E., Kwong G. K., Millikin J. C., Finnerty J. R.2007aThe cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evdence from the starlet sea anemone, Nematostella vetensis. Genome Biol. 7, R64 (doi:10.1186/gb-2006-7-7-r64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F., Mazza M. E., Pang K., Matus D. Q., Baxevanix A. D., Martindale M. Q., Finnerty J. R.2007bPre-Bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE 2, e153 (doi:10.1371/journal.pone.0000153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K., Schmid V.2005Evolution of striated muscle: jellyfish and the origin of triploblasty. Dev. Biol. 282, 14–26 (doi:10.1016/j.ydbio.2005.03.032) [DOI] [PubMed] [Google Scholar]
- Seipel K., Schmid V.2006Mesodermal anatomies in cnidarian polyps and medusae. Int. J. Dev. Biol. 50, 589–599 (doi:10.1387/ijdb.062150ks) [DOI] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B. M., Vervoort M.2007Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7, 33 (doi:10.1186/1471-2148-7-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Cole C. N., McPeek M. A., Peterson K. J.2006The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. (Mol. Dev. Evol.) 306B, 575–588 (doi:10.1002/jez.b.21118) [DOI] [PubMed] [Google Scholar]
- Slack J. M. W., Holland P. W. H., Graham C. F.1993The zootype and the phylotypic stage. Nature 361, 490–492 (doi:10.1038/361490a0) [DOI] [PubMed] [Google Scholar]
- Sperling E. A., Pisani D., Peterson K. J.2007Poriferan paraphyly and its implications for Precambrian palaeobiology. In The rise and fall of the Ediacaran biota (eds Vickers-Rich P., Komarower P.), vol. Special Publication 286, pp. 355–368.London, UK: Geological Society [Google Scholar]
- Srivastava M., et al. 2008The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 (doi:10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- Stollenwerk A., Schoppmeier M., Damen W. G. M.2003Involvement of Notch and Delta genes in spider segmentation. Nature 423, 863–865 (doi:10.1038/nature01682) [DOI] [PubMed] [Google Scholar]
- Tautz D.2004Segmentation. Dev. Cell 7, 301–312 (doi:10.1016/j.devcel.2004.08.008) [DOI] [PubMed] [Google Scholar]
- Technau U., et al. 2005Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633–639 (doi:10.1016/j.tig.2005.09.007) [DOI] [PubMed] [Google Scholar]
- Valentine J. W., Collins A. G., Meyer C. P.1994Morphological complexity increase in metazoans. Paleobiology 20, 131–142 [Google Scholar]
- Valentine J. W., Jablonski D., Erwin D. H.1999Fossils, molecules and embryos: new perspectives on the Cambrian explosion. Development 126, 851–859 [DOI] [PubMed] [Google Scholar]
- Wheeler B. M., Heimberg A. M., Moy V. N., Sperling E. A., Holstein T. W., Heber S., Peterson K. J.2009The deep evolution of metazoan microRNAs. Evol. Develop 11, 50–68 (doi:10.1111/j.1525-142X.2008.00302.x) [DOI] [PubMed] [Google Scholar]
- Xiao S. H., Laflemme M.2009On the eve of animal radiation: phylogeny, ecology and evolution of the Ediacara biota. Trends Ecol. Evol. 24, 31–40 (doi:10.1016/j.tree.2008.07.015) [DOI] [PubMed] [Google Scholar]