We recently described the high-resolution X-ray structure of a helical bundle composed of eight copies of the β-peptide Zwit-1F (Figure 1A,B).1 Like many proteins in Nature, the Zwit-1F octamer contains parallel and antiparallel helices, extensive inter-helical electrostatic interactions, and a solvent-excluded hydrophobic core. Here we explore the stability of the Zwit-1F octamer in solution using circular dichroism (CD) spectroscopy, analytical ultracentrifugation (AU), differential scanning calorimetry (DSC), and NMR. These studies demonstrate that the thermodynamic and kinetic properties of Zwit-1F closely resemble those of natural α-helical bundle proteins.
Figure 1.
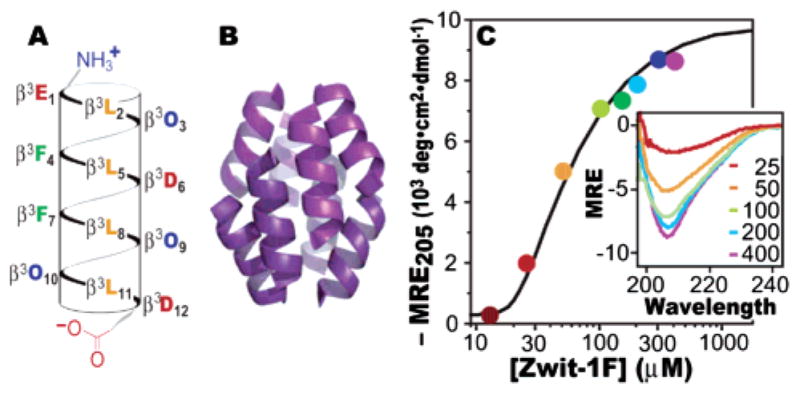
(A) Helical net representation of the Zwit-1F monomer. β3-Amino acids are designated by the single letter corresponding to the equivalent α-amino acid. O signifies ornithine. (B) Zwit-1F octamer structure determined by X-ray crystallography.1 (C) Plot of MRE205 as a function of [Zwit-1F] fit to a monomer–octamer equilibrium. Inset: CD spectra (MRE in units of 103 deg·cm2·dmol−1) at the indicated [Zwit-1F] (μM).
CD spectroscopy indicates that Zwit-1F is minimally 314-helical in dilute solution (as judged by the molar residue ellipticity at 205 nm, MRE205)2 but undergoes a large increase in helical structure between 20 and 200 μM (Figure 1C). The concentration dependence of MRE205 fits a monomer–octamer equilibrium with an association constant of 4.0 × 1030 M−7 (ln Ka = 70.5 ± 1.9).3 This value matches the result of AU analysis, which fits a monomer–octamer equilibrium with ln Ka = 71.0 ± 0.9.3 Taken together, the AU and CD data support a model in which the unfolded Zwit-1F monomer is in equilibrium with the folded octamer.4
Few known natural proteins assemble as octamers. Examples include the histones5 (hetero-octamer), TATA binding protein6 (octamer in 1 M KCl), and the well-characterized hemerythrin (ln Ka = 84).7 Although Zwit-1F is less stable than hemerythrin, it is smaller in mass (13.1 vs 110 kDa) and interaction surface area (7000 vs 15 000 Å2).1,8 To compare the stability of Zwit-1F to that of proteins of diverse size and stoichiometry, we calculated the free energy of association per Å2 of buried surface area (ΔGarea). The ΔGarea of Zwit-1F is higher than that of hemerythrin, the tetrameric aldolase, and natural helical bundle proteins GCN4 and ROP (Table 1). In fact, ΔGarea for Zwit-1F is close to the average value (7.0 ± 2.8 cal·mol−1·A−2) observed for protein complexes burying at least 1000 Å2 of surface area upon association.9,10 This comparison implies that the lower affinity of Zwit-1F is due to its small size and not an inherent instability of β3-peptide complexes.
Table 1.
Comparison of Protein Association Parametersa
| protein (stoichiometry) | MWmonomer | ΔGarea |
|---|---|---|
| Zwit-1F (8) | 1.6 kDa | 5.9 |
| hemerythrin (8) | 13.8 kDa | 3.37 |
| aldolase (4) | 39.2 kDa | 3.911 |
| GCN4 (2) | 4.0 kDa | 4.812 |
| ROP (2) | 7.2 kDa | ≥3.013 |
ΔGarea values in units of cal· mol−1·Å−2. Interaction surface areas and ΔGarea calculated as described in Supporting Information.
Temperature-dependent CD studies (Figure 2A) show Zwit-1F to exhibit a concentration-dependent Tm, an inherent property of protein quaternary structure.14 The Zwit-1F Tm, which increases from 57 °C at 50 μM to 95 °C at 300 μM, is comparable to Tm values of thermostable proteins such as ubiquitin (Tm = 90 °C) and bovine pancreatic trypsin inhibitor (Tm = 101 °C).15 The Zwit-1F Tm is significantly higher than the Tm of GCN4 (41–78 °C at 1–880 μM)16 and ROP (58–71 °C at 0.5–160 μM).17 We note, however, that the unfolding of Zwit-1F is less cooperative: the width of the temperature derivative of the CD signal at half-maximum is 40 versus 20 °C for GCN4 or 15 °C for ROP.16,17
Figure 2.
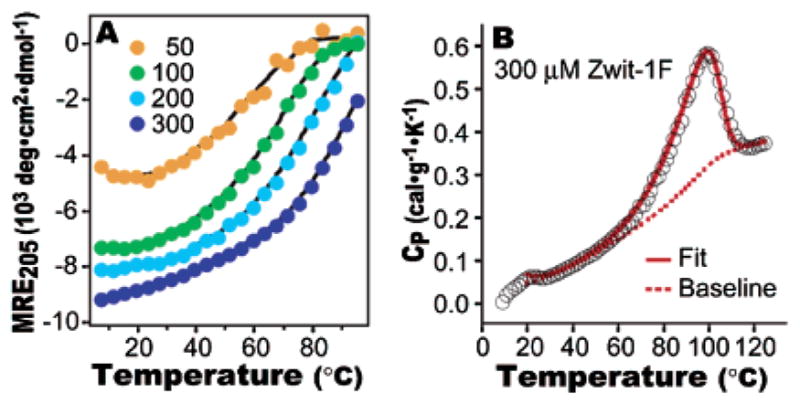
(A) Temperature-dependent CD analysis of Zwit-1F. Plot of MRE205 as a function of temperature at the indicated Zwit-1F concentration (μM). (B) DSC analysis of Zwit-1F unfolding fit to a subunit dissociation model. Raw data are shown as black circles.3
A high Tm is not a definitive measurement of thermodynamic stability, so DSC was used to further characterize Zwit-1F unfolding (Figure 2B). At 300 μM concentration (where Zwit-1F is 87% octameric), the temperature-dependent heat capacity (CP) peaks near the Tm identified by CD. This peak is embedded in a sloping baseline (∂Cp/∂T = 5.1 cal·mol−1·K−2) 3.1 mcal·g−1·K−2) that is similar to the CP versus temperature plot of monomeric β3-peptides, for which no cooperative unfolding peak has yet been observed.2 For most natural proteins, (∂Cp/∂T) is about 1 mcal·g−1·K−2 in the folded state,15 but GCN4 (∂Cp/∂T = 3.6 mcal·g−1·K−2)16 and some ROP mutants (∂Cp/∂T = 4–5 mcal·g−1·K−2)13 have sharply sloped pretransition baselines like Zwit-1F.
The DSC data fit well to a process defined by a two-state transition with dissociation of eight subunits using the program EXAM.3,18 The fitted enthalpy and heat capacity change per mole octamer are 107.4 ± 0.3 kcal·mol−1 and 1.4 ± 0.1 kcal·mol−1·K−1, respectively. Substituting these values into the Gibbs–Helmholz equation3 yields an equilibrium constant of 5.3 × 1031 (ln K = 73.3 ± 1.4) at 25 °C, in excellent agreement with values derived from CD and AU data. The integrated calorimetric unfolding enthalpy (ΔHCal) for Zwit-1F is 7.2 cal·g−1, within the range observed for natural globular proteins (5.2–11.8 cal·g−1),19,20 but somewhat lower than GCN4 (7.7 cal·g−1)21 and ROP (9.5 cal·g−1).17
The NMR spectra of many well-folded natural and designed proteins are characterized by differentiated amide resonances and slow hydrogen/deuterium exchange.22 The amide N–H resonances in the 1H spectrum of Zwit-1F, under conditions where the sample is 97% octameric, span 1.4 ppm (Figure 3A). While this span is narrower than that observed in the NMR spectra of large proteins such as α-lactalbumin (3 ppm), it is comparable to that seen for coiled-coil proteins GCN4 and ROP (1.3 and 2.2 ppm, respectively).13,23,24 In contrast to Zwit-1F, the amide resonances of the poorly folded, monomeric β-peptide Acid-1YA2,11 span only 0.5 ppm.3 These results indicate that the Zwit-1F fold in solution creates distinct electronic environments for the amide backbone protons.
Figure 3.
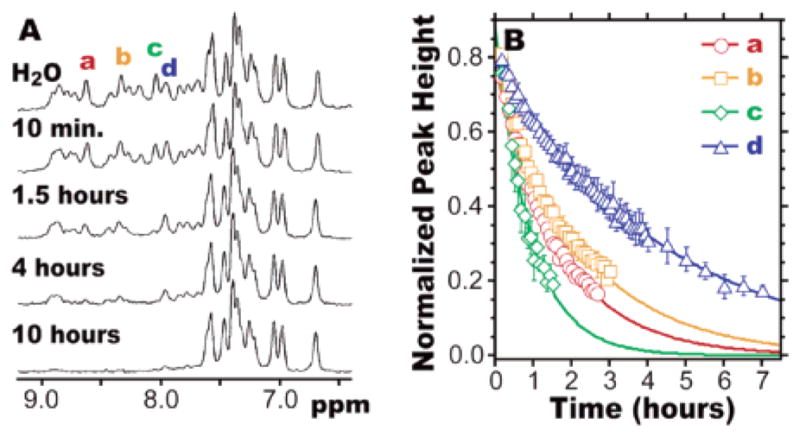
(A) 500 MHz 1H NMR spectra of 1.5 mM Zwit-1F, acquired in phosphate-buffered “H2O” (9:1 H2O/D2O) or at the indicated times after reconstitution of a lyophilized Zwit-1F sample in phosphate-buffered D2O. (B) Peak heights of the indicated resonances (normalized to the peak at 6.70 ppm) fit to exponential decays.3 Bars indicate standard error.
Participation in a hydrogen bond can protect an amide N–H from exchange with bulk solvent; since exchange occurs from the unfolded state, a slow amide exchange rate constant (kex) correlates with protein stability in solution.22 Exchange is often characterized by a protection factor (P) equal to krc/kex, where krc is the rate constant for exchange of a random coil amide N–H under similar conditions. When a lyophilized sample of Zwit-1F is redissolved at 1.5 mM concentration in D2O, 9 of 14 resolvable peaks require more than 4 h to become indistinguishable from baseline. The time dependence of exchange corresponds to exchange rate constants between 0.6 × 10−4 and 2.9 × 10−4 s−1. Using β-alanine (βG in our nomenclature) as a random coil model,3,25 these values of kex correspond to a protection factor of 2 × 104 for Zwit-1F. Thus, amide protons in Zwit-1F are less protected than those in large protein cores, where P ≥ 105.22,26 However, the protection factor for Zwit-1F, like the span of amide resonances, is comparable to ROP (105 at 250 μM)13 and GCN4 (104 at 1.0 mM).23,24 Acid-1YA2,11 undergoes amide N–H exchange in less than 10 min, showing that slow exchange requires a stable β-peptide fold.3
The biophysical experiments presented here describe the thermodynamic and kinetic stability of the Zwit-1F octamer in solution. The data allow us to quantify the similarity of Zwit-1F to GCN4 and ROP, two small, well-folded α-amino acid helix bundle proteins. In fact, the Tm, ΔGarea, and ΔHCal for Zwit-1F are even comparable to much larger natural proteins. Taken together with the recent high-resolution structure of Zwit-1F,1 these studies show that β-amino acid heteropolymers can assemble into quaternary complexes that resemble natural proteins in both solid-state structure and solution-phase stability. We note that our characterizations do not preclude some molten globule character of the Zwit-1F core in solution.27 Nonetheless, these studies establish Zwit-1F as a remarkably protein-like stepping stone in the path toward fully synthetic mimics of biological molecules.
Supplementary Material
Acknowledgments
This work was supported by NIH and NFCR. We thank Dr. J. Hoch and Z. Sutter (UConn Health Center) for access to a Microcal VP-DSC microcalorimeter, and Dr. F. Schwarz (NIST) for the program EXAM.
Footnotes
Supporting Information Available: Experimental procedures, Table 1 calculations, and data fits (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Daniels DS, Petersson EJ, Qiu XJ, Schepartz A. J Am Chem Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CD extrema between 205 and 215 nm have been attributed previously to the 314-helix. For details, see: Seebach D, Beck AK, Bierbaum DJ. Chem Biodiversity. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087.
- 3.See Supporting Information.
- 4.The MRE205 concentration dependence of Zwit-1F also fits a monomer–hexamer equilibrium, as did previous AU data. Although the Zwit-1F X-ray structure shows an octamer, we cannot currently exclude the presence of hexameric forms in solution.
- 5.Baxevanis AD, Godfrey JE, Moudrianakis EN. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 6.Daugherty MA, Brenowitz M, Fried MG. J Mol Biol. 1999;285:1389–1399. doi: 10.1006/jmbi.1998.2427. [DOI] [PubMed] [Google Scholar]
- 7.Langerman NR, Klotz IM. Biochemistry. 1969;8:4746–4757. doi: 10.1021/bi00840a014. [DOI] [PubMed] [Google Scholar]
- 8.Holmes MA, Stenkamp RE. J Mol Biol. 1991;220:723–737. doi: 10.1016/0022-2836(91)90113-k. [DOI] [PubMed] [Google Scholar]
- 9.Nooren IMA, Thornton JM. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 10.Brooijmans N, Sharp KA, Kuntz ID. Proteins: Struct, Funct, Genet. 2002;48:645–653. doi: 10.1002/prot.10139. [DOI] [PubMed] [Google Scholar]
- 11.Tolan DR, Schuler B, Beernink PT, Jaenicke R. Biol Chem. 2003;384:1463–1471. doi: 10.1515/BC.2003.162. [DOI] [PubMed] [Google Scholar]
- 12.Wendt H, Baici A, Bosshard HR. J Am Chem Soc. 1994;116:6973–6974. [Google Scholar]
- 13.Munson M, Balasubramanian S, Fleming KG, Nagi AD, Obrien R, Sturtevant JM, Regan L. Protein Sci. 1996;5:1584–1593. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturtevant JM. Annu Rev Phys Chem. 1987;38:463–488. [Google Scholar]
- 15.Makhatadze GI, Privalov PL. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 16.Dragan AI, Privalov PL. J Mol Biol. 2002;321:891–908. doi: 10.1016/s0022-2836(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 17.Steif C, Weber P, Hinz HJ, Flossdorf J, Cesareni G, Kokkinidis M. Biochemistry. 1993;32:3867–3876. doi: 10.1021/bi00066a005. [DOI] [PubMed] [Google Scholar]
- 18.Kirchhoff WM. NIST Technical Note. 1993:1401. [Google Scholar]
- 19.Dragan AI, Li ZL, Makeyeva EN, Milgotina EI, Liu YY, Crane-Robinson C, Privalov PL. Biochemistry. 2006;45:141–151. doi: 10.1021/bi051705m. [DOI] [PubMed] [Google Scholar]
- 20.Makhatadze GI, Kim KS, Woodward C, Privalov PL. Protein Sci. 1993;2:2028–2036. doi: 10.1002/pro.5560021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson KS, Vinson CR, Freire E. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001.See Supporting Information for comparison to GCN4 ΔHCal from Dragan AI, Privalov PL. J Mol Biol. 2002;321:891–908. doi: 10.1016/s0022-2836(02)00699-x.
- 22.Krishna MMG, Hoang L, Lin Y, Englander SW. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea EK, Lumb KJ, Kim PS. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 24.Lumb KJ, Kim PS. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 25.Glickson JD, Applequist J. J Am Chem Soc. 1971;93:3276–3281. doi: 10.1021/ja00742a030. [DOI] [PubMed] [Google Scholar]
- 26.Schulman BA, Redfield C, Peng ZY, Dobson CM, Kim PS. J Mol Biol. 1995;253:651–657. doi: 10.1006/jmbi.1995.0579. [DOI] [PubMed] [Google Scholar]
- 27.Although our DSC measurements support a two-state model, broad transitions of this type have been observed for proteins in molten globule states ( Moosavi-Movahedi AA, Chamani J, Gharanfoli M, Hakimelahi GH. Thermochim Acta. 2004;409:137–144. and designed proteins can become more structured in the solid state due to intermolecular crystal contacts Hill CP, Anderson DH, Wesson L, DeGrado WF, Eisenberg D. Science. 1990;249:543. doi: 10.1126/science.2382133. A standard test for molten globule states is binding of the fluorophore ANS Goto Y, Fink AL. Biochemistry. 1989;28:945–952. doi: 10.1021/bi00429a004.), but this test is not feasible for Zwit-1F.3 Experiments to examine the Zwit-1F structure using NMR relaxation techniques are in progress.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


