Abstract
Most current thinking about evolution is couched in the concept of trees. The notion of a tree with recursively bifurcating branches representing recurrent divergence events is a plausible metaphor to describe the evolution of multicellular organisms like vertebrates or land plants. But if we try to force the tree metaphor onto the whole of the evolutionary process, things go badly awry, because the more closely we inspect microbial genomes through the looking glass of gene and genome sequence comparisons, the smaller the amount of the data that fits the concept of a bifurcating tree becomes. That is mainly because among microbes, endosymbiosis and lateral gene transfer are important, two mechanisms of natural variation that differ from the kind of natural variation that Darwin had in mind. For such reasons, when it comes to discussing the relationships among all living things, that is, including the microbes and all of their genes rather than just one or a select few, many biologists are now beginning to talk about networks rather than trees in the context of evolutionary relationships among microbial chromosomes. But talk is not enough. If we were to actually construct networks instead of trees to describe the evolutionary process, what would they look like? Here we consider endosymbiosis and an example of a network of genomes involving 181 sequenced prokaryotes and how that squares off with some ideas about early cell evolution.
Keywords: phylogeny, networks, genomics
1. Introduction
Some evolutionary relationships are well described by a series of recursive bifurcations—a tree. The phylogeny of birds, fish or mammals are examples. As the lineages split, so do the gene histories, leading to the expectation that different genes for such groups should tend to give roughly the same phylogeny, provided that molecular phylogeny generally works (Landan & Graur 2008), and provided that recurrent genome duplications, as are common among eukaryotes (Scannell et al. 2006), have not led to rampant (hidden) paralogy. Xenology is quite rare in eukaryotes (e.g. Rumpho et al. 2008) but, among the microbes, evolutionary relationships can entail cell mergers (endosymbiosis), donation and acquisition of genes, such that within a single organism or genome different genes can have fundamentally different histories.
For example, when we retrace the evolutionary process of lineage splittings down into the origin of different algal groups possessing plastids surrounded by three or four membranes, we are confronted with the process of secondary endosymbiosis, where cellular individuals of highly disparate eukaryotic lineages have merged on at least three different occasions to bring forth novel algal lineages at very high taxonomic levels (Stoebe & Maier 2002; Lane & Archibald 2008). This is sketched in figure 1, where the chromists and alveolates are drawn as separate groups as recent findings suggest (Frommolt et al. 2008; Sanchez-Puerta & Delwiche 2008). Going back further, the origin of the plant lineage is attributable to the symbiosis of a cyanobacterium with a eukaryotic host, another cellular merger in the phylogeny of life (Gould et al. 2008). Going back further still, the origin of mitochondria at the origin of known eukaryotes is yet another decisive cellular merger (Dyall et al. 2004; van der Giezen et al. 2005; Embley & Martin 2006). At each such symbiotic merger, genes are transferred from symbionts to the chromosomes of their host, a process called endosymbiotic gene transfer (Martin et al. 1993, 1998, 2002; Timmis et al. 2004). Further back still, among free-living prokaryotes, from which the ancestors of plastids and mitochondria stem, lateral gene transfer has resulted in distributions of genes across prokaryotic chromosomes that do not strictly correspond to a hierarchical classification or any single bifurcating tree (Doolittle 1999; Doolittle & Bapteste 2007; McInerney et al. 2008).
Figure 1.
A schematic depiction of cell evolution connecting prokaryotes and eukaryotes and eukaryotes with complex plastids. Diversification of groups is symbolized by triangles, but branching patterns for groups is not. Thin lines subtending each triangle indicate that only two cellular partners (one cell per thin line each) participate in any given endosymbiosis. Origins symbolizes the origins of prokaryotes from the elements on early Earth, eubacteria and archaebacteria are prokaryotes, the rest of the lineages are eukaryotes. See text for more details.
Ho hum, one might say, we knew that, so what's new? Maybe the more important question is not what's new, but what isn't new? What isn't new is that despite knowing that many processes in microbial evolution are not tree-like in nature, biologists still tend to use the metaphor of trees to conceive, discuss and represent the process of the overall relatedness of things (Ciccarelli et al. 2006). That is not terribly surprising because most, but not all, approaches to describing the evolutionary process involve phylogeny at the computer in some form, and phylogenetic work at the computer generally produces trees, because trees are the most simple way to model protein divergence (a node), evolution (a branch) and homology (a clade) but they contain no information as for the type of homology (e.g. orthology, xenology, etc.). There are some exceptions, the directed cyclic graphs used by Rivera & Lake (2004) to generate a ring instead of a tree being one, and there are others (Beiko et al. 2005; Kunin et al. 2005; Dagan et al. 2008; Lima-Mendez et al. 2008). But by and large, biologists tend to have a tree in mind when they approach the issue of evolutionary relatedness, also among microbes, and they (we) tend to use programs that generate trees, and therefore they (we) tend to see trees as the result of investigation on the topic. Many graphical representations of microbial evolution that incorporate LGT and/or that depict endosymbiosis have been published (Doolittle 1999; Martin 1999; Brown 2003; Huang & Gogarten 2006; McInerney et al. 2008) but, like figure 1, which schematically depicts how endosymbiosis runs contrary to the notion of a strictly bifurcating tree, they tend to involve something like an artist's impression of the evolutionary process that takes into account many different kinds of observations. It would probably help matters, that is, it would probably help us as evolutionary biologists studying microbial evolution, to convey in a more objective and scientific manner to ourselves and to non-specialists alike, if we could produce as a computer-generated printed product of our current understanding of microbial evolution. That would require computer-based methods that would allow us to depict non tree-like processes for the simple purpose of having a better level of congruence between what we think is actually going on in microbial evolution in nature (based on observations) and how we model it. If microbial evolution is not tree-like in salient aspects, then we need tools to investigate it that do not force the data into the straightjacket of a tree.
2. Networks
Phylogenetic relationship can be modelled as graphs, in which the species (or genes) are represented by vertices and their evolutionary relationship are represented by edges. By graph theory definitions, a phylogenetic tree is a connected, acyclic, directed (and sometime also rooted) graph (Harary 1969). But there are some alternatives to trees. Networks are one such kind of alternative. Using the same mathematical model, if we allow the graph to be cyclic, then we get a phylogenetic network (Huson & Bryant 2006). Hence, we could construct an evolutionary graph of shared genes among prokaryotic genomes in which the nodes (or vertices) of the graph represent sequenced genomes with the edges between nodes representing shared genes. If all of the gene inheritance were vertical, then we should obtain a tree. If there are lateral components of inheritance, then the network should recover and depict them, too.
A problem arises though, in that it is not as simple as it might seem at first sight to discern between vertical inheritance and lateral transfer. Any gene tree that is assumed to be an accurately inferred gene tree but is also discordant with the a priori expected relationships for the taxon labels (the species containing the respective gene), disregarding for the moment the issue of whence those a priori expectations stem, can readily be explained by assuming some number of ancient gene duplications and differential loss. But each time we assume a duplication and a loss to explain discordant branches, we are assuming the presence of an additional gene in the genome ancestral to the species under study. That is fine for one or two genes, or maybe a dozen or maybe a hundred. If, however, we have to add that kind of corollary assumption to every prokaryotic gene and its tree, then the size of the ancestral genome that results from those corollary assumptions begins to burgeon and quickly reaches an untenable size, that is, it becomes the genome of Eden, as Doolittle et al. (2003) put it. That logical constraint turns out to be a very useful tool, it turns out, in our efforts to understand gene transfer and chromosome evolution, as we briefly explain in the following.
We recently undertook an endeavour to describe prokaryote genome evolution in terms of networks (Dagan et al. 2008). In essence, we assorted 539 723 protein coding genes among 181 sequenced prokaryote genomes into 54 349 families using the standard MCL algorithm. Many of those families have a very patchy distribution, that is, members of many families are found in a few genomes from different taxonomic groups. If we make the extreme and testable assumption that there has been no LGT in the evolution of those genes among the 160 eubacteria and 21 archaebacteria sampled, then the distributions of those genes shared across more than one genome would be governed by lineage specific gene origin and gene loss only. That assumption can be tested by comparing the distribution of inferred ancestral genome sizes under such assumptions with the modern distribution of contemporary genome sizes, measured in gene families to see if they are significantly different, which they are (Dagan & Martin 2007a). A premise underlying that test is that there is no a priori reason to expect that prokaryotic genome sizes in the past were fundamentally different from those observed today. If we assume that there is no LGT then we are also assuming that all gene trees are compatible, and each gene is present in the genome ancestral to its first appearance in the evolution of the genomes we are considering. Thus gene distributions alone demand a certain amount of LGT among prokaryotic genomes, at least approximately 1 LGT per gene family per gene family lifespan, because too much vertical inheritance leads us into the genome of Eden problem (Dagan & Martin 2007a). Allowing LGT reduces the inferred size of ancestral genomes, but allowing too much LGT reduces their size to distributions that are once again significantly different from modern genome sizes, but too small (the genome of Lilliput) rather than too large.
The constraint of ancestral genome size opens two inroads to studying genome evolution. First, it permits estimates for how much LGT has gone on in prokaryote evolution (Dagan & Martin 2007a). Those estimates are attained without comparing gene trees and furthermore by assuming all gene trees to be compatible, hence they constitute minimum lower bound estimates. Second, it permits us to address genome evolution in terms of evolutionary networks consisting of vertically and laterally inherited genes. How? Given an assumed (or inferred) phylogeny for any given component of the genomes in question, then each of the ancestral nodes in that phylogeny corresponds to a genome-sized collection of genes. The constraint of genome size provides a criterion to decide whether a gene is present at a given ancestral node, that is, present in an inferred ancestral genome, or not. That is important because if we have a criterion for deciding which genes are present at which nodes, then shared genes across nodes correspond to edges in a network, and we can construct an evolutionary network that captures both vertical and horizontal components of gene inheritance, as illustrated in figure 2.
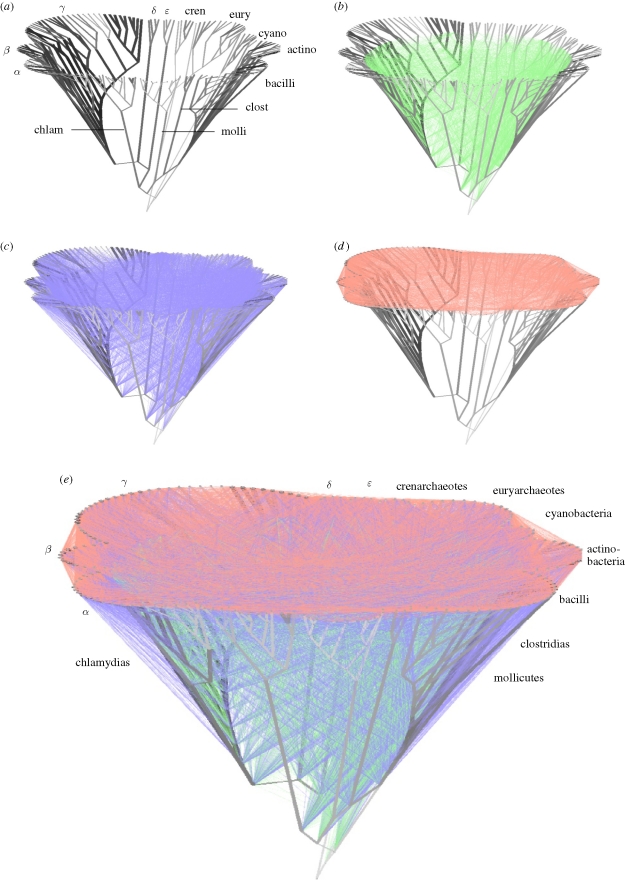
Figure 2.
A network representation of vertical inheritance and lateral exchange among prokaryotes. (a–d) The individual components from which (e), modified from Dagan et al. (2008), was constructed. Prokaryotic groups sampled are indicated, greek letters designate proteobacterial subdivisions. Note that in (c), where lateral edges connecting internal nodes to external nodes are shown, some genes seem to be transferred from ‘ancient’ nodes to more modern nodes, whereas the lateral edge specifies the group of taxa from within which the donor or recipient is inferred. Similar applies to (b) for edges connecting internal nodes. See text for more details.
Figure 2a shows an assumed phylogeny for 181 genomes and corresponds to the topology and species designations shown in fig. 3a and the supplementary material of Dagan et al. (2008). The tree that we use as a vertical backbone was not just assumed from thin air, rather it was constructed from analyses of the rRNA operon assuming monophyly for the prokaryotic taxa shown, but its specific branching order might as well just have been assumed, for two reasons. First, there is currently little evidence to suggest that any genes in prokaryote genomes have strictly co-evolved with the rRNA operon over the whole of evolutionary time (Bapteste et al. 2008), hence even if we had the right rRNA tree, there remains the more pressing question of ‘for what would it be a proxy?’ (Doolittle & Bapteste 2007). Second, for 181 genomes there are 3.6 × 10379 possible trees, and the chances of getting the right tree are comfortingly negligible (by comparison there are about 1080 protons in the universe, very close to the number of trees for 60 genomes). Nonetheless, we can work with that assumed tree and specify as its root the branch between eubacteria and archaebacteria, because that is where genome similarity as measured in shared proportions of shared genes would place the root (Dagan & Martin 2007a), notwithstanding other suggestions as to where the root might be (see the contribution by Lake et al. 2009). Then, given the genome of Eden constraint, we can draw an edge between all nodes that are connected by a shared gene, that is, nodes that are connected by the presence of a member of one of our 54 349 protein families, which would give us a network of genomes.
Before drawing such a network, there is a matter to consider concerning the congruence between the edges to be drawn in the figure (shared genes) and the process they are intended to represent (LGT). If we infer that there was only one LGT in the history of a given gene family, then there is only one lateral edge connecting the nodes bearing that gene. In that case, there is a 1 : 1 correspondence between the number of lateral edges and the number of LGTs. But if three nodes need to be connected by lateral edges, then there are three edges that connect them but there are only two LGT events at the minimum need to be assumed, which applies to 27 per cent of the genes in the present example. Similarly, if four nodes need to be connected by lateral edges, then there are six edges that can connect them, but only three LGT events are needed to explain the gene distribution. Kunin et al. (2005) dealt with this problem by assigning weights to lateral edges corresponding to their probabilities of 2/3, 3/6, etc. We dealt with it by taking 1000 replicate samples from the matrix representation of the lateral network in which superfluous edges are randomly deleted, such that the number of lateral edges and the number of LGT events exactly correspond (Dagan et al. 2008). Using that procedure, we can plot the lateral edges onto figure 2a, which represents vertical inheritance among genomes, and furthermore depict lateral transfer among genomes as well, which was one aim of our undertaking to use networks for describing genome evolution.
Among the 1000 replicates, there are 2330 ± 16 lateral edges that connect internal nodes to internal nodes (figure 2b), 5886 ± 20 lateral edges that connect internal nodes to external nodes (figure 2c) and 4046 ± 16 lateral edges that connect external nodes to external nodes (figure 2d). Each of these edges corresponds to a lateral gene transfer, and if we plot all edges in one figure, the result is that shown in figure 2e. We designate the network as a minimal lateral network because the procedures that were used to determine gene presence or absence at nodes entail two simplifying assumptions that severely underestimate the amount of LGT that has actually gone on among genomes: (i) we assume that all genes are orthologous, that is, that all multiple occurrences of a gene family in a genome are assumed to be the result of recent gene duplications within that genome, and (ii) we assume that all gene trees for all families are compatible. Those are rather severe assumptions, but they do deliver estimates for the minimum LGT rate and the minimum number of LGT events to be plotted in the network.
3. Including eukaryotes in the network
Ideally, one would like to see eukaryotes and prokaryotes in the same network of shared genes and it can be expected that such graphs will eventually emerge. But current gene sharing networks encompass only prokaryotes (Kunin et al. 2005; Dagan et al. 2008) or phage (Lima-Mendez et al. 2008). They depict genome evolution among prokaryotes as a process of donor–recipient relationships that has been more or less continuous over evolutionary time, with genes acquired by conjugation (plasmids), transduction (phages), transformation (natural competence) (Thomas & Nielsen 2005) or gene transfer agents (Lang & Beatty 2007) but also being transmitted vertically by the process of chromosome replication as well, in agreement with some current views the process of microbial genome evolution (Doolittle & Bapteste 2007). While these four mechanisms of gene spread among prokaryotes just mentioned are very well characterized at the molecular and genetic level, similar genetically and molecular defined mechanisms have not been characterized among eukaryotes. Thus it would seem that there is a big difference between prokaryotes and eukaryotes concerning the prevalence, mechanisms and biological significance of lateral gene transfer. Indeed, it is not unusual to find that three strains of the same prokaryotic species such as E. coli might share less than 40 per cent of their genes in common (Welch et al. 2002), while sequencing a representative for a eukaryotic lineage, such as Entamoeba, might reveal only 1–2% of the genome consisting of genes that might have been specifically acquired in that lineage (Loftus et al. 2005). Clearly, the frequency and impact of LGT in prokaryote and eukaryote genomes is different.
But at the same time, a particular kind of gene transfer among eukaryotes, namely gene transfer from organelles, or endosymbiotic gene transfer (Martin et al. 1993), represents a very important source of genetic novelty among the eukaryotes (Timmis et al. 2004; Lane and Archibald 2008). Gene transfer from organelles sets eukaryotes apart from prokaryotes, which in contrast to eukaryotes lack organelles descended from free-living prokaryotes. That is not to say that no prokaryotes harbour prokaryotic endosymbionts, for there are two such examples known (Wujek 1979; van Dohlen et al. 2001), but there are no prokaryotes known to harbour double-membrane bounded organelles, raising the question of what is an endosymbiont and what is an organelle (Cavalier-Smith & Lee 1985). A practical distinction between the two is whether the endosymbiont has evolved a protein import apparatus, as in the case of chloroplasts (Kanalon & McFadden 2008), mitochondria (Dolezal et al. 2006) and secondary plastids (Hempel et al. 2007), in which case it would qualify as an organelle, or not, in which case it is best called an endosymbiont (Theissen & Martin 2006).
Endosymbionts living in the cytosol are very common among eukaryotes today and probably have been throughout evolution (Dagan & Martin 2007b), but endosymbiotic associations that give rise to organelles are not common at all. Available evidence indicates that there was only one origin of plastids from cyanobacteria (Gould et al. 2008), and only one origin of mitochondria from proteobacteria (see contribution by Embley in this volume), as sketched in figure 1. Once every 4 Gyr is rare. Both symbioses entailed the origin of a specific protein-import machinery. Both entailed the origin of a novel taxon at the highest levels (known plants and known eukaryotes). Both entailed a symbiosis of one cell within another, each possessing a genome's worth of genes. If an endosymbiont lyses, its chromosome is free to recombine with that of its host, if the host lyses, the symbiosis is over, hence the transfer of genes is generally unidirectional from endosymbiont to host, which can be seen as a ratchet mechanism (Doolittle 1998). We can see the workings of endosymbiotic gene transfer in eukaryote genomes today. We can see that bulk recombination is involved, as the 367 kb insertion of the complete mitochondrial genome in Arabidopsis and the 121 kb insertion of the complete chloroplast genome attest (Huang et al. 2005). The mechanism of insertional recombination involves non-homologous end joining (Hazcani-Covo & Covo 2008). Gene transfer from transformed mitochondria and from transformed plastids can be demonstrated in the lab (Thorsness & Fox 1990; Huang et al. 2003) and there is increasing interest in the role of stress factors, such as oxidative stress, that might promote the rate incorporation of organelle sequences in nuclear genomes over recent evolutionary time (Cullis et al. 2008). Given the ease and frequency with which genes are transferred from organelles to the nucleus, the question arises as to why there are any genes left in organelles at all, and despite many different proposals to account for this observation, only one really fills the bill, namely that of Allen (1993, 2003), who suggested that organelles have retained genomes in order to allow redox-dependent regulation of gene expression within individual organelles that possess bioenergetic membranes. This proposal is strongly supported by recent characterization of proteins involved in redox-regulated plastid gene regulation (Puthiyaveetil et al. 2008) and would furthermore directly account for the lack of DNA in hydrogenosomes, anaerobic forms of mitochondria that generate energy via substrate level phosphorylation, and hence lack membrane-associated electron transport (Müller 2007).
There is also evidence for the workings of endosymbiotic gene transfer early in evolution as well. In plants, estimates for the fraction of genes acquired from the ancestor of plastids range from approximately 15 to 20 per cent of nuclear protein coding genes, with systematic underestimations owing to the difficulties of phylogenetic inference with poorly conserved sequences figuring prominently in the issue (Deusch et al. 2008). In eukaryotes that never possessed plastids, such as yeast, the majority of genes having homologues among prokaryotes are more similar to eubacterial homologues than they are to archaebacterial homologues and the former are generally involved in metabolic functions (operational genes) while the latter are generally involved in information storage and expression (informational genes) (Rivera et al. 1998; Esser et al. 2004; Rivera & Lake 2004).
The generally surprising observation that eukaryotes possess a majority of eubacterial genes (Martin et al. 2007) is distinctly at odds with the view that eukaryotes are sisters of archaebacteria, but it is readily accounted for under endosymbiotic models for the origin of eukaryotes (Pisani et al. 2007), if we allow for the very real possibility that there was a substantial quantity of endosymbiotic gene transfer subsequent to the origin of mitochondria. That brings us to the question of which genes, exactly, the ancestor of mitochondria, or the ancestor of plastids for that matter, possessed? We can phrase that question another way, and in the specific context of this paper, namely, what is the relationship of figure 1 to figure 2? Both figures purport to represent something that most people who will ever read this paper generally accept, namely that plastids and mitochondria really are descended from free living endosymbionts (figure 1) and that prokaryotes really do redistribute their genes across chromosomes over time (figure 2). If we add to that the recognition that many genes in eukaryote genomes really do stem from those two endosymbionts via endosymbiotic gene transfer, then which genes did those endosymbionts harbour in their chromosomes at the time when they became endosymbionts?
If we take the evidence seriously that prokaryotes really do pass their genes around over time, as we should, then it would appear that the collection of genes possessed by the ancestor of mitochondria is probably best preserved in its most contiguous form among eukaryote genomes, rather than among prokaryote genomes. This issue has been around for about 10 years (Martin 1999; Esser et al. 2007) but for the most part it has been disregarded, with some exceptions (Gross et al. 2008). For example, Huang et al. (2005) recently reported that there are some genes the plants and chlamydias share more or less specifically, and they suggested that this constitutes evidence for the participation of an additional endosymbiont, a chlamydial one, at the origin of plastids. But if we let go of the notion that the chromosomes of prokaryotic ‘lineages’ are static collections of genes that have co-evolved in a linked manner within the same chromosome over billions of years (Doolittle 1999), as data from genomes suggests that we should (Doolittle & Bapteste 2007), then we can contrast two ways of looking at the chlamydia data as an example of many similar sorts of observations emerging from genomes: (i) is it more reasonable to assume that a gene or group of genes can be used as a proxy for the existence of an additional endosymbiont in the plant lineage? Put another way, does every gene, in the extreme, serve as a proxy for the expected patterns of sequence similarity for the rest of the genes present in a given chromosome at a given point in time? Or (ii) are prokaryotic chromosomes, including those related to the ancestors of organelles, really ‘fluid’ structures, with genes coming in and going out over time? In our view, the latter question is much closer to being a formulation to which we could respond with a straightforward ‘yes’ and feel comfortable saying so.
It will probably take some time before LGT among prokaryotes (figure 2) and the endosymbiotic origins of chloroplasts and mitochondria (figure 1) can be reconstructed at the computer in a unified framework that starts with genome sequences and ends up with a network that is both readily printable and readily interpretable. It will take longer still before the secondary endosymbioses can be included in such an endeavour, because the data coming from those genomes are painting an increasingly complex picture (Frommolt et al. 2008; Sanchez-Puerta & Delwiche 2008). Apropos complexity, as the tsunami of data from eukaryote genome projects rolls in, it is being churned through various alignment and phylogeny pipelines and many of the trees so produced are showing unusual branching patterns or unusual sequence similarities. This has led to a situation where many reports for LGT among eukaryotes are emerging, the most spectacular being the initial claim for several hundred laterally acquired bacterial genes in the human genome, which turned out not to be true (Salzberg et al. 2001; Stanhope et al. 2001). However, because eukaryotes, in contrast to prokaryotes (Thomas & Nielsen 2005; Lang & Beatty 2007), lack genetically and molecularly well-defined mechanisms of gene transfer across species boundaries, the search for mechanisms to explain the presence of odd branching or otherwise unexpected sequences has been expanded to include mere physical contact between organisms (Keeling & Palmer 2008) or even LGT via meteorites (Bergthorsson et al. 2003), to highlight one prominent example. Such suggestions leave us less than comfortable.
In addition to the lack of molecularly characterized mechanisms, another contrast of LGT among prokaryotes to reports of eukaryote-to-eukaryote LGTs is that the latter all too often entail oddly branching copies of highly similar genes (Keeling & Palmer 2008) but without any corresponding effects for organismic ecology, whereas LGT among prokaryotes can, and often does, transform the overall physiology of an organism (Kennedy et al. 2001; Boucher et al. 2003; Mongodin et al. 2005) with dramatic and obvious consequences for its ecology and evolution. In that vein, chloroplasts and mitochondria also transformed the physiology of their hosts through endosymbiosis and donated some fundamentally new genes to their hosts (for example, for photosynthesis and mitochondrial ATP synthesis), not just divergent copies of the same ones.
Thus, LGT among prokaryotes and gene transfer in the context of endosymbiosis can be correlated to changes in ecology and physiology, but most of the reports for ‘odd-branch’ LGT among eukaryotes cannot (Keeling & Palmer 2008). This is not to say that eukaryotes never acquire genes from other eukaryotes. But the ‘odd branch’ approach to LGT has some hefty caveats because there are lots of genes out there in the databases and there are thousands of alignments and trees that can be made from them. Some of those trees will have high support values for artefactual branches for reasons intrinsic to the computational process of phylogenetic reconstruction (Delsuc et al. 2003; Bapteste et al. 2008; Shavit et al. 2007), and even the random choice of whether we align amino acids in a protein sequence from N-terminus to C-terminus or in the reverse order can exert a dramatic influence on phylogenetic and phylogenomic results (Landan & Graur 2007; Deusch et al. 2008). Such issues still loom somewhat over investigations of LGT that are based in tree comparisons alone and where the inference of LGT can account for differences in observed branching patterns, but little else.
4. Warning: many people disagree with some views expressed here
We have presented two figures here to illustrate our current views on early evolution from the standpoint of endosymbiosis (figure 1) and LGT (figure 2). Figure 1 might be more controversial than figure 2 in various aspects and we feel obliged to point out that many scientists would staunchly disagree with aspects of the sketch presented in figure 1, hence a few words seem in order to justify why we drew it the way we did. In figure 1, we have sketched the origin of the host lineage for the origin of mitochondria as an archaebacterium outright, because it precludes the notion that nucleated but mitochondrion-lacking cells (archezoa) ever existed (Embley & Martin 2006) in agreement with some recent analyses based on supertrees (Pisani et al. 2007) and based on careful phylogenetic studies of informational genes (Cox et al. 2008). Some would staunchly disagree, maintaining that there are indeed eukaryotes around that never possessed mitochondria (Margulis et al. 2007), that the host that acquired the mitochondrion was a eubacterium (de Duve 2007), or that the common ancestry of mitochondria and hydrogenosomes is somehow tenuous (de Duve 2007; Margulis et al. 2007). We politely disagree, and will not argue their case here. We have indicated a later origin of eukaryotes than of prokaryotes, consistent with microfossil evidence suggesting their later emergence (Knoll et al. 2006; Rasmussen et al. 2008), and this runs contrary to views, with which we disagree, that eukaryotes represent a lineage that is as old as or older than prokaryotes (Kurland et al. 2007). We have drawn the root in figure 1 between archaebacteria and eubacteria, with which many scientists would also disagree, maintaining that archaebacteria arose via mutations from a bona fide eubacterium (Cavalier-Smith 2002) or that prokaryotes are derived from eukaryotes (Glansdorff et al. 2008) or that other placements of the root are preferable (see contribution by Lake et al. 2009). Again, we disagree and do not argue the opposing views.
Our placement of the root is consistent with geochemical evidence for the antiquity of both prokaryotic groups (Nisbet & Sleep 2001; Ueno et al. 2006) and with the observation that the two main groups of prokaryotes are deeply divergent, not only at the level of their cell wall and membrane constituents (Martin & Russell 2003), but also at the level of processes so basic as DNA maintenance (Koonin & Martin 2005). Also, we have drawn the base of figure 1 to suggest that the first prokaryotes might have arisen from something that looks like a hydrothermal vent, which need not be true, but there are enough similarities between energy-releasing geochemical reactions involving H2 and CO2 at some modern hydrothermal vents and energy releasing biological reactions involving H2 and CO2 among some modern microbes to pursue the idea further (Martin et al. 2008). Many scientists would disagree with the view that hydrothermal vents had anything to do with the origin of life (Orgel 2008).
Finally, there is the matter that we have not suggested any branching orders for either prokaryotic groups or eukaryotic groups in figure 1, other than implying that the organelle-generating symbioses among eukaryotes correspond to a relative temporal sequence. Among the prokaryotes, we have schematically indicated some kind of metabolic diversification (colours), but without suggesting what the order of appearance for different metabolic types might be. There is quite a lot of phylogenomic and phylogenetic work devoted to the relative branching orders of prokaryotic groups, and serious efforts have been undertaken to link that branching order to geochemical evidence and dates, for example in Battistuzzi et al. (2004) and Gribaldo & Brochier-Armanet (2006). Other efforts have focused inferring geological history from phylogenetic trees (Ciccarelli et al. 2006). But a general problem arises in such studies. In order to construct a tree for all groups, one has to have genes that are present in all groups, and this usually boils down to the ribosomal proteins or their superoperon (Hansmann and Martin 2000) or what has been called ‘the core’ (Charlebois & Doolittle 2004). The problem is that it is difficult to demonstrate that sequences differences or branching patterns in ‘the core’, should it evolve as a coherent unit in the first place (Bapteste et al. 2008), serve as a good predictor for which, what kind of, and how many genes we are likely to find in the remainder of the chromosome surrounding that core. For example, methanogens and archaeal halophiles have related and similar cores (Gribaldo & Brochier-Armanet 2006), but methanogens are strictly anaerobic chemolithoautotrophs while halophiles are (usually) aerobic heterotrophs with light-harnessing abilities (Kennedy et al. 2001; Boucher et al. 2003), while Salinibacter has a core similar to the eubacterial Bacteroides/Chlorobi group, but a physiology and gene collection reminiscent of archaeal halophiles (Mongodin et al. 2005). That example is certainly not new to anyone, but it perhaps illustrates the point that sequence similarities within the core are not a good proxy for what is likely to be found in the rest of the genome. Eukaryotes are another such example, the archaebacterial nature of their genetic apparatus does not predict the eubacterial nature of their energy metabolism, but some endosymbiotic models for the origin of mitochondria that entail gene transfers from symbiont to host do (Pisani et al. 2007).
5. Conclusion
The problems relating to the notion that the evolution of all living things can be represented by a tree have been well put by others (Doolittle 1999; Brown 2003; Doolittle & Bapteste 2007; McInerney et al. 2008), and we broadly agree with that view. The main non tree-like processes to deal with seem to be LGT among prokaryotes and gene transfer from organelles (endosymbiotic gene transfer) among eukaryotes. The onus of offering alternatives would appear to be upon those of us who are saying that the tree metaphor is inadequate. Networks are an alternative that can be used in the case of prokaryotes (Dagan et al. 2008). It is obvious that there exists some amount of vertical inheritance via chromosome replication and segregation as well as some amount of lateral inheritance via other means among prokaryotes; hence the network approach to genome evolution should depict both. If we approach the problem of describing the overall course of prokaryote genome evolution from the standpoint of shared genes among genomes rather than shared phylogeny of some core, as recent studies of phage evolution have (Lima-Mendez et al. 2008), then we are taking steps away from the familiar conceptual environment of trees and into the less well-charted territory of evolutionary processes that cannot be modelled by a tree, but might better fit the process of prokaryote genome evolution as it occurs in nature.
Acknowledgements
We thank the German Israeli Foundation (T.D.), the German Research Foundation, the European Research Council and the BMBF for financial support.
Footnotes
One contribution of 11 to a Theme Issue ‘The network of life: genome beginnings and evolution’.
References
- Allen J. F.1993Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631 (doi:10.1006/jtbi.1993.1210) [DOI] [PubMed] [Google Scholar]
- Allen J. F.2003The function of genomes in bioenergetic organelles. Phil. Trans. R. Soc. B 358, 19–37 (doi:10.1098/rstb.2002.1191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E., Susko E., Leigh J., Ruiz-Trillo I., Bucknam J., Doolittle W. F.2008Alternative methods for concatenation of core genes indicate a lack of resolution in deep nodes of the prokaryotic phylogeny. Mol. Biol. Evol. 25, 83–91 (doi:10.1093/molbev/msm229) [DOI] [PubMed] [Google Scholar]
- Battistuzzi F. U., Feijao A., Hedges S. B.2004A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4, 44 (doi:10.1186/1471-2148-4-44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko R. G., Harlow T. J., Ragan M. A.2005Highways of gene sharing in prokaryotes. Proc. Natl Acad. Sci. USA 102, 14 332–14 337 (doi:10.1073/pnas.0504068102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U., Adams K. L., Thomasson B., Palmer J. D.2003Widespread horizontal gene transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (doi:10.1038/nature01743) [DOI] [PubMed] [Google Scholar]
- Boucher Y., Douady C. J., Papke R. T., Walsh D. A., Boudreau M. E., Nesbo C. L., Case R. J., Doolittle W. F.2003Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37, 283–328 (doi:10.1146/annurev.genet.37.050503.084247) [DOI] [PubMed] [Google Scholar]
- Brown J. R.2003Ancient horizontal gene transfer. Nat. Rev. Genet. 4, 121–132 (doi:10.1038/nrg1000) [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.2002The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 52, 7–76 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T., Lee J. J.1985Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J. Protozool. 32, 376–379 [Google Scholar]
- Charlebois R. L., Doolittle W. F.2004Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14, 2469–2477 (doi:10.1101/gr.3024704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F. D., Doerks T., von Mering C., Creevey C. J., Snel B., Bork P.2006Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (doi:10.1126/science.1123061) [DOI] [PubMed] [Google Scholar]
- Cox C. J., Foster P. G., Hirt R. P., Harris S. R., Embley T. M.2008The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA 105, 20 356–20 361 (doi:10.1073/pnas.0810647105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis C. A., Vorster B. J., Van Der Vyver C., Kunert K. J.2008Transfer of genetic material between the chloroplast and nucleus: how is it related to stress in plants? Ann. Bot. 103, 625–633 (doi:10.1093/aob/mcn173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T., Martin W.2007aAncestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl Acad. Sci. USA 104, 870–875 (doi:10.1073/pnas.0606318104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T., Martin W.2007bTesting hypotheses without considering predictions. BioEssays 29, 500–503 (doi:10.1002/bies.20566) [DOI] [PubMed] [Google Scholar]
- Dagan T., Artz-Randrup Y., Martin W.2008Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl Acad. Sci. USA 105, 10 039–10 044 (doi:10.1073/pnas.0800679105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C.2007The origin of eukaryotes: a reappraisal. Nat. Rev. Genet. 8, 395–403 (doi:10.1038/nrg2071) [DOI] [PubMed] [Google Scholar]
- Delsuc F., Phillips M. J., Penny D.2003Hexapod origins: Monophyletic or paraphyletic? Science 301, 1482 (doi:10.1126/science.1086558) [DOI] [PubMed] [Google Scholar]
- Deusch O., Landan G., Roettger M., Gruenheit N., Kowallik K. V., Allen J. F., Martin W., Dagan T.2008Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 25, 748–761 (doi:10.1093/molbev/msn022) [DOI] [PubMed] [Google Scholar]
- Dolezal P., Likic V., Tachezy J., Lithgow T.2006Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 (doi:10.1126/science.1127895) [DOI] [PubMed] [Google Scholar]
- Doolittle W. F.1998You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trend. Genet. 14, 307–311 [DOI] [PubMed] [Google Scholar]
- Doolittle W. F.1999Phylogenetic classification and the universal tree. Science 284, 2124–2128 (doi:10.1126/science.284.5423.2124) [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Bapteste E.2007Pattern pluralism and the Tree of Life hypothesis. Proc. Natl Acad. Sci. USA 104, 2043–2049 (doi:10.1073/pnas.0610699104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., Boucher Y., Nesbo C. L., Douady C. J., Andersson J. O., Roger A. J.2003How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Phil. Trans. R. Soc. B 358, 39–58 (doi:10.1098/rstb.2002.1185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall S. D., Brown M. T., Johnson P. J.2004Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 (doi:10.1126/science.1094884) [DOI] [PubMed] [Google Scholar]
- Embley T. M., Martin W.2006Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (doi:10.1038/nature04546) [DOI] [PubMed] [Google Scholar]
- Esser C., et al. 2004A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (doi:10.1093/molbev/msh160) [DOI] [PubMed] [Google Scholar]
- Esser C., Martin W., Dagan T.2007The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol. Lett. 3, 180–184 (doi:10.1098/rsbl.2006.0582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommolt R., Werner S., Paulsen H., Goss R., Wilhelm C., Zauner C., Maier U. G., Grossman A. R., Bhattacharya D., Lohr M.2008Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol. Biol. Evol. 25, 2653–2667 (doi:10.1093/molbev/msn206) [DOI] [PubMed] [Google Scholar]
- Glansdorff N., Xu Y., Labedan B.2008The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct. 3, 29 (doi:10.1186/1745-6150-3-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. B., Waller R. R., McFadden G. I.2008Plastid evolution. Annu. Rev. Plant. Biol. 59, 491–517 (doi:10.1146/annurev.arplant.59.032607.092915) [DOI] [PubMed] [Google Scholar]
- Gribaldo S., Brochier-Armanet C.2006The origin and evolution of Archaea: a state of the art. Phil. Trans. R. Soc. B 361, 1007–1022 (doi:10.1098/rstb.2006.1841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J., Meurer J., Bhattacharya D.2008Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol. Biol. 8, 117 (doi:10.1186/1471-2148-8-117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmann S., Martin W.2000Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes. Int. J. Syst. Evol. Microbiol. 50, 1655–1663 [DOI] [PubMed] [Google Scholar]
- Harary F.1969Graph Theory Reading, MA: Perseus Books Publishing L. L. C [Google Scholar]
- Hazkani-Covo E., Covo S.2008Numt-mediated double strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 4, e1000237 (doi:10.1371/journal.pgen.1000237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel F., Bozarth A., Sommer M. S., Zauner S., Przyborski J. M., Maier U. G.2007Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol. Chem. 388, 899–906 (doi:10.1515/BC.2007.119) [DOI] [PubMed] [Google Scholar]
- Huang J. L., Gogarten J. P.2006Ancient horizontal gene transfer can benefit phylog phylogenetic enetic reconstruction. Trends Genet. 22, 361–366 (doi:10.1016/j.tig.2006.05.004) [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Ayliffe M. A., Timmis J. N.2003Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422, 72–76 (doi:10.1038/nature01435) [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Gruenheit N., Ahmadinejad N., Timmis J. N., Martin W.2005Mutational decay and age of chloroplast and mitochondrial genomes transferred recently to angiosperm nuclear chromosomes. Plant Physiol. 138, 1723–1733 (doi:10.1104/pp.105.060327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D.2006Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (doi:10.1093/molbev/msj030) [DOI] [PubMed] [Google Scholar]
- Kalanon M., McFadden G. I.2008The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112 (doi:10.1534/genetics.107.085704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J., Palmer J. D.2008Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (doi:10.1038/nrg2386) [DOI] [PubMed] [Google Scholar]
- Kennedy S. P., Ng W. V., Salzberg S. L., Hood L., DasSarma S.2001Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11, 1641–1650 (doi:10.1101/gr.190201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A. H., Javaux E. J., Hewitt D., Cohen P.2006Eukaryotic organisms in Proterozoic oceans. Phil. Trans. R. Soc. B 361, 1023–1038 (doi:10.1098/rstb.2006.1843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Martin W.2005On the origin of genomes and cells within inorganic compartments. Trends Genet. 21, 647–654 (doi:10.1016/j.tig.2005.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Goldovsky L., Darzentas N., Ouzounis C. A.2005The net of life: reconstructing the microbial phylogenetic network. Genome Res. 15, 954–959 (doi:10.1101/gr.3666505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. G., Canback B., Berg O. G.2007The origins of modem proteomes. Biochimie 89, 1454–1463 (doi:10.1016/j.biochi.2007.09.004) [DOI] [PubMed] [Google Scholar]
- Lake J. A., Skophammer R. G., Herbold C. W., Servin J. A.2009Genome beginnings: rooting the tree of life. Phil. Trans. R. Soc. B 364, 2177–2185 (doi:10.1098/rstb.2009.0035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G., Graur D.2007Heads or tails: A simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 24, 1380–1383 (doi:10.1093/molbev/msm060) [DOI] [PubMed] [Google Scholar]
- Landan G., Graur D.2008Characterization of pairwise and multiple sequence alignment errors. Gene, PMID: 18614299. Epub ahead of print [DOI] [PubMed] [Google Scholar]
- Lane C. E., Archibald J. M.2008The eukaryotic tree of life: endosymbiosis takes its TOL. Trends. Ecol. Evol. 23, 268–275 (doi:10.1016/j.tree.2008.02.004) [DOI] [PubMed] [Google Scholar]
- Lang A. S., Beatty J. T.2007Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 15, 54–62 (doi:10.1016/j.tim.2006.12.001) [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G., Van Helden J., Toussaint A., Leplae R.2008Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25, 762–777 (doi:10.1093/molbev/msn023) [DOI] [PubMed] [Google Scholar]
- Loftus B., et al. 2005The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868 (doi:10.1038/nature03291) [DOI] [PubMed] [Google Scholar]
- Margulis L., Chapman M., Dolan M. F.2007Semes for analysis of evolution: de Duve's peroxisomes and Meyer's hydrogenases in the sulphurous Proterozoiceon. Nat. Rev. Genet. 8, 1 (doi:10.1038/nrg2071-c1) [DOI] [PubMed] [Google Scholar]
- Martin W.1999Mosaic bacterial chromosomes—a challenge en route to a tree of genomes. BioEssays 21, 99–104 (doi:10.1002/(SICI)1521-1878(199902)21:2<99::AID-BIES3>3.0.CO;2-B) [DOI] [PubMed] [Google Scholar]
- Martin W., Russell M.2003On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil. Trans. R. Soc. Lond. B 358, 59–85 (doi:10.1098/rstb.2002.1183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Brinkmann H., Savona C., Cerff R.1993Evidence for a chimaeric nature of nuclear genomes: Eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc. Natl Acad. Sci. USA 90, 8692–8696 (doi:10.1073/pnas.90.18.8692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Stoebe B., Goremykin V., Hansmann S., Hasegawa M., Kowallik K. V.1998Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (doi:10.1038/30234) [DOI] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D.2002Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA 99, 12 246–12 251 (doi:10.1073/pnas.182432999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Dagan T., Koonin E. V., Dipippo J. L., Gogarten J. P., Lake J. A.2007The evolution of eukaryotes. Science 316, 542–543 [DOI] [PubMed] [Google Scholar]
- Martin W., Baross J., Kelley D., Russell M. J.2008Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6, 805–814 (doi:10.1038/nrmicro1991) [DOI] [PubMed] [Google Scholar]
- McInerney J. O., Cotton J. A., Pisani D.2008The prokaryotic tree of life: past, present … and future? Trends Ecol. Evol. 23, 276–281 (doi:10.1016/j.tree.2008.01.008) [DOI] [PubMed] [Google Scholar]
- Mongodin E. F., et al. 2005The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl Acad. Sci. USA 102, 18 147–18 152 (doi:10.1073/pnas.0509073102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.2007The road to hydrogenosomes. In Origin of Mitochondria and Hydrogenosomes. (eds Martin W. F., Müller M.), pp. 1–12 Heidelberg, Germany: Springer-Verlag [Google Scholar]
- Nisbet E. G., Sleep N. H.2001The habitat and nature of early life. Nature 409, 1083–1091 (doi:10.1038/35059210) [DOI] [PubMed] [Google Scholar]
- Orgel L. E.2008The implausibility of metabolic cycles on the prebiotic earth. PLoS Biol. 6, e18 (doi:10.1371/journal.pbio.0060018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D., Cotton J. A., McInerney J. O.2007Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol. Biol. Evol. 24, 1752–1760 (doi:10.1093/molbev/msm095) [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S., et al. 2008The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl Acad. Sci. USA 105, 10 061–10 066 (doi:10.1073/pnas.0803928105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen B., Fletcher I. R., Brocks J. J., Kilburn M. R.2008Reassessing the first occurrence of eukaryotes and cyanobacteria. Nature 455, 1101–1105 (doi:10.1038/nature07381) [DOI] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A.2004The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431, 152–155 (doi:10.1038/nature02848) [DOI] [PubMed] [Google Scholar]
- Rivera M. C., Jain R., Moore J. E., Lake J. A.1998Genomic evidence for two functionally distinct gene classes. Proc. Natl Acad. Sci. USA 95, 6239–6244 (doi:10.1073/pnas.95.11.6239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho M. E., Worful J. M., Lee J., Kannan K., Tyler M. S., Bhattacharya D., Moustafa A., Manhart J. R.2008Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl Acad. Sci. USA 105, 17 867–17 871 (doi:10.1073/pnas.0804968105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S. L., White O., Peterson J., Eisen J. A.2001Microbial genes in the human genome: lateral transfer or gene loss? Science 292, 1903–1906 (doi:10.1126/science.1061036) [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V., Delwiche C. F.2008A hypothesis for plastid evolution in chromalveolates. J. Phycol. 44, 1097–1107 (doi:10.1111/j.1529-8817.2008.00559.x) [DOI] [PubMed] [Google Scholar]
- Scannell D. R., Byrne K. P., Gordon J. L., Wong S., Wolfe K. H.2006Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345 (doi:10.1038/nature04562) [DOI] [PubMed] [Google Scholar]
- Shavit L., Penny D., Hendy M. D., Holland B. R.2007The problem of rooting rapid radiations. Mol. Biol. Evol. 24, 2400–2411 (doi:10.1093/molbev/msm178) [DOI] [PubMed] [Google Scholar]
- Stanhope M. J., Lupas A., Italia M. J., Koretke K. K., Volker C., Brown J. R.2001Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411, 940–944 (doi:10.1038/35082058) [DOI] [PubMed] [Google Scholar]
- Stoebe B., Maier G.2002One, two, three: nature's toolbox for building plastids. Protoplasma 219, 123–130 (doi:10.1007/s007090200013) [DOI] [PubMed] [Google Scholar]
- Theissen U., Martin W.2006The difference between organelles and endosymbionts. Curr. Biol. 16, R1016–R1017 (doi:10.1016/j.cub.2006.11.020) [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D.1990Escape of DNA from the mitochondria to the nucleus in the yeast Saccharomyces cerevisiae. Nature 346, 376–379 (doi:10.1038/346376a0) [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Nielsen K. M.2005Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (doi:10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- Timmis J. N., Ayliffe M. A., Huang C. Y., Martin W.2004Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (doi:10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
- Ueno Y., Yamada K., Yoshida N., Maruyama S., Isozaki Y.2006Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440, 516–519 (doi:10.1038/nature04584) [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Tovar J., Clark C. G.2005Mitochondrion-derived organelles in protists and fungi. Int. Rev. Cytol. 244, 175–225 [DOI] [PubMed] [Google Scholar]
- von Dohlen C. D., Kohler S., Alsop S. T., McManus W. R.2001Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature 412, 433–436 (doi:10.1038/35086563) [DOI] [PubMed] [Google Scholar]
- Welch R. A., et al. 2002Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA 99, 17 020–17 024 (doi:10.1073/pnas.252529799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wujek D. E.1979Intracellular bacteria in the blue-green-alga Pleurocapsa minor. Trans. Am. Micros. Soc. 98, 143–145 (doi:10.2307/3225953) [Google Scholar]