Abstract
Folded polymers are used in Nature for virtually every vital process. Nonnatural folded polymers, or foldamers, have the potential for similar versatility, and the design and refinement of such molecules is of considerable current interest. Here we report a complete and systematic analysis of the relationship between side chain structure and the 14-helicity of a well-studied class of foldamers, β3-peptides, in water. Our experimental results (1) verify the importance of macrodipole stabilization for maintaining 14-helix structure, (2) provide comprehensive evidence that β3-amino acids branched at the first side chain carbon are 14-helix-stabilizing, (3) suggest a novel role for side chain hydrogen bonding as an additional stabilizing force in β3-peptides containing β3-homoserine or β3-homothreonine, and (4) demonstrate that diverse functionality can be incorporated into a stable 14-helix. Gas- and solution-phase calculations and Monte Carlo simulations recapitulate the experimental trends only in the context of oligomers, yielding insight into the mechanisms behind 14-helix folding. The 14-helix propensities of β3-amino acids differ starkly from the α-helix propensities of analogous α-amino acids. This contrast informs current models for α-helix folding, and suggests that 14-helix folding is governed by radically different biophysical forces than is α-helix folding. The ability to modulate 14-helix structure through side chain choice will assist rational design of 14-helical β-peptide ligands for macromolecular targets.
Folded polymers are used in Nature for virtually every vital process, from catalysis to information storage, from cellular signaling to molecular transport. Nonnatural folded polymers, or foldamers,1 have the potential for similar versatility, and the design and refinement of such molecules is of considerable current interest.1-3 Previous studies of natural polymers – peptides, proteins, and nucleic acids – have shown that polymer folding is an extremely subtle process, governed by countless interactions among the backbone, side chains, and solvent. Nevertheless, control of foldamer structure is crucial if these molecules are to realize their full potential as tools in biology and medicine.
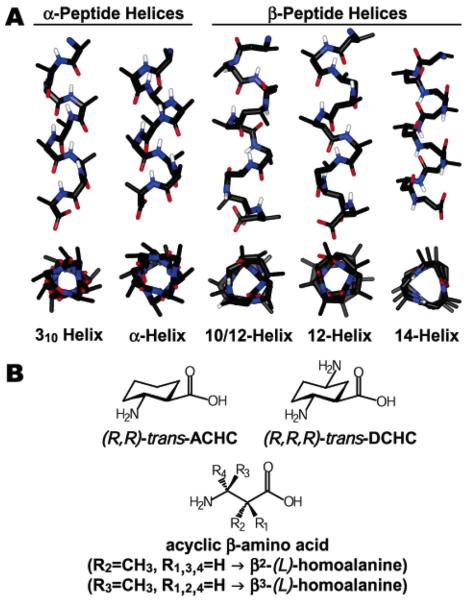
A key element of foldamer design is therefore the ability to predict the folded structure of a given backbone and modulate this structure by altering the backbone and/or side chains.1,3 In this respect, β-peptides are perhaps the best-characterized foldamers, as they populate a wide array of secondary structures including helices, sheets, and reverse turns.1,4,5 β-peptide helices are named for the number of atoms in a hydrogen-bonded ring and include the 10-helix, the 10/12-helix, the 12-helix, and the 14-helix (Figure 1A). Helix type is largely determined by choice of β-amino acid monomer: cyclic ring constraints within the monomer of four atoms, five atoms, or six atoms promote the 10-helix, 12-helix, and 14-helix, respectively,6-9 while acyclic, monosubstituted residues (β2- and β3-residues) tend to fold into 14-helices, or 10/12 helices if patterned as alternating β2/β3 residues (Figure 1B).10-15 Thus, control over preferred helical secondary structure can be achieved via judicious choice of substitution pattern.
Figure 1.
(A) Helix classes formed by α-peptides and β-peptides. Carbon atoms are shown in black, nitrogens, in blue, oxygens, in red, and amide hydrogens, in white. Other hydrogen atoms have been omitted for clarity. (B) β-amino acid monomers that promote the formation of unique helices.
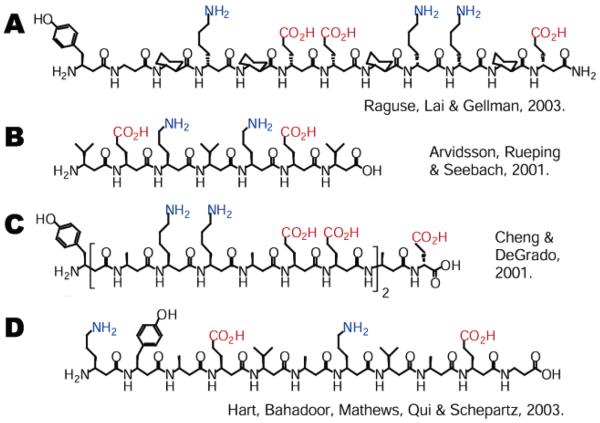
Early β-peptides that possessed helical structures in organic solvents were relatively insoluble or poorly structured in aqueous solution.16-18 Gellman and co-workers addressed this problem by introducing an additional amino group into the 14-helix-promoting cyclic residue trans-2-aminocyclohexanecarboxylic acid (ACHC) to produce trans-2,5-diaminocyclohexanecarboxylic acid (DCHC) (Figure 1B).18 It has been shown that ACHC and/or DCHC residues can promote helix formation within a larger β-peptide of acyclic residues (Figure 2A).9,19-21 Oligomers containing these cyclic residues can be soluble and helical in water but lack versatility because the cyclic β-amino acids are not easily functionalized.18, 22
Figure 2.
β3-Peptides that populate 14-helical structures in water. Coloring has been used to accentuate (i, i + 3) salt-bridges. (A) β-Dodecamer utilizing cyclic and acyclic residues reported by Raguse et al.20 (B) Salt-bridge-stabilized β3-heptapeptide with appreciable 14-helical CD signature in water reported by Arvidsson et al.24 (C) Salt-bridge-stabilized 15-mer β3-peptide reported by Cheng and DeGrado.25 (D) Macrodipole-stabilized β3-undecapeptide reported by Hart et al.26
β-peptides consisting solely of the more synthetically accessible and diverse β3-residues present a different obstacle: while these oligomers are generally 14-helical in methanol and soluble in water, they are poorly structured in aqueous solution.15,16,23 A 14-helical turn is characterized by a hydrogen bond between an amide proton at residue (i) and a backbone carbonyl at residue (i + 2),4 positioning side chains three residues apart directly in line when viewed along the helix axis (Figure 1A). This unique side chain alignment was exploited by several researchers to increase the extent of secondary structure of 14-helical β3-peptides in water. Seebach reported in 2001 that a β3-heptapeptide (Figure 2B) containing two pairs of oppositely charged residues (β3-homoornithine and β3-homoglutamic acid) at adjacent (i, i + 3) positions was completely 14-helical in methanol, as evidenced by CD spectroscopic data and NMR structural data.24 This oligomer was also water-soluble, and while previous β3-peptides had shown little evidence of structure in aqueous solution, the salt-bridge-stabilized β3-heptapeptide showed a 14-helical CD signature in water. Also in 2001, Cheng and DeGrado independently reported a 15-mer β3-peptide (Figure 2C) patterned with oppositely charged residues in an analogous way.25 Cheng and Degrado's β3-peptide was longer than Seebach's β3-peptide and contained a greater number of salt-bridges, so it is not surprising that the former possessed greater 14-helix structure as judged by CD analysis. Both β3-peptides contained oppositely charged residues, and thus the potential to form salt-bridges, on two of the three helical faces, and it was demonstrated in each case that screening of the charges (by varying salt concentration or pH of the buffer) reduced overall structure. Taken together, these studies highlight the powerful stabilizing role of electrostatic interactions within a 14-helix.
More recently, we have shown that the extent of 14-helicity in β3-peptides in water can be enhanced in a different but complementary way by neutralization of the 14-helix macrodipole.26 β3-Undecapeptides with charged side chains placed to minimize the overall 14-helix macrodipole (Figure 2D) showed increased 14-helix structure in water relative to analogous β-peptides that lacked such design elements. Enhancing 14-helix stability in this way restricts residue choice on only one of the three helical faces, allowing residues on the other two faces to be used for other purposes, such as the recognition of macromolecular targets. This work also demonstrated that β3-homophenylalanine, β3-homoserine, and β3-homoisoleucine are well-tolerated in several positions along the helix. Of particular note was the separate observation that β3-homoisoleucine was stabilizing relative to β3-homoalanine, a direct confirmation of previous observations that residues branched at the first side chain carbon promote 14-helix formation.17,27,28
In this work we explore more rigorously the 14-helix propensities of β3-amino acids with proteinogenic side chains. β-Peptides can form stable helical structures with many fewer residues than α-peptides, yet evidence suggests that the intrinsic helix propensities of β3-amino acids are weak relative to those of α-amino acids.4,25 Thus, a broad, single-substitution host–guest study of the kind used to delineate the α-helix propensities of α-amino acids29-31 seemed ideal for the exploration of the relative 14-helix propensities of β-amino acids. In addition, since the intrinsic α-helix propensities of α-amino acids have been probed so thoroughly, it was logical to think a meaningful comparison could be made across these two similar polymers and secondary structures. Finally, knowledge of intrinsic 14-helix propensities is crucial to the incorporation of tailored function into our 14-helical designs.32
Results
Experimental Design and Host Peptide Development
Our experiments were guided by pioneering host–guest studies of the intrinsic α-helix propensities of α-amino acids.30,31 In these studies, a reference or “host” peptide is designed with several important features, including high water solubility, a residue that facilitates rapid and accurate concentration determination, availability of an appropriate site or sites for substitution of “guest” side chains, control of side chain–side chain interactions within the host, and elimination of potential side chain–side chain interactions between the host peptide and the guest side chain.31 Moreover, the reference and test peptides should possess helical contents between 20% and 80% to ensure that the properties of the host peptide are sensitive to changes induced by the guest side chain.29 The α-helix propensities of natural and nonnatural α-amino acids have been evaluated in this way using various reference peptides, including one containing mostly alanine,30 a known helix-promoting residue, one that uses side chain–side chain salt-bridges to stabilize the helix,33,34 and one that uses both alanine residues and side chain–side chain interactions for helix stabilization.35,36
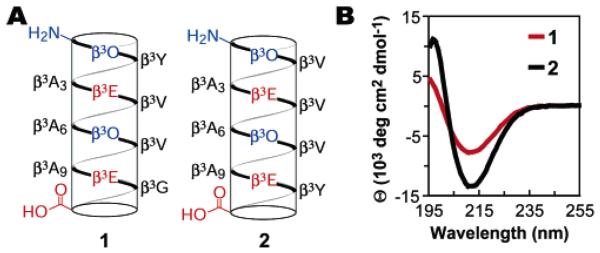
The previously studied β-peptide 1 contains salt-bridges oriented to minimize the 14-helix macrodipole (Figure 3A).26 Initially, circular dichroism (CD) spectroscopy was used to characterize its structure. While CD data on β-peptides must be interpreted carefully,37 it is reasonable to assume that, for β3-peptides in particular, changes in intensity of the 14-helical signature correlate to relative changes in overall mean 14-helical population.4,20,38-40 The CD spectrum of 1 in water (Figure 3B) is indicative of a 14-helix structure, with a characteristic minimum near 214 nm and a maximum near 195 nm. The secondary structure of 1 in CD3OH was further explored using 2-D NMR spectroscopy, and all backbone NOEs consistent with 14-helix structure were observed, except those obscured by resonance overlap. No NOEs inconsistent with 14-helix structure were observed.26
Figure 3.
(A) Helical net diagrams of β-peptides 1 and 2. β3X refers to a β3-amino acid with side chain analogous to the α-amino acid with the common one-letter code X. Coloring has been used to emphasize electrostatic stabilization features. (B) Circular dichroism spectra of β-peptides 1 (red) and 2 (black) at 100 μM and 80 μM, respectively, in PBC (1 mM sodium phosphate/borate/citrate, pH 7.0) at 25 °C.
Although 1 populated a 14-helical structure in CD3OH, CD spectroscopy in aqueous solution suggested an overall helical content between 27% and 37%, falling short of the roughly 50% helical level required for an ideal host peptide.29 β-Peptide 2 (Figure 3A) was designed with two changes that were expected to increase 14-helicity into the desired range: the β3-homoglycine residue, which we surmised would destabilize the 14-helix, was replaced with a potentially more stabilizing β3-homovaline residue,17,27,28 and the β3-homotyrosine residue was moved to the C-terminus following common practice for α-helical host αpeptides.29 The CD spectra of β-peptides 1 and 2 in water (Figure 3B) demonstrate the large effect of these alterations, with a 78% decrease in the mean residue ellipticity minimum at 214 nm (Θ214 = −7450 and −13 320 deg cm−2 dmol−1 for 1 and 2, respectively).
β-Peptide 2 was an excellent reference peptide for a more extensive host–guest analysis for several reasons. First, 2 possesses roughly 48–67% overall helical content, within the most sensitive region for detecting net changes.29 Second, the β3-homoalanines at the 3, 6, and 9 positions of 2 provided excellent points for side chain substitution. The methyl side chain is ideal for substitution, and because each substituent's (i + 3) and/or (i − 3) neighbors are restricted to methyl groups, guest side chain–host side chain interactions are minimized. Third, by substituting into these three positions individually, three independent assessments of helix propensity can be made: one near the N-terminus, one in a central position, and one near the C-terminus.
Host–Guest Analysis
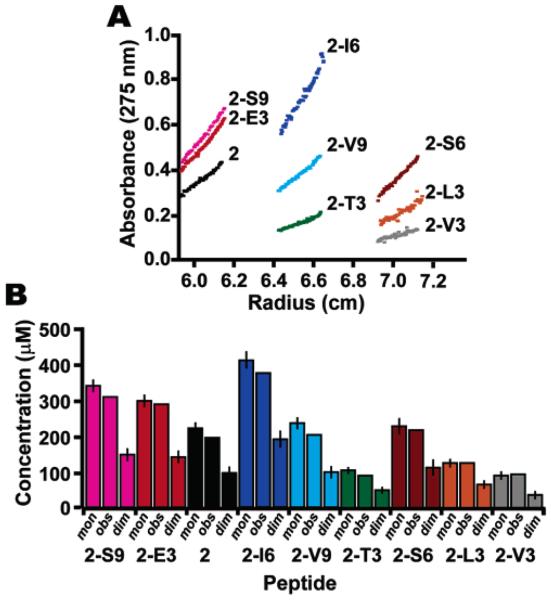
β-Peptide 2 was used as a reference peptide for a host–guest analysis of the 14-helix propensities of a wide variety of β3-amino acids. The 28 β3-peptides used for the analysis represent the host peptide and 27 variants in which each of three positions was substituted with each of nine different proteinogenic side chains. These side chains varied widely in functionality and included charged, aliphatic, polar, and aromatic groups. In the following discussion, the terms “stabilizing” and “destabilizing” will be used in reference to β3-homoalanine (the residue present in the host peptide 2) unless otherwise noted, and percent changes in 14-helicity will be calculated based on the intensity of CD minima near 214 nm relative to that of the host peptide. Each peptide was analyzed by CD in PBC buffer at neutral pH, at 25 °C, over a range of concentrations. A subset of nine peptides was also characterized by analytical ultracentrifugation (AU) to determine the oligomeric state. The molecules chosen for AU analysis represent a variety of substitutions among the three positions, while also including those peptides found to have slightly concentration-dependent CD spectra. The AU data, as well as material balance calculations based on the AU data, are shown in Figure 4. While curve fits for monomer and dimer molecular weights were equally valid, a characteristic of low molecular weight compounds, the material balance calculations41 show that all peptides tested by AU were monomeric at concentrations ranging from 80 to 400 μM.
Figure 4.
Analytical ultracentrifugation data (A) and material balances (B) for the following: 2-S9 (pink), 2-E3 (red), 2 (black), 2-I6 (blue), 2-V9 (cyan), 2-T3 (green), 2-S6 (brown), 2-L3 (orange), and 2-V3 (gray). Calculated concentrations are based on material balances for a monomer fit (mon) or a dimer fit (dim) to the AU data. The observed concentration is shown for comparison (obs).
Charged Side Chains Modulate 14-Helix Stability in a Position-Dependent Way
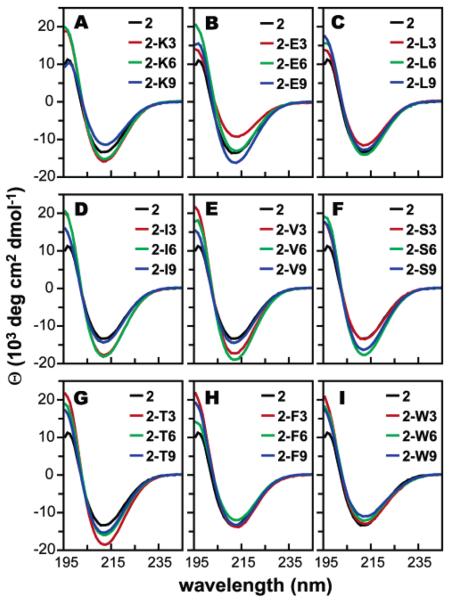
We have recently shown that, as with α-helices, interactions with the helix macrodipole are a significant factor in 14-helix stability.26 Thus, one might expect the relative 14-helix propensities of charged residues to be position-dependent, with positively charged residues exerting a stabilizing effect near the N-terminus and a destabilizing effect near the C-terminus, and vice versa for negatively charged residues. To test these predictions, β3-homoglutamic acid and β3-homolysine were substituted individually at each of the three positions within 2. The CD spectra for peptides 2-K3, 2-K6, 2-K9, 2-E3, 2-E6, and 2-E9 are shown in Figure 5A and B. In agreement with expectation, when located near the N-terminus (position 3), β3-homolysine increases the extent of 14-helix structure by 20% and β3-homoglutamic acid decreases the extent of 14-helix structure by 31%. When located near the C-terminus (position 9), β3-homolysine diminishes the extent of 14-helix structure by 13% and β3-homoglutamic acid increases the extent of 14-helix structure by 21%. In a more central position (position 6), β3-homolysine is moderately stabilizing (15% increase) and β3-homoglutamic acid is neutral to slightly destabilizing.
Figure 5.
Circular dichroism spectra of β3-peptide variants of 2 containing substitutions at position 3 (red), 6 (green), or 9 (blue) with the following: (a) β3-homolysine (2-K3, 2-K6, 2-K9), (b) β3-homoglutamic acid (2-E3, 2-E6, 2-E9), (c) β3-homoleucine (2-L3, 2-L6, 2-L9), (d) β3-homoisoleucine (2-I3, 2-I6, 2-I9), (e) β3-homovaline (2-V3, 2-V6, 2-V9), (f) β3-homoserine (2-S3, 2-S6, 2-S9), (g) β3-homothreonine (2-T3, 2-T6, 2-T9), (h) β3-homophenylalanine (2-F3, 2-F6, 2-F9), or (i) β3-homotryptophan (2-W3, 2-W6, 2-W9). Spectra were acquired in PBC buffer at 25 °C and a β3-peptide concentration of 80 μM.
Aliphatic Side Chains
The effects of aliphatic side chains on the 14-helix stability of 2 were examined next. It has been observed in other contexts that β3-homovaline and β3-homoisoleucine tend to increase the extent of 14-helix structure, whereas β3-homoleucine decreases the extent of 14-helix structure.17,26-28 β-peptides 1 and 2 were each designed with three β3-homovaline residues for this reason. Our host–guest study directly addresses the question of whether side chains branched at the first side chain carbon are 14-helix stabilizing. We individually substituted the iso-propyl (2-V3, 2-V6, 2-V9), iso-butyl (2-I3, 2-I6, 2-I9), and sec-butyl (2-L3, 2-L6, 2-L9) side chains to examine the effects of branching on overall 14-helicity.
The CD spectra of 2-V3, 2-V6, 2-V9, 2-I3, 2-I6, 2-I9, 2-L3, 2-L6, and 2-L9 are shown in Figure 5C–E. In accord with previous observations, the sec-butyl side chain of β3-homoleucine is either 14-helix-destabilizing (by 12% at position 3) or neutral (at positions 6 and 9). However, the iso-propyl and iso-butyl side chains of β3-homovaline and β3-homoisoleucine are 14-helix-stabilizing at all positions. The effect is greatest in central and N-terminal positions, where a β3-homovaline or β3-homoisoleucine residue lowers the mean residue ellipticity minimum from −13 320 deg cm−2 dmol−1 (for the host β-peptide 2) to −17 440 and below, as low as −19 130 deg cm−2 dmol−1 for β-peptide 2-V6. These values represent increases in mean 14-helix structure of 31% to 44% relative to the host peptide. The consistently large increases observed for individual substitutions of aliphatic side chains branched at the first side chain carbon directly confirm that these residues are particularly 14-helix-promoting.
Polar Side Chains
To further explore 14-helix propensities of β3-amino acids, two polar residues were chosen for substitution. The hydroxymethyl side chain of β3-homoserine and the 1-hydroxyethyl side chain of β3-homothreonine seemed suitable and had the added benefit of testing the effect of branching at the first side chain carbon within a different context. The CD spectra for peptides 2-S3, 2-S6, 2-S9, 2-T3, 2-T6, and 2-T9 are shown in Figure 5F and G. Unexpectedly, the β3-homoserine substitution is stabilizing at positions 6 and 9 (by 34% and 24%, respectively) but neutral at position 3. β3-Homothreonine, by contrast, is stabilizing at all three positions, lowering the mean residue ellipticity minimum near 214 nm as low as −18 720 deg cm−2 dmol−1 for 2-T3, corresponding to a 41% increase in 14-helical structure. The changes upon substitution with β3-homothreonine are similar to those seen for other side chains branched at the first carbon, providing further evidence that these side chains are particularly 14-helix-promoting. However, since the unbranched hydroxymethyl side chain appears to be stabilizing to an equal or greater extent than the branched side chain, and since this effect is position-dependent, there may be another stabilizing interaction present involving the hydroxyl group.
Aromatic Side Chains
Extensive analyses of known protein–protein interfaces have shown that large hydrophobic residues occur with higher frequency in surface recognition patches.42 These residues are also commonly involved in important pairwise interactions that stabilize protein–protein complexes.43 Thus, the ability to incorporate large hydrophobic residues is critical to the use of β-peptides as peptide and protein mimics. Three large aliphatic residues had already been examined, so to test the 14-helix propensity of large aromatic residues β3-homophenylalanine and β3-homotryptophan were substituted into β-peptide 2. The CD spectra for the β-peptides with aromatic substitutions are shown in Figure 5H and I. The benzyl side chain (analogous to that of phenylalanine) was 14-helix neutral in positions 3 and 9 and destabilized the 14-helix structure by only 10% in the central position. The methyl-indole side chain (analogous to that of tryptophan) was neutral at position 3 and slightly destabilizing in the other positions (up to 17%). These results provide optimism that even bulky aromatic side chains will be tolerated within the context of a reasonably stable β-peptide 14-helix.
Electrostatic Screening
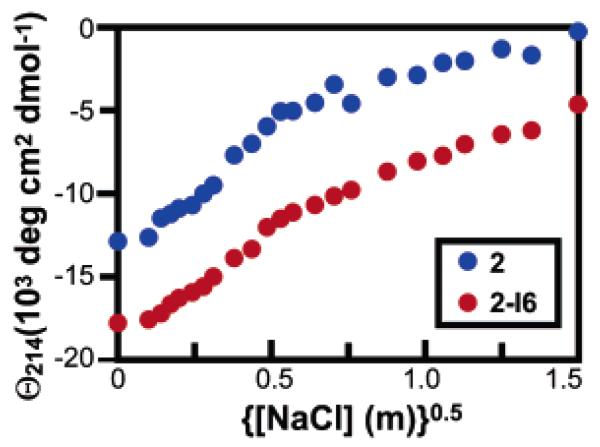
Debye–Hückel theory states that the energy of ion–ion electrostatic interactions should scale negatively with the square root of the salt concentration in molality.44 Previously, β-peptides with 14-helical structure stabilized by electrostatic interactions have been shown to unfold in the presence of high salt, in accordance with this principle.25 To provide additional evidence that electrostatic interactions contribute to the stabilities of the β-peptides studied here, we monitored the effects of increasing concentrations of NaCl on the structure of peptides 2 and 2-I6. Figure 6 illustrates the relationship between the mean residue ellipticity at 214 nm (Θ214) of these two β-peptides and salt concentration from 0 to 2.25 M. The shallow sigmoidal shape of the curves has been observed in other salt-bridge-stabilized β-peptides25 and demonstrates that screening of electrostatic interactions destabilizes the 14-helix. Interestingly, the curves for peptides 2 and 2-I6 are roughly parallel, implying that the methyl to iso-butyl substitution stabilizes the 14-helical fold in a manner unrelated to electrostatics and that even when electrostatic interactions are highly screened, the iso-butyl side chain can stabilize a 14-helical structure. Thus, the effects of salt-bridging and branched side chain substitution are independent. This observation is consistent with a model in which 14-helix folding is noncooperative,45 so that destabilization of one portion of the helix does not affect the stability of the rest of the helix.
Figure 6.
Mean residue ellipticity at 214 nM of 2-I6 (blue) and 2 (red) plotted against square root of salt concentration in molality. Spectra were acquired in PBC buffer at 25 °C and a β3-peptide concentration of 80 μM.
Conformational Analysis of β3-Amino Acids
The CD data presented above provides a glimpse into the propensities of individual β3-amino acids for stabilizing a 14-helix structure. We wondered whether the observed helix propensities could be accounted for by the conformational preferences of each individual β3-amino acid. Using gas-phase energy optimizations and the BOSS 4.5 program (a program for semiempirical statistical mechanics and energy minimization calculations),46 we analyzed seven dipeptides of the general form acetyl-β3XNHCH3, where β3X was one of β3-homoglycine (β3G), β3-homoalanine (β3A), β3-homoleucine (β3L), β3-homoisoleucine (β3I), β3-homovaline (β3V), β3-homoserine (β3S), or β3-homothreonine (β3T). The minimizations were performed using starting conformations corresponding to all possible combinations of values of the three backbone dihedral angles ϕ (N–C3), θ (C3–C2), and ψ (C2-C′), each spanning the −180° to 180° range in 30° intervals. From each of these 1331 conformations, the molecules were allowed to optimize their geometry to find local energy minima. The calculated relative energies for low-energy conformations of β3G and β3A, the only residues studied previously, agree well with published ab initio results at the MP2/6-31G(d) level,47,48 even though the quantum mechanical calculations used a terminal amide cap instead of a methyl amide.
As in similar studies of α-amino acids, most of the stable conformations located, including the global minimum of each molecule, were stabilized by intramolecular hydrogen bonds. These interactions close a six- or eight-membered hydrogen-bonded ring, to form the C6 or C8 conformation, respectively.49 Local energy minima were found at backbone dihedral angles (ϕ, θ, ψ) near (−100°, −60°, −90°) for all dipeptides, which is close to the canonical 14-helix conformation.4,5 However, these conformations possessed energies 5 to 10.5 kcal mol−1 above the global minimum. To obtain better resolution of the conformational space, we built three-dimensional Ramachandran plots50 for each dipeptide listed above. In this set of calculations, the three backbone dihedrals were scanned from −180° to 180° in 15° intervals (12 167 different starting points for each dipeptide) and fixed during energy minimizations, allowing all other degrees of freedom to vary. Despite the increased resolution of this analysis, no other minima were found. Taken together, these data imply that the experimentally observed 14-helix propensities cannot be explained by the different conformational preferences of individual β3-amino acids.
Conformational Analysis of β3-Oligopeptides
We focused next on β3-oligopeptides, to assess the role of interresidue interactions in 14-helix stabilization. To this end, we energy-minimized 25 β3-oligopeptides with the general sequence acetyl-(β3A)m-β3X-(β3A)n-NHCH3, m + n = 11, where β3X was one of β3A, β3L, β3I, β3V, β3S, or β3T at positions 3, 6, 9, or 12. The relative energies of these β3-oligopeptides were examined using a variety of starting conformations. The three backbone dihedral angles (ϕ, θ, ψ) for all residues were initially set to values corresponding to the C6 (110°, 60°, 180°), C8 (−66°, −45°, 95°), β-sheet (180°, 180°, 180°), 12-helix (−90°, 90°, −110°), or 14-helix (−155°,60°, −135°) conformations. These starting points were all minimized in the gas phase, and their energies of solvation in water were calculated using the Generalized Born/Surface Area (GB/SA) model51 in a manner that differentiates native folds from decoys,52 although previous applications of this technique employed an equivalent formulation of the same principle. Table 1 lists the minimized β-peptide energies relative to each individual β-peptide's lowest-energy conformation (ΔE). These energies predict that the 14-helix should be the lowest-energy conformation for β3-oligopeptides, followed by the 12-helix, which matches experimental observations.
Table 1.
Minimized β3-Oligopeptide Energies in Vacuo and in Aqueous Solution
| acetyl-(β3A)12-NHCH3 |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| starting conformation |
C6 | β | C8 | 12h | 14h | ||||||||||||||||
| gas phasea | 71.2 | 137.9 | 51.9 | 17.1 | 0.0 | ||||||||||||||||
| aqueousb | 64.2 | 77.0 | 54.5 | 37.9 | 0.0 | ||||||||||||||||
|
| |||||||||||||||||||||
| substituted position |
|||||||||||||||||||||
| substituted residue |
starting conformation |
3 |
6 |
9 |
12 |
||||||||||||||||
| C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | ||
|
| |||||||||||||||||||||
| β3L | gas phase | 77.8 | 128.8 | 51.8 | 16.8 | 0.0 | 72.7 | 128.8 | 51.8 | 16.9 | 0.0 | 81.8 | 126.4 | 50.6 | 16.2 | 0.0 | 82.4 | 141.2 | 51.3 | 16.3 | 0.0 |
| aqueous | 67.0 | 80.7 | 54.4 | 37.1 | 0.0 | 65.4 | 81.2 | 54.7 | 37.8 | 0.0 | 70.1 | 80.3 | 53.9 | 38.1 | 0.0 | 71.0 | 79.9 | 54.5 | 37.6 | 0.0 | |
| β3I | gas phase | 88.7 | 139.2 | 52.1 | 18.4 | 0.0 | 83.1 | 139.5 | 52.4 | 19.3 | 0.0 | 84.2 | 139.4 | 52.5 | 19.0 | 0.0 | 76.8 | 138.9 | 51.2 | 18.5 | 0.0 |
| aqueous | 68.7 | 78.0 | 54.9 | 39.0 | 0.0 | 67.4 | 78.6 | 55.4 | 40.2 | 0.0 | 68.7 | 78.2 | 55.1 | 39.7 | 0.0 | 68.1 | 77.9 | 55.0 | 39.6 | 0.0 | |
| β3V | gas phase | 85.2 | 139.1 | 52.5 | 17.9 | 0.0 | 83.9 | 138.4 | 51.5 | 18.1 | 0.0 | 88.3 | 139.2 | 52.2 | 18.7 | 0.0 | 83.1 | 139.0 | 51.2 | 18.8 | 0.0 |
| aqueous | 66.1 | 78.3 | 55.6 | 39.0 | 0.0 | 68.0 | 78.1 | 55.1 | 39.7 | 0.0 | 70.6 | 78.2 | 55.0 | 39.3 | 0.0 | 71.0 | 78.2 | 55.1 | 40.3 | 0.0 | |
| β3S | gas phase | 79.4 | 138.3 | 52.4 | 18.0 | 0.0 | 70.5 | 137.0 | 51.3 | 16.2 | 0.0 | 78.4 | 137.5 | 51.9 | 16.5 | 0.0 | 74.7 | 134.2 | 46.4 | 12.3 | 0.0 |
| aqueous | 66.2 | 75.6 | 51.5 | 36.2 | 0.0 | 63.1 | 75.3 | 51.7 | 36.0 | 0.0 | 66.7 | 75.5 | 52.1 | 36.2 | 0.0 | 66.3 | 74.6 | 50.7 | 35.9 | 0.0 | |
| β3T | gas phase | 82.0 | 138.4 | 51.5 | 17.2 | 0.0 | 80.2 | 137.6 | 50.6 | 15.9 | 0.0 | 72.5 | 138.0 | 51.4 | 16.3 | 0.0 | 71.3 | 134.8 | 46.4 | 12.0 | 0.0 |
| aqueous | 67.8 | 75.7 | 50.1 | 35.3 | 0.0 | 67.9 | 76.2 | 50.7 | 36.0 | 0.0 | 63.8 | 76.3 | 51.2 | 36.2 | 0.0 | 64.4 | 75.3 | 50.9 | 35.5 | 0.0 | |
Energies are calculated in gas phase in kcal mol−1 (Eint) and are normalized to the lowest-energy conformation for each individual β-peptide.
Energies are the sum of Eint and ΔGGB/SA and are normalized to the lowest-energy conformation for each individual β-peptide.
The results from energy minimizations report on the lowest-energy structures of each β-peptide at 0 K. While they can predict relative energies for conformations of a single β-peptide, the energies are not truly comparable across different peptides and different substitutions. To obtain a more general basis for comparison, Monte Carlo (MC) simulations at 25 °C using the GB/SA solvation model were performed, using each of the minima obtained through energy minimization as starting conformations. For each simulation the equilibration period spanned 8 million configurations, after which data were collected and averaged for 2 million configurations more. The average energies for each MC ensemble, shown in Table 2, confirm that the 14-helix is the most stable conformation for all β3-oligopeptides studied. The 12-helix is, on average, 33 kcal mol−1 higher in energy, but still stable, while all the other starting conformations (C6, C8, and β-sheet) folded to other, more compact structures. Comparing energies for substituted β-peptides with those of the simulated host, acetyl-(β3A)12-NHCH3, reveals that the β3S and β3T substitutions are most 14-helix-stabilizing in these simulations, followed by β3V and then β3I, β3A, and β3L.
Table 2.
β3-Oligopeptide Average Energies in Aqueous Solution in MC Simulations
| acetyl-(β3-A)12-NHCH3 |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| starting conformation |
C6 | b | C8 | 12h | 14h | ||||||||||||||||
| aqueousa | 69.5 | 38.3 | 44.7 | 65.9 | 36.2 | ||||||||||||||||
| substituted position |
|||||||||||||||||||||
| substituted residue |
starting conformation |
3 |
6 |
9 |
12 |
||||||||||||||||
| C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | C6 | β | C8 | 12h | 14h | ||
|
| |||||||||||||||||||||
| β3-L | aqueous | 77.7 | 66.1 | 56.3 | 72.3 | 39.2 | 77.7 | 68.0 | 58.4 | 73.0 | 30.1 | 70.1 | 37.5 | 48.3 | 70.9 | 34.8 | 64.2 | 66.5 | 47.9 | 73.5 | 40.1 |
| β3-I | aqueous | 70.4 | 55.3 | 45.1 | 73.6 | 35.5 | 68.5 | 53.1 | 55.2 | 66.6 | 35.5 | 68.9 | 61.5 | 41.1 | 63.4 | 30.3 | 74.5 | 52.6 | 47.1 | 70.7 | 34.1 |
| β3-V | aqueous | 66.2 | 68.3 | 39.3 | 69.1 | 27.8 | 61.9 | 58.2 | 44.2 | 55.9 | 18.7 | 70.2 | 65.5 | 37.2 | 66.5 | 30.8 | 62.5 | 52.2 | 39.8 | 57.9 | 17.0 |
| β3-S | aqueous | 49.2 | 50.5 | 24.7 | 46.6 | 16.9 | 44.6 | 38.7 | 32.1 | 44.6 | 16.4 | 48.8 | 55.2 | 21.6 | 41.2 | 9.3 | 50.8 | 21.7 | 30.1 | 38.9 | 10.6 |
| β3-T | aqueous | 39.7 | 10.9 | 15.9 | 29.5 | 4.3 | 34.5 | 32.4 | 19.4 | 29.3 | 8.6 | 35.0 | 26.6 | 17.2 | 31.4 | 2.1 | 39.2 | 10.2 | 19.5 | 29.8 | −2.4 |
Energies are calculated as Eint+ΔGGB/SA in kcal·mol−1 over 2 million configurations.
Structural Properties of the Simulated β3-Oligopeptides
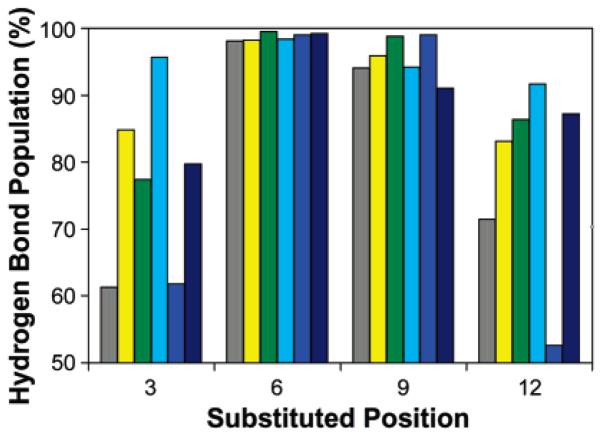
The 14-helix content of each simulated β3-oligopeptide can be estimated directly from the MC simulation data by quantifying structural properties, specifically the hydrogen bond populations and average torsional angles. Significantly, the average population of the N-terminal hydrogen bond over all the simulations is only 39%, while the average population of the C-terminal hydrogen bond is 77%. On this basis it would be predicted that stabilizing effects would be more pronounced at positions near the N-terminus. This prediction matches our CD results, with the sole exception of β3-oligopeptides containing a β3S residue. We also compared the hydrogen bond populations near each substitution site in each of the β3-oligopeptides studied, calculated as the average population of the (i − 2 f →), (i − 1 → i + 1), and (i → i + 2) hydrogen bonds surrounding a substitution at position (i) over 2 million configurations (Figure 7). In all cases there are only small effects at the middle positions, presumably because at these positions the hydrogen bond populations are already high in the all-β3A peptide. Even so, several trends can be discerned by comparing the relative hydrogen bond populations. Substitution of β3L within an all-β3A peptide has little or no effect on hydrogen bond populations at positions 3, 6, and 9 and a slight decrease at the C-terminal position 12. By contrast, single substitutions of β3I, β3V, and β3S show large increases in hydrogen bond population at the N-terminus and moderate increases at the C-terminus, while β3-T moderately increases hydrogen bond populations at both termini. These observations match the CD data very well.
Figure 7.
Hydrogen bond populations near substitution sites in Monte Carlo simulations of 14-helical oligo-β3-homoalanine peptides individually substituted at the indicated position with the following: β3-homoalanine (gray), β3-homoserine (yellow), β3-homothreonine (green), β3-homoisoleucine (cyan), β3-homoleucine (blue), or β3-homovaline (navy blue). Hydrogen bond population near a substitution site was calculated as the average population of the (i − 2 → i), (i − 1 → i + 1), and (i → i + 2) hydrogen bonds (when available) surrounding substitution position (i) over 2 million configurations.
Interestingly, the Monte Carlo simulations indicate that, within a fully 14-helical conformation, the side chain hydroxyl groups of β3S and β3T can hydrogen bond to the (i + 2) carbonyl oxygen when available. When substituted at internal positions 6 and 9, β3S and β3T formed these side chain hydrogen bonds in 68% and 35% of the configurations sampled, respectively. This observation implies β3S is more likely to form these stabilizing H-bonds within a fully 14-helical context and may explain the unexpected stabilizing effects observed by CD upon β3S substitution.
Another noticeable structural difference among the MC simulations of the 25 peptides is in the average backbone θ torsion (N–C3–C2–C′) at the substituted position. Within the context of a β3-oligopeptide, the average θ torsional angles for β3L (48°) and β3S (49°) residues are very similar to values for β3A in the same positions (52°), and these three deviate significantly from the optimal gauche torsion found for energy-minimized dipeptides (60°). β3I (60°) and β3V (59°) promote torsions very close to optimal, while β3T has a torsional angle that is intermediate (54°). The distorted torsions appear to allow better interaction of the (i, i + 3) side chains at the expense of a slight distortion of the helical axis, which does not seem to affect the hydrogen bond distances. Overall, the hydrogen bond populations and torsional geometries of singly substituted oligo-β3-homoalanine help to explain the general trends observed in the CD data, as well as confirm that 14-helix propensities appear to arise from interresidue interactions.
Discussion
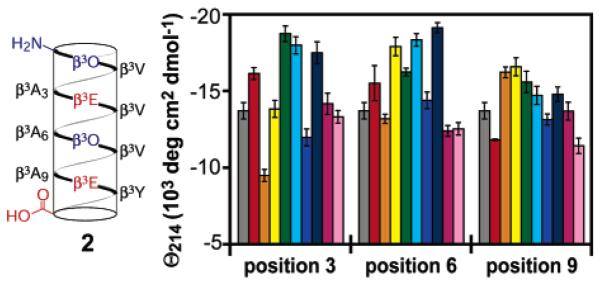
The overall results from the host-guest analysis are summarized in Figure 8. The wide range of CD intensities observed indicate strong thermodynamic preferences among β3-amino acids for or against a 14-helix structure. One of the goals of this study was to compare 14-helix propensities of β3-amino acids to the α-helix propensities of the corresponding α-amino acids, and the breadth of the host–guest analysis allows a meaningful and direct comparison. In addition, the results confirmed existing observations, such as the stabilizing effects of side chains branched at the first carbon, and informed new hypotheses, such as the possibility of 14-helix stabilization via side chain hydrogen bonding for β-peptides containing β3-homoserine or β3-homothreonine.
Figure 8.
Summary of the host–guest analysis of β3-amino acid 14-helicity. Plot shows the negative mean residue ellipticity at 214 nm of variants of β-peptide 2 (shown at left, and in chart at right in gray) substituted at positions 3, 6, and 9 with the following: β3-homoglutamic acid (red), β3-homolysine (orange), β3-homoserine (yellow), β3-homothreonine (green), β3-homoisoleucine (cyan), β3-homoleucine (blue), β3-homovaline (navy blue), β3-homophenylalanine (purple), and β3-homotryptophan (pink). Circular dichroism was performed at a β-peptide concentration of 80 μM in PBC buffer, pH 7.0, at 25 °C.
Comparison of the Helix Propensities of α- and β-Amino Acids
As observed for α-helices over 15 years ago,53 charged residues can stabilize or destabilize the 14-helix depending on their location relative to the N- and C-termini. These effects result from electrostatic interactions that diminish or intensify the 14-helix macrodipole. A similar effect is also observed when charged groups are located at helix termini: R-helices are stabilized by capped (uncharged) termini,53,54 while 14-helices are stabilized by free (charged) termini.25,26 The extent of structure stabilization observed herein due to charge-macrodipole interactions is similar in magnitude (stabilization or destabilization of ~10% relative to uncharged side chains near termini when measured by CD at pH = 7.0) to that seen for α-helices,53,54 suggesting that the magnitude of the 14-helix macrodipole (which has not yet been measured experimentally) may be similar to, or greater than, that of the α-helix macrodpole.55 Our simulations agree with this prediction, with calculated dipole moments of 45.4 and 56.9 D for oligoalanine- and oligo-β3-homoalanine-based peptides, respectively. Because short, isolated α-helices are not well-folded under the conditions used to study 14-helices (25 °C in aqueous buffer), a more quantitative comparison of charged effects is difficult. Overall, it appears that charge-macrodipole interactions play similar roles in α-helix and 14-helix stabilization and would be predicted to have a similar effect on other foldameric, amide-based hydrogen-bonded helices. It is interesting to note that β3-homolysine is more 14-helix-stabilizing than β3-homoglutamic acid in the center of the β-peptide 14-helix. It is unclear whether this difference is due to electrostatics or to some other intrinsic side chain property, though in α-helical models lysine has been consistently more helix-stabilizing than glutamic acid.31
Among noncharged side chains the 14-helix propensities of β3-amino acids contrast starkly to the α-helix propensities of their α-amino acid counterparts. The methyl side chain (alanine) is among the most α-helix-promoting but is one of the least 14-helix-stabilizing side chains in the present study. The same is true for the sec-butyl side chain (leucine and β3-homoleucine). Meanwhile, the iso-butyl side chain of isoleucine, which has moderate α-helix propensity, the iso-propyl side chain of valine, which has very low α-helix propensity, and the 1-hydroxyethyl side chain of threonine, which also has very low α-helix propensity, are all highly 14-helix-stabilizing relative to the methyl side chain. The broad discrepancies among individual side chain propensities for α-helices and 14-helices imply that the folding of these secondary structures may be governed by very different biophysical forces and speak to the mechanism underlying intrinsic helix propensities. While the rank order for α-helix propensities of α-amino acids is generally agreed upon,31 the rationale for this ranking remains controversial. Burial of hydrophobic surface area against the side of the helix,56 shielding of backbone hydrogen bonds from solvent,57-59 backbone electrostatics,60 and side chain conformational entropy,61,62 among other principles, have been advanced as the dominant basis for intrinsic α-helix propensities. Because aliphatic and small polar side chains show such radical differences in their observed α-helical and 14-helix propensities, the underlying cause of intrinsic propensities must be due primarily to interactions with the helix backbone and/or other side chains and not due to attributes such as conformational entropy or steric effects, whose scope is largely intraresidue. This hypothesis is borne out by the computational result that recapitulates 14-helix propensities of β-amino acids only within the context of oligomers. Further, because the 1-hydroxyethyl, iso-propyl, iso-butyl, and sec-butyl side chains do not follow the same pattern for both helices, a closer look must be taken at some recent theories.56-59 If propensity should scale with buried hydrophobic surface area upon helix formation, then calculations should reveal that the 1-hydroxyethyl, iso-propyl, and iso-butyl side chains bury more surface area than the sec-butyl side chain in a 14-helix context but less surface area in an α-helix context. A similar test could be performed for determining the extent of backbone hydrogen bond shielding afforded by these four side chains. It will be interesting to see whether further host-guest analysis, coupled with more rigorous simulations of β-peptide 14-helices, will be able to shed some light on this continuing debate.
Stabilizing Effect of Side Chains Branched at the First Carbon
Observations from as far back as 1999 imply that β3-amino acids branched at the first carbon are 14-helix-stabilizing.17,27 A recent study has examined the relative effects of multiple β3-homoleucines or β3-homovalines on the 14-helicity of a β3-decapeptide, albeit one with potentially repulsive (i, i + 3) side chain–side chain interactions.28 The present study examines the relative effects of methyl, iso-propyl, iso-butyl, and sec-butyl side chains on 14-helicity within the context of an already 14-helical scaffold and directly demonstrates that a single side chain branched at the first carbon can have a large stabilizing effect on overall 14-helicity. Calculations on singly substituted oligo-β3-homoalanine peptides also predict this effect, while no such effects are seen upon an extended conformational analysis of monomeric β3-amino acids. Thus, the stabilizing effects of side chains branched at the first carbon are not due to an intrinsic preference for the 14-helix torsional conformation within the monomer but instead are dependent upon interactions (direct or indirect) that involve other residues along the 14-helix.
Large Hydrophobic Side Chains Are Well-Tolerated
Large hydrophobic side chains are crucial for the development of functional, 14-helical β-peptides for two reasons. First, natural protein–protein interfaces contain a relatively high proportion of large, hydrophobic residues.42,63 Second, positioning these residues at (i, i + 3) positions within a 14-helical context would generate an extensive, continuous hydrophobic surface with the potential to bind a target with high affinity.32 β3-Homovaline and β3-homoisoleucine are 14-helix-promoting, and substitutions of β3-homoleucine, β3-homophenylalanine, and β3-homotryptophan reveal that these large hydrophobic β3-amino acids are neutral or only slightly destabilizing relative to β3-homoalanine. This finding highlights the robustness and versatility of the 14-helical host peptide and gives a first inkling of its suitability as a scaffold for molecular recognition.
Simulations Reproduce the Effects of Single β3-Amino Acid Substitutions
If the differences in the CD spectra of the peptides are proportional to their 14-helix content, increases of up to 30% can be observed upon substitution of a single β3-amino acid. For a two-state equilibrium the difference in free energy between structures that are 50% and 80% helical is only 0.82 kcal mol−1 at 25 °C, a rather challenging goal for computer simulations. While such resolution could be achieved by free-energy perturbation methods,64 the analysis would require a large number of very time-intensive simulations to reproduce the structural trends observed. Instead we attempted to discern through careful simulation if computational models can produce qualitative trends in secondary structure preferences among the β3-amino acids studied by CD spectroscopy.
Energy minimization of dipeptides (capped monomers) did not reproduce the experimental trends in 14-helix propensities. The energy minima calculated for dipeptides imply that the monomers do not even intrinsically prefer the 14-helical conformation (as is the case with α-amino acids and the α-helix49). Moreover, the gaps between the 14-helical local minima and the global minima are actually largest for residues for which 14-helix stabilization (relative to β3A) was observed by CD. Thus, the observed trends in 14-helix propensity cannot be explained by examining relative energies of different conformations of monomeric β3-amino acids.
It is only when medium-range interactions are taken into account that our calculations are able to reproduce the trends we observe experimentally. Energy minimization of a set of 25 β3-peptide 12-mers made up of all β3A and one substituted β3-amino acid clearly indicates that the 14-helix is stable even in vacuo. It was the lowest-energy minimum for all the molecules tested, being an average of 16 kcal mol−1 below the next lowest minimum, the 12-helix. The effect of solvent increases this gap to 37 kcal mol−1, which makes sense, since although the 12-helix is longer than the 14-helix, it is calculated to have a smaller dipole moment (42 D on the average versus 57 D for the 14-helix) presumably due to having fewer unfulfilled hydrogen bonds at each end. Monte Carlo simulations using these lowest-energy conformations as a starting point provided a more realistic look at β3-peptide energetics. The ensemble average energies of the substituted oligo-β3A-peptides ranked the β3-amino acids in the following order, from most 14-helix-stabilizing to least: β3T > β3S > β3V > β3I ≥ β3A > β3L. This ranking roughly fits the CD data and indicates that careful simulation can indeed reproduce the experimentally observed 14-helix propensities of β3-amino acids.
While equilibrium theory dictates that the relative populations of different conformations should be a function of their relative free energies, a comprehensive comparison of energetics would require exhaustive sampling of the relevant conformational space. By contrast, the overall 14-helix content of each individual β-peptide can be estimated by examining its structural fluctuations over the 2 million Monte Carlo configurations. For instance, for all the β3-oligopeptides simulated, the N-terminal hydrogen bond is only about half as populated as the C-terminal hydrogen bond, implying that stabilizing effects should be diminished at the C-terminus. This prediction is supported by the CD data (Figure 8). More generally, 14-helix propensities of the β3-amino acids can be ranked by hydrogen bond population as follows: β3I > β3V ≈ β3S ≥ β3T > β3A ≥ β3L. This ranking matches the CD data well. Average torsional angles for the substituted oligo-β3A-peptides indicate that distortions from the optimal 14-helix geometry, which give the helix axis a slight bend, may aid interresidue interactions without compromising hydrogen bonding. Overall, the experimental evidence supports our simulations, which appears to accurately model the interresidue interactions involved in 14-helix formation and side-chain-mediated stabilization.
Simulations Explain the Effects of Polar Substitutions
Unexpectedly, the CD data indicate that β3-homoserine is at least as stabilizing as β3-homothreonine, despite a lack of side chain branching. However, serine is not particularly α-helix-stabilizing, and threonine is among the least α-helix-stabilizing α-amino acids. It has been proposed that polar groups can promote or detract from the secondary structure, depending upon the relative energetics of hydrogen bonding in the unfolded and folded states.31 If this is the case, the 1-hydroxyethyl and hydroxymethyl side chains must make more favorable contacts in the 14-helical context relative to an unfolded β-peptide (or fewer unfavorable contacts in the unfolded state relative to the folded state) than they make in the α-helical context relative to an unfolded α-peptide. Interestingly, in our Monte Carlo simulations the side chain hydroxyl groups of β3S and β3T appeared to make hydrogen bond contacts to the (i + 2) carbonyl oxygen when available. When substituted at internal positions (positions 6 and 9), β3S and β3T formed these side chain hydrogen bonds in 68% and 35% of the 2 million configurations sampled, respectively. The fact that β3S may have a high propensity to form 14-helix-stabilizing hydrogen bonds may explain its unexpectedly high propensity for promoting 14-helix structure. More high resolution structural data will undoubtedly shed light on the mechanism behind the unexpected 14-helix propensities of polar β3-amino acids.
A Scaffold for Molecular Recognition
The host–guest analysis as a whole validates peptide 2 as an excellent folded scaffold for molecular recognition. At its present length it is able to present up to four side chains in a linear arrangement, though it is yet unknown whether the helix propensity effects of multiple substitutions will be cooperative or simply additive. The scaffold might easily be extended in increments of six residues (to retain the favorable salt-bridging and charge-macrodipole interactions) or be further substituted on the β3-homovaline-bearing face, yielding an even larger surface area for incorporation of function. Even so, three residues may be enough to recognize some important protein targets, especially considering β3-amino acids have few limitations on side chain functionality. Future work will target important cellular proteins using this scaffold in order to evaluate 14-helical β-peptides as α-helix mimetics.32
Experimental Section
General
Fmoc-protected α-amino acids, PYBOP, HOBt, and Wang resin were purchased from Novabiochem (San Diego, CA). Dimethylformamide (DMF), N-methyl-2-pyrrolidone (NMP), N-methyl morpholine (NMM), trifluoroacetic acid (TFA), and piperidine were purchased from American Bioanalytical (Natick, MA). All other reagents were purchased from Sigma-Aldrich. Certain Fmoc-β3-(l)-amino acids were purchased from Peptech Corp. (Cambridge, MA), although most were synthesized from enantiomerically pure α-amino acid precursors via the Arndt–Eistert procedure.15
β3-Peptide Synthesis, Manual Procedure
Some β3-peptides were synthesized manually, in a glass peptide synthesis vessel with fritted glass at the top and bottom and a sidearm for addition of reagents (Ace Glass, Vineland, NJ). Peptides were synthesized on a 30 or 50 μmol scale using Wang resin and Fmoc-protected β3-amino acid monomers, using standard Fmoc strategy. Wang resin was loaded as described.26 Peptide elongation and cleavage were performed essentially as described.26
β3-Peptide Synthesis, Semiautomated Procedure
Some β3-peptides were synthesized on a Symphony/Multiplex automated peptide synthesizer (Protein Technologies, Tuscon, AZ) using standard Fmoc strategy. Peptides were synthesized on a 25 μmol scale using Wang resin loaded as described.26 One cycle of peptide elongation consisted of the following steps: resin was washed with N-methyl-2-pyrrolidone (NMP) (3 × 30 s), deprotected with 20% piperidine/DMF (1 × 2 min, 2 × 8 min), washed with NMP (6 × 30 s), coupled for 30 min with 3 equiv of the appropriate β3-amino acid and 3 equiv of PYBOP, 3 equiv of HOBt, and 8 equiv of DIEA, washed once with NMP (1 × 30 s), capped for 20 min with 6% v/v acetic anhydride, 6% v/v NMM in NMP, and then washed with NMP (2 × 30 s). The β3-amino acid (3 equiv, 75 μmol), PYBOP (39.0 mg, 75 μmol), and HOBt (11.4 mg, 75 μmol) were weighed out previously and dissolved in 5 mL of NMP immediately prior to coupling. Diisopropylethylamine (DIEA) (8 equiv, 35 μL) was added immediately prior to adding the reagents to the resin through the peptide synthesis vessel sidearm. Cleavage of peptides synthesized semiautomatically was performed as follows: the resin was washed with NMP (8 × 30 s), washed with methylene chloride (8 × 30 s), dried 20 min under N2, cleaved for 2 h with cleavage reagent (3% v/v water, 3% v/v triisopropylsilane in TFA), and then washed once with cleavage reagent. The cleaved peptide was collected and concentrated by rotary evaporation, reconstituted in H2O/CH3CN (1: 1), and analyzed for purity by HPLC and MALDI-TOF mass spectrometry.
β-Peptide Purification and Characterization
Peptides were purified by reversed-phase HPLC. Identity and purity of compounds were assessed by analytical HPLC and MALDI-TOF (matrix-assisted laser desorption–ionization time-of-flight) mass spectrometry on a Voyager (Applied Biosystems) MALDI-TOF spectrometer with a 337 nm laser using the α-cyano-4-hydroxycinnaminic acid matrix. The instrument was calibrated using adrenocorticotropic hormone (ACTH) (M + H+ = 2093.1) and angiotensin I (M + H+ = 1296.7). Following purification, peptides were immediately lyophilized, kept at −20 °C, and reconstituted just prior to use. Theoretical and observed molecular weights for each peptide are listed in Table 3.
Table 3.
Theoretical and MALDI-TOF MS-Observed Molecular Weights for Each β3-Peptide Studied
| β-peptide | formula | mass calcd (M + H+) |
masses found |
|---|---|---|---|
| 2 | C64H109N13O17 | 1332.6 | 1331.3 (M + H+), 1353.4 (M + Na+) |
| 2-K3 | C67H116N14O17 | 1389.7 | 1388.2 (M + H+), 1410.2 (M + Na+) |
| 2-K6 | C67H116N14O17 | 1389.7 | 1390.6 (M + H+), 1412.6 (M + Na+) |
| 2-K9 | C67H116N14O17 | 1389.7 | 1388.8 (M + H+), 1410.7 (M + Na+) |
| 2-E3 | C66H111N13O19 | 1390.7 | 1388.2 (M + H+), 1410.3 (M + Na+) |
| 2-E6 | C66H111N13O19 | 1390.7 | 1389.4 (M + H+), 1411.4 (M + Na+) |
| 2-E9 | C66H111N13O19 | 1390.7 | 1389.0 (M + H+), 1411.0 (M + Na+) |
| 2-S3 | C64H109N13O18 | 1348.6 | 1347.9 (M + H+), 1369.9 (M + Na+) |
| 2-S6 | C64H109N13O18 | 1348.6 | 1347.5 (M + H+), 1369.5 (M + Na+) |
| 2-S9 | C64H109N13O18 | 1348.6 | 1347.5 (M + H+), 1369.5 (M + Na+) |
| 2-T3 | C65H111N13O18 | 1362.7 | 1360.8 (M + H+), 1382.8 (M + Na+) |
| 2-T6 | C65H111N13O18 | 1362.7 | 1361.3 (M + H+), 1383.4 (M + Na+) |
| 2-T9 | C65H111N13O18 | 1362.7 | 1361.6 (M + H+), 1383.6 (M + Na+) |
| 2-I3 | C67H115N13O17 | 1374.7 | 1374.9 (M + H+), 1396.8 (M + Na+) |
| 2-I6 | C67H115N13O17 | 1374.7 | 1374.0 (M + H+), 1396.0 (M + Na+) |
| 2-I9 | C67H115N13O17 | 1374.7 | 1374.1 (M + H+), 1396.0 (M + Na+) |
| 2-V3 | C66H113N13O17 | 1360.7 | 1359.3 (M + H+), 1381.3 (M + Na+) |
| 2-V6 | C66H113N13O17 | 1360.7 | 1360.0 (M + H+), 1382.0 (M + Na+) |
| 2-V9 | C66H113N13O17 | 1360.7 | 1359.5 (M + H+), 1381.6 (M + Na+) |
| 2-L3 | C67H115N13O17 | 1374.7 | 1373.0 (M + H+), 1395.0 (M + Na+) |
| 2-L6 | C67H115N13O17 | 1374.7 | 1374.0 (M + H+), 1396.0 (M + Na+) |
| 2-L9 | C67H115N13O17 | 1374.7 | 1373.9 (M + H+), 1395.9 (M + Na+) |
| 2-F3 | C70H113N13O17 | 1408.7 | 1407.8 (M + H+), 1429.8 (M + Na+) |
| 2-F6 | C70H113N13O17 | 1408.7 | 1407.0 (M + H+), 1429.0 (M + Na+) |
| 2-F9 | C70H113N13O17 | 1408.7 | 1408.0 (M + H+), 1430.1 (M + Na+) |
| 2-W3 | C72H114N14O17 | 1447.8 | 1446.0 (M + H+), 1468.0 (M + Na+) |
| 2-W6 | C72H114N14O17 | 1447.8 | 1444.9 (M + H+), 1467.0 (M + Na+) |
| 2-W9 | C72H114N14O17 | 1447.8 | 1445.3 (M + H+), 1467.3 (M + Na+) |
β-Peptide 1
Characterized as described.26
Circular Dichroism
Circular dichroism (CD) spectra between 195 and 260 nm were acquired with an Aviv 202 CD spectrometer at 25 °C using a 2 mm path length quartz cell (Hellma, Plainview, NY). Samples were prepared by dissolving lyophilized, HPLC-purified peptide in PBC buffer (1 mM each phosphoric acid, boric acid, and citric acid, pH adjusted with NaOH to 7.0). The concentration of the sample was determined by measuring absorbance at 280 nm; this analysis assumes that the extinction coefficient of a β-peptide containing a single β3-homotyrosine is equal to that of an α-peptide containing a single α-tyrosine (1300 M−1 cm−1 at 280 nm), as used previously to estimate β-peptide concentration.25,28,65 Peptide was diluted to 80 μM in PBC buffer, and serial dilutions were then made to generate solutions of 40, 20, and 10 μM. Three scans of each peptide solution were taken, with 2 s averaging times and a 2 nm bandwidth. These scans were averaged, and a blank buffer spectrum was subtracted to generate the corrected spectra. This full procedure was performed three independent times to ensure accuracy and repeatability, and these three spectra were averaged to generate the final spectrum for each peptide at each concentration. No data smoothing was used at any step. CD signal was converted into mean residue ellipticity ([Θ], deg cm2 dmol−1) using the equation:
where Ψ is raw ellipticity in degrees, res is the number of residues, l is path length in decimeters, and c is molar concentration. The concentration of each CD sample was verified after analysis by repeating the UV absorbance measurement. Aromatic side chains are known to contribute to the far-UV CD of model α-helical peptides 66 in a manner highly dependent on local backbone conformation.67 It is not yet known whether these contributions are sizable for β3-peptide 14-helices, though any future corrections for the C-terminal β3-homotyrosine will likely be small and uniform within the series. Contributions from the β3-homophenylalainine and β3-homotryptophan residues used as guest side chains might more significantly affect our conclusions regarding β-peptides containing these residues, but any future corrections are unlikely to be of a magnitude of more than ±5%.66,67 Experiments with constrained peptides have shown that fully 14-helical, short β-peptides may have mean residue ellipticity minima at 214 nm as low as Θ214 = −20 000 deg cm2 dmol−1, whereas longer β-peptides (about 15 residues) may have minima as low as −28 000 deg cm2 dmol−1 (for amino acids with S chirality at the substituted backbone carbon; R chirality reverses the handedness of the helix, and a maximum of 20 000 deg cm2 dmol−1 or higher is seen).4
The effects of electrostatic screening were monitored by acquiring CD spectra of 80 μM peptide at concentrations of NaCl from 0 to 2.25 M. Salt concentration was varied by stepwise addition of high-salt buffer to a sample of peptide within the CD cell, followed by mixing by aspiration and a 20 min equilibration. Spectra were taken at 25 °C using a 2 nm bandwidth and 5 s averaging time, and results were adjusted for cumulative changes in salt concentration, peptide concentration, and total volume.
Analytical Ultracentrifugation
Measurements were made using an Optima XLI analytical ultracentrifuge from Beckman-Coulter (Fullerton, CA). Samples were prepared by dissolving HPLC-purified peptide in PBC buffer and were centrifuged to equilibrium at 25 °C at 50 000 rpm in six-channel, carbon-epoxy composite centerpieces supplied by Beckman. Equilibrium was assessed by the absence of significant change in radial concentration gradients in scans at 14 and 16 h. Data were analyzed by fitting the data to the equation for a single ideal species using Igo-Pro (Wavemetrics, Lake Oswego OR). The equation is
where:
C(r,ro) = concentration (any units) of sedimenting species at radial positions r, ro cm from the center of rotation.
ν̄ = partial specific volume of sedimenting species (cc/g).
ρ = density of supporting buffer (g/cc).
ω = angular velocity of rotor (radians/s).
Mn = “Molar” molecular weight of sedimenting species (g/mol).
R = Gas constant (8.315 × 107 ergs K−1 mol−1).
T = Temperature (K).
Peptide partial specific volumes were assumed for simplicity to be the same as the average value calculated for previously studied β-peptides: 0.785 cm3/g.26 Because of cross-correlation of molecular weight with baseline values, curve fits were insensitive to variations in this value. In fact, equally good curve fits could be obtained by assuming either monomer or dimer molecular weights. This ambiguity is an unavoidable consequence of the low curvature exhibited in the concentration profiles of the low molecular weight compounds. To distinguish monomers from higher order aggregated species as the dominant population, curve fits were performed using integral multiples of the sequence-calculated molecular weights. This fixes the molecular weight of the presumed species, which also fixes the baseline (absorbance at zero concentration). Then, one can integrate over the net absorbance profile, to obtain the calculated average concentration in the cell. Here, the average concentrations predicted by fixing the molecular weights to those of the monomers are larger and more consistent with known concentrations than those predicted by fixing the molecular weights to dimer values. This is because the higher molecular weights predict higher curvature which, for the same data set, can only be attained by increasing the baseline values and hence reducing the calculated average concentrations. This represents a useful material balance criterion previously developed to study binding of low and high molecular weight compounds.41 Error in the calculated values (shown in Figure 4) represents the sum of uncertainties in the determination of the outer radius of the cell compartment and uncertainties arising from baseline values as reported from the curve-fitting algorithm.
Computational Analysis
All the dipeptide minimizations were done with the BOSS 4.5 program,68 modified so that the bonding list was taken from the input Z-matrix and not recalculated based on interatomic distances. All the calculations for oligo-β-peptides were carried out with the development version of MCPRO 1.6869 which includes the GB/SA treatment for solvent. The OPLS-AA force field49 was used throughout this study, augmented with backbone torsional parameters developed specifically for β-peptides in our laboratories based on high level ab initio calculations.70 In all cases the torsion angles around the peptide bond (ω) were initially set to 180° but allowed to vary freely. All possible degrees of freedom were allowed to vary in all calculations, with the exception of the dipeptide three-dimensional Ramachandran plots where the backbone dihedrals were fixed. Due to convergence problems, particularly with conformations that have very close nonbonded contacts, the dipeptide minimizations were done in triplicate with the BFGS, Fletche-Powell, and Powell algorithms, and the resulting conformer with the lowest energy was used. The oligopeptide optimizations were started with 250 steps of steepest descent minimization before switching to a conjugate gradients algorithm until convergence was reached. This treatment was not sufficient for 5 of the cases, which had to be optimized using 5000 steepest-descent steps before switching to conjugate gradients. No cutoff was used (i.e., all nonbonded pairs evaluated), and convergence criterion was set to 0.1 cal mol−1 for all minimizations. MC simulations were done using W. C. Still's GB/SA solvation model.51
Conclusions
The present study has revealed β3-peptides with unprecedented overall 14-helicities in water. These β-peptides are made from the synthetically accessible and diverse β3-amino acids and require no ring constraints to achieve folding. The most 14-helical β-peptide generated, 2-V6, has a mean residue ellipticity minimum at 214 nm (MRE214) of −19 130 ± 340 deg cm2 dmol−1 in aqueous solution, indicating a mean helix structure of up to 95%. This rivals the values observed for the most 14-helical β-peptides observed to date in methanol or in micelles, even those with cyclic residues.7,27,71 The 14-helix propensities of β3-amino acids observed in this study contrast sharply with the α-helix propensities of corresponding α-amino acids. These data, along with other evidence, demonstrate that 14-helix folding is governed by radically different biophysical forces than α-helix folding. Finally, the present study belies the assumption that unconstrained β3-amino acids cannot form highly structured 14-helices in water and demonstrates that diverse functionality can be incorporated into such a stable helix through simple substitution. Using β-peptide 2 as a scaffold, it may soon be possible to rationally design 14-helical β-peptide ligands for a variety of macromolecular targets.32
Acknowledgment
This work was supported by the NIH (GM 65453 to A.S. and GM 032136 to W.L.J.), by the National Foundation for Cancer Research, and in part by a grant to Yale University, in support of A.S., from the Howard Hughes Medical Institute. J.A.K. was supported by a National Science Foundation Predoctoral Fellowship and by an NIH Training Grant in Biophysics (GM 08283-11-15). We thank Deping Wang for providing the parameter files for one dipeptide and Jakob Ulmschneider for the initial setup of GB/SA calculations.
References
- 1.Gellman SH. Acc. Chem. Res. 1998;31(4):173–180. [Google Scholar]
- 2.Patch JA, Barron AE. Curr. Opin. Chem. Biol. 2002;6(6):872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 3.Hill JH, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem. Rev. 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 4.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101(10):3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 5.DeGrado WF, Schneider JP, Hamuro Y. J. Pept. Res. 1999;54(3):206–217. doi: 10.1034/j.1399-3011.1999.00131.x. [DOI] [PubMed] [Google Scholar]
- 6.Claridge TDW, Goodman JM, Moreno A, Angus D, Barker SF, Taillefumier C, Watterson MP, Fleet GWJ. Tetrahedron Lett. 2001;42(25):4251–4255. [Google Scholar]
- 7.Appella DH, Christianson LA, Klein DA, Powell DR, Huang XL, Barchi JJ, Gellman SH. Nature. 1997;387(6631):381–384. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 8.Appella DH, Christianson LA, Karle IL, Powell DR, Gellman SH. J. Am. Chem. Soc. 1996;118(51):13071–13072. [Google Scholar]
- 9.Appella DH, Barchi JJ, Durell SR, Gellman SH. J. Am. Chem. Soc. 1999;121(10):2309–2310. [Google Scholar]
- 10.Seebach D, Abele S, Gademann K, Guichard G, Hintermann T, Jaun B, Matthews JL, Schreiber JV. Helv. Chim. Acta. 1998;81(5):932–982. [Google Scholar]
- 11.Seebach D, Gademann K, Schreiber JV, Matthews JL, Hintermann T, Jaun B, Oberer L, Hommel U, Widmer H. Helv. Chim. Acta. 1997;80(7):2033–2038. [Google Scholar]
- 12.Seebach D, Schreiber JV, Abele S, Daura X, van Gunsteren WF. Helv. Chim. Acta. 2000;83(1):34–57. [Google Scholar]
- 13.Guichard G, Abele S, Seebach D. Helv. Chim. Acta. 1998;81(2):187–206. [Google Scholar]
- 14.Seebach D, Ciceri PE, Overhand M, Jaun B, Rigo D, Oberer L, Hommel U, Amstutz R, Widmer H. Helv. Chim. Acta. 1996;79(8):2043–2066. [Google Scholar]
- 15.Seebach D, Overhand M, Kuhnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Helv. Chim. Acta. 1996;79(4):913–941. [Google Scholar]
- 16.Etezady-Esfarjani T, Hilty C, Wuthrich K, Rueping M, Schreiber J, Seebach D. Helv. Chim. Acta. 2002;85(5):1197–1209. [Google Scholar]
- 17.Gung BW, Zou D, Stalcup AM, Cottrell CE. J. Org. Chem. 1999;64(7):2176–2177. [Google Scholar]
- 18.Appella DH, LePlae PR, Raguse TL, Gellman SH. J. Org. Chem. 2000;65(15):4766–4769. doi: 10.1021/jo000277h. [DOI] [PubMed] [Google Scholar]
- 19.Wang XF, Espinosa JF, Gellman SH. J. Am. Chem. Soc. 2000;122(19):4821–4822. [Google Scholar]
- 20.Raguse TL, Lai JR, Gellman SH. J. Am. Chem. Soc. 2003;125(19):5592–5593. doi: 10.1021/ja0341485. [DOI] [PubMed] [Google Scholar]
- 21.LePlae PR, Fisk JD, Porter EA, Weisblum B, Gellman SH. J. Am. Chem. Soc. 2002;124(24):6820–6821. doi: 10.1021/ja017869h. [DOI] [PubMed] [Google Scholar]
- 22.Wipf P, Wang XD. Tetrahedron Lett. 2000;41(45):8747–8751. [Google Scholar]
- 23.Abele S, Guichard G, Seebach D. Helv. Chim. Acta. 1998;81(12):2141–2156. [Google Scholar]
- 24.Arvidsson PI, Rueping M, Seebach D. Chem. Commun. 2001;(7):649–650. [Google Scholar]
- 25.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2001;123(21):5162–5163. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 26.Hart SA, Bahadoor ABF, Matthews EE, Qiu XYJ, Schepartz A. J. Am. Chem. Soc. 2003;125(14):4022–4023. doi: 10.1021/ja029868a. [DOI] [PubMed] [Google Scholar]
- 27.Hamuro Y, Schneider JP, DeGrado WF. J. Am. Chem. Soc. 1999;121:12200–12201. [Google Scholar]
- 28.Raguse TL, Lai JR, Gellman SH. Helv. Chim. Acta. 2002;85(12):4154–4164. [Google Scholar]
- 29.Rohl CA, Baldwin RL. Energetics of Biological Macromolecules, Pt B. 1998;295:1–26. [Google Scholar]
- 30.Chakrabartty A, Kortemme T, Baldwin RL. Protein Sci. 1994;3(5):843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakrabartty A, Baldwin RL. Adv. Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 32.Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J. Am. Chem. Soc. 2004;126(31):9468–9. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 33.Gans PJ, Lyu PC, Manning MC, Woody RW, Kallenbach NR. Biopolymers. 1991;31(13):1605–1614. doi: 10.1002/bip.360311315. [DOI] [PubMed] [Google Scholar]
- 34.Lyu PC, Liff MI, Marky LA, Kallenbach NR. Science. 1990;250(4981):669–673. doi: 10.1126/science.2237416. [DOI] [PubMed] [Google Scholar]
- 35.Park SH, Shalongo W, Stellwagen E. Biochemistry. 1993;32(47):12901–12905. doi: 10.1021/bi00210a044. [DOI] [PubMed] [Google Scholar]
- 36.Park SH, Shalongo W, Stellwagen E. Biochemistry. 1993;32(27):7048–7053. doi: 10.1021/bi00078a033. [DOI] [PubMed] [Google Scholar]
- 37.Glattli A, Daura X, Seebach D, van Gunsteren WF. J. Am. Chem. Soc. 2002;124(44):12972–12978. doi: 10.1021/ja020758d. [DOI] [PubMed] [Google Scholar]
- 38.Park JS, Lee HS, Lai JR, Kim BM, Gellman SH. J. Am. Chem. Soc. 2003;125(28):8539–8545. doi: 10.1021/ja034180z. [DOI] [PubMed] [Google Scholar]
- 39.Arvidsson PI, Frackenpohl J, Seebach D. Helv. Chim. Acta. 2003;86(5):1522–1553. [Google Scholar]
- 40.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2002;124(39):11564–11565. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]
- 41.Arkin M, Lear JD. Anal. Biochem. 2001;299(1):98–107. doi: 10.1006/abio.2001.5396. [DOI] [PubMed] [Google Scholar]
- 42.Jones S, Thornton JM. Proc. Natl. Acad. Sci. U.S.A. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glaser F, Steinberg DM, Vakser IA, Ben-Tal N. Proteins. 2001;43(2):89–102. [PubMed] [Google Scholar]
- 44.Creighton TE. Proteins. 2nd ed. W. H. Freeman and Co.; New York: 1993. [Google Scholar]
- 45.Gademann K, Jaun B, Seebach D, Perozzo R, Scapozza L, Folkers G. Helv. Chim. Acta. 1999;82(1):1–11. [Google Scholar]
- 46.Jorgensen WL. The Encyclopedia of Computational Chemistry. Vol. 5. John Wiley & Sons Ltd; Athens, USA: 1998. BOSS – Biochemical and Organic Simulation System; pp. 3281–3285. xd4 Ed.xd5 Λxd4 Eds.xd5. [Google Scholar]
- 47.Wu YD, Wang DP. J. Am. Chem. Soc. 1999;121(40):9352–9362. [Google Scholar]
- 48.Wu YD, Wang DP. J. Am. Chem. Soc. 1998;120(51):13485–13493. [Google Scholar]
- 49.Jorgensen WL, Maxwell DS, TiradoRives J. J. Am. Chem. Soc. 1996;118(45):11225–11236. [Google Scholar]
- 50.Ramachandran GN, Ramakrishnan C, Sasisekharan V. J. Mol. Biol. 1963;7(1):95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 51.Qiu D, Shenkin PS, Hollinger FP, Still WC. J. Phys. Chem. A. 1997;101(16):3005–3014. [Google Scholar]
- 52.Felts AK, Gallicchio E, Wallqvist A, Levy RM. Proteins. 2002;48(2):404–422. doi: 10.1002/prot.10171. [DOI] [PubMed] [Google Scholar]
- 53.Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Nature. 1987;326(6113):563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- 54.Fairman R, Shoemaker KR, York EJ, Stewart JM, Baldwin RL. Proteins. 1989;5(1):1–7. doi: 10.1002/prot.340050102. [DOI] [PubMed] [Google Scholar]
- 55.Lockhart DJ, Kim PS. Science. 1992;257(5072):947–951. doi: 10.1126/science.1502559. [DOI] [PubMed] [Google Scholar]
- 56.Blaber M, Zhang XJ, Matthews BW. Science. 1993;260(5114):1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 57.Garcia AE, Sanbonmatsu KY. Proc. Natl. Acad. Sci. U.S.A. 2002;99(5):2782–2787. doi: 10.1073/pnas.042496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo PZ, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 1999;96(9):4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Avbelj F, Luo PZ, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 2000;97(20):10786–10791. doi: 10.1073/pnas.200343197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avbelj F, Moult J. Biochemistry. 1995;34(3):755–764. doi: 10.1021/bi00003a008. [DOI] [PubMed] [Google Scholar]
- 61.Creamer TP, Rose GD. Proteins. 1994;19(2):85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 62.Creamer TP, Rose GD. Proc. Natl. Acad. Sci. U.S.A. 1992;89(13):5937–5941. doi: 10.1073/pnas.89.13.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma BY, Elkayam T, Wolfson H, Nussinov R. Proc. Natl. Acad. Sci. U.S.A. 2003;100(10):5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kollman P. Chem. Rev. 1993;93(7):2395–2417. [Google Scholar]
- 65.Cantor CR, Schimmel PR. Biophysical Chemistry. II. W. H. Freeman and Co.; New York: 1998. [Google Scholar]
- 66.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Biochemistry. 1993;32(21):5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 67.Bhattacharjee S, Toth G, Lovas S, Hirst JD. J. Phys. Chem. B. 2003;107(33):8682–8688. [Google Scholar]
- 68.Jorgensen WL. BOSS 4.5. Yale University; New Haven, CT 06520: [Google Scholar]
- 69.Jorgensen WL. MCPRO 1.68, development Version. Yale University; New Haven, CT 06520: [Google Scholar]
- 70.Chandrasekhar J, Saunders M, Jorgensen WL. J. Comput. Chem. 2001;22(14):1646–1654. [Google Scholar]
- 71.Barchi JJ, Huang XL, Appella DH, Christianson LA, Durell SR, Gellman SH. J. Am. Chem. Soc. 2000;122(12):2711–2718. [Google Scholar]