Abstract
Golgi body-mediated signaling has been linked to its fragmentation and regeneration during the mitotic cycle of the cell. During this process, Golgi-resident proteins are released to the cytosol and interact with other signaling molecules to regulate various cellular processes. Acyl-coenzyme A binding domain containing 3 protein (ACBD3) is a Golgi protein involved in several signaling events. ACBD3 protein was previously known as peripheral-type benzodiazepine receptor and cAMP-dependent protein kinase associated protein 7 (PAP7), Golgi complex-associated protein of 60 kDa (GCP60), Golgi complex-associated protein 1 (GOCAP1), and Golgi phosphoprotein 1 (GOLPH1). In this review, we present the gene ontology of ACBD3, its relations to other Acyl-coenzyme A binding protein (ACBP) domain containing proteins, and its biological function in steroidogenesis, apoptosis, neurogenesis, and embryogenesis. We also discuss the role of ACBD3 in asymmetric cell division and cancer. New findings about ACBD3 may help understand this newly characterized signaling molecule and stimulate further research into its role in molecular endocrinology, neurology, and stem cell biology.
Keywords: ACBD3, PAP7, GOCAP1, GCP60, cell signaling, steroidogenesis, cell differentiation
1. Introduction
The Golgi apparatus is composed of flattened fluid-filled sacs that control the flow of molecules in a cell. In many instances, cell signaling requires the availability of Golgi apparatus resident proteins during the cell cycle. These proteins become available through Golgi reorganization. The molecular mechanism of Golgi fragmentation remains unknown [1]. The Golgi apparatus serves as a signaling platform, involved in many signaling pathways. The Golgi apparatus plays important roles in diverse cellular processes and in the regulation of down-stream events.
Accumulated evidence suggests that the Golgi resident protein, acyl-coenzyme A binding domain containing 3 protein (ACBD3), is involved in the maintenance of Golgi apparatus structure in general [2-4]. Actually this protein was previously known as peripheral-type benzodiazepine receptor (PBR) and cAMP-dependent protein kinase (PKA) associated protein 7 (PAP7), Golgi complex-associated protein of 60 kDa (GCP60), Golgi complex-associated protein 1 (GOCAP1; OMIM # 606809), and Golgi phosphoprotein 1 (GOLPH1; OMIM # 606809) in biomedical scientific literature and/or public databases. ACBD3 plays a central role in several cellular signaling cascades after its release from the Golgi apparatus. ACBD3 is shown to be involved in cAMP-dependent steroidogenesis, apoptosis, iron homeostasis, and determination of neural progenitor cell fate [3,5-7]. In this article, we present the gene structure of ACBD3. We discuss its relationship to other functional domain-related proteins, its biological function, and implications in cell signaling, particularly in embryonic development and cancer research.
2. Golgi fragmentation and cell cycle
The mammalian Golgi body originates from a highly organized ribbon structure located close to the nucleus. The Golgi body fragments, a common feature of a number of physiological processes, from the large ribboned structure into small vesicular and tubular compartments during cell mitosis and apoptosis [8,9]. During mitosis, to ensure that the daughter cells share similar amount of cytoplasmic organelles, in addition to genetic material, the Golgi membrane morphology of maternal cells changes extensively (Fig. 1). The phosphorylation of several proteins, including golgin-84, GM130 and GRASP-65 as well as the inactivation of small GTPases10–13, has been proposed to be the trigger of this process. The mechanism underlying these events is not fully understood. The C-terminal-binding protein/brefeldin A-ADP ribosylated substrate (CtBP3/BARS) appears to be an important factor in Golgi fragmentation where it seems to interact with lipids and proteins in lipid vesicle formation [10-12]. However, caspase-dependent cleavage of golgins, such as giantin, golgin-160 and p115 fragmentation, can induce the Golgi fragmentation during apoptosis [13-15]. Nuclear translocation of the golgin-160 fragment is regulated via its interaction with ACBD3 [16,17].
Fig. 1.
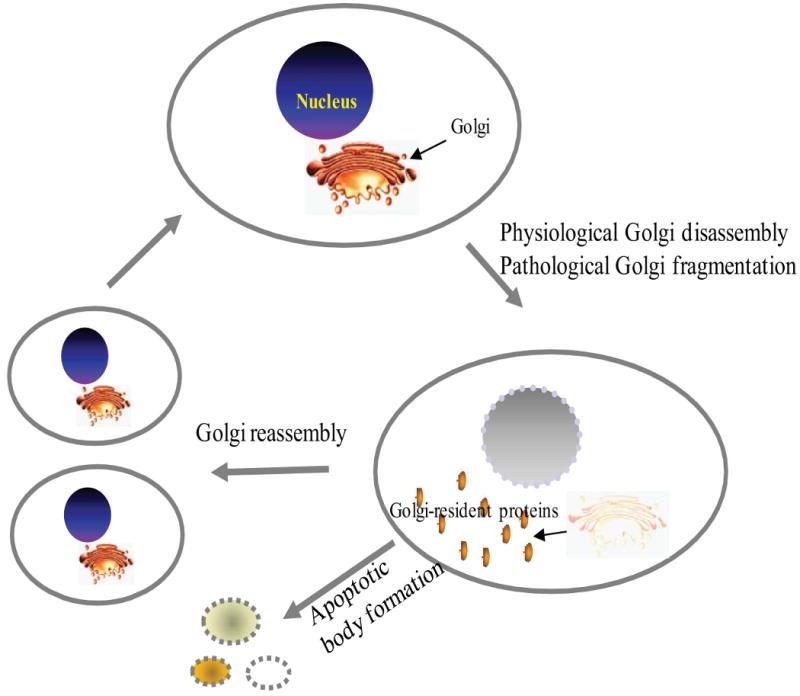
Schematic illustration of Golgi apparatus fragmentation and reassembly during cell mitosis and apoptosis. Many reasons can lead to Golgi fragmentation, but it can be generally classified as physiological Golgi disassembly (Mitosis) or pathological Golgi fragmentation due to apoptosis, presence of misfolded proteins, neurodegenerative diseases, Chlamydia infection, etc.
After Golgi fragmentation, many Golgi-residing proteins, such as heterotrimeric G proteins, phosphoinositide 3-kinases (PI3Ks), endothelial nitric oxide synthase (eNOS), and CDC42, are released. These proteins are involved in many cellular processes, including cell proliferation and apoptosis signaling pathways (see [18]). A number of recently published reports, summarized in this review, suggest that ACBD3 functions as a signaling molecule in several different cellular signaling pathways.
3. Nomenclature and classification
ACBD3 was first identified in 2001 by Li et al. and Sohda et al. as PAP7 and GCP60 proteins, respectively [3,5,16,19]. Since this gene plays a role in many biological functions, the proposed functional nomenclatures, PAP7 and GCP60, did not encapsulate the diverse functions of this molecule. A structure-based nomenclature was believed to better reflect the general characteristics of the protein, while avoiding association of the protein with any one specific function. The name ACBD3 was adopted by the HUGO Gene Nomenclature Committee as the formal name for PAP7 and GCP60 (http://www.genenames.org).
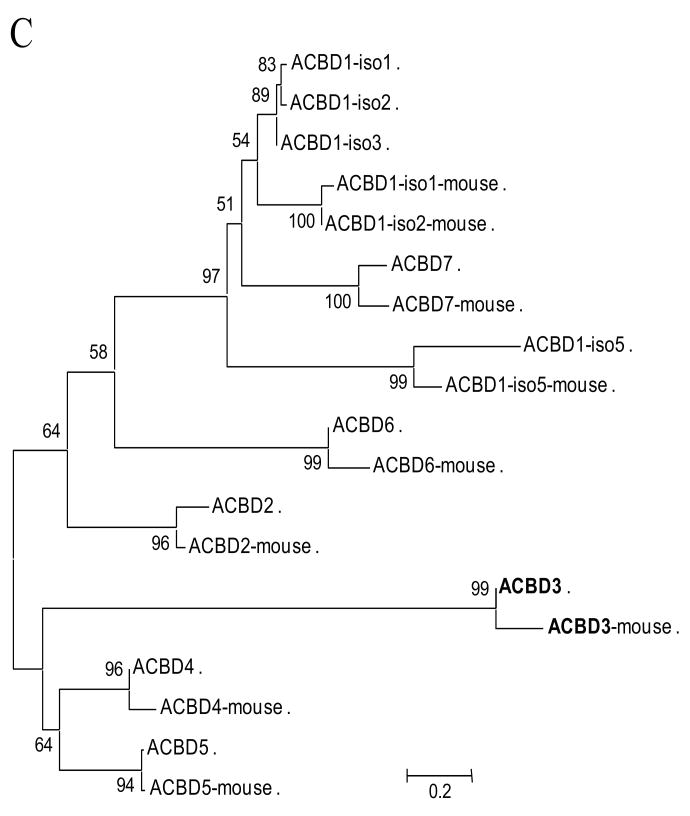
Several acyl-CoA-binding protein (ACBP) domain-containing proteins have been identified. These include ACBD1 (previously known as a diazepam binding inhibitor, DBI and ACBP), ACBD2 (peroxisomal D3,D2-enoyl-CoA isomerase, PECI), ACBD4, ACBD5, ACBD6, and ACBD7 (Table 1). The domain shared among these proteins is acyl-coenzyme A binding domain (ACBD) at the N-terminal end. However, some additional prominent domains were observed in their C-terminal ends, such as enoyl-CoA somerase/hydratase domain in the C-terminus of ACBD2, two ankyrin motifs at the C-terminus of ACBD6 and one GOLD (for Golgi dynamics) domain at the C-terminus of ACBD3 (Fig. 2). The sequence and structural information obtained clearly suggest that ACBD3 possesses a conserved ACBP domain similar to that found in six other ACBP domain-containing proteins.
Table 1.
Comparison of ACBP domain-containing proteins in humans.
| Approved Gene Symbol |
Approved Gene Name* |
No. AA** |
MW*** (kDa) |
pI**** | Location | Sequence Accession IDs |
Previous Symbols |
Aliases | Functions | Diseases |
|---|---|---|---|---|---|---|---|---|---|---|
| DBI | diazepam binding inhibitor (GABA receptor modulator, acyl-Coenzyme A binding protein) | 87 | 10.04 | 6.12 | 2q12-q21 | L76366, NM_020548 | ACBP, ACBD1 | steroidogenesis, neurotransmitter | ||
| PECI | peroxisomal D3,D2-enoyl-CoA isomerase | 364 | 40.18 | 8.93 | 6p24.3 | AF069301, M_006117 | ACBD2, DRS1, HCA88 | peroxisomal fatty acid metabolism | hepatocellular carcinoma, renal-cell carcinoma | |
| ACBD3 | acyl-Coenzyme A binding domain containing 3 | 528 | 60.59 | 5.02 | 1q41 | AB043587, NM_022735 | PAP7, GOLPH1, GOCAP1 | PAP7, GCP60 | apoptosis, steroidogenesis, iron-homeostasis, neurogenesis | cancers, tumorigenesis, drug response |
| ACBD4 | acyl-Coenzyme A binding domain containing 4 | 305 | 34.76 | 5.56 | 17q21.31 | BC029164, NM_024722 | FLJ13322 | valproic acid-induced | ||
| ACBD5 | acyl-Coenzyme A binding domain containing 5 | 490 | 54.79 | 5.13 | 10p12.1 | AF505653, NM_145698 | DKFZp434A2417, KIAA1996 | cell differentiation | ||
| ACBD6 | acyl-Coenzyme A binding domain containing 6 | 282 | 31.15 | 5 | 1q25.1 | BC006505, NM_032360 | MGC2404 | hematopoiesis, vessel development | ||
| ACBD7 | acyl-Coenzyme A binding domain containing 7 | 88 | 9.79 | 6.26 | 10 | AK095538 | FLJ38219, bA455B2.2 | tissue-specific expression |
the names listed are according to the HUGO Gene Nomenclature Committee (http://www.genenames.org/aboutHGNC.html).
Number of amino acids;
Molecular mass;
Theoretical isoelectric point (pH units).
Fig. 2.
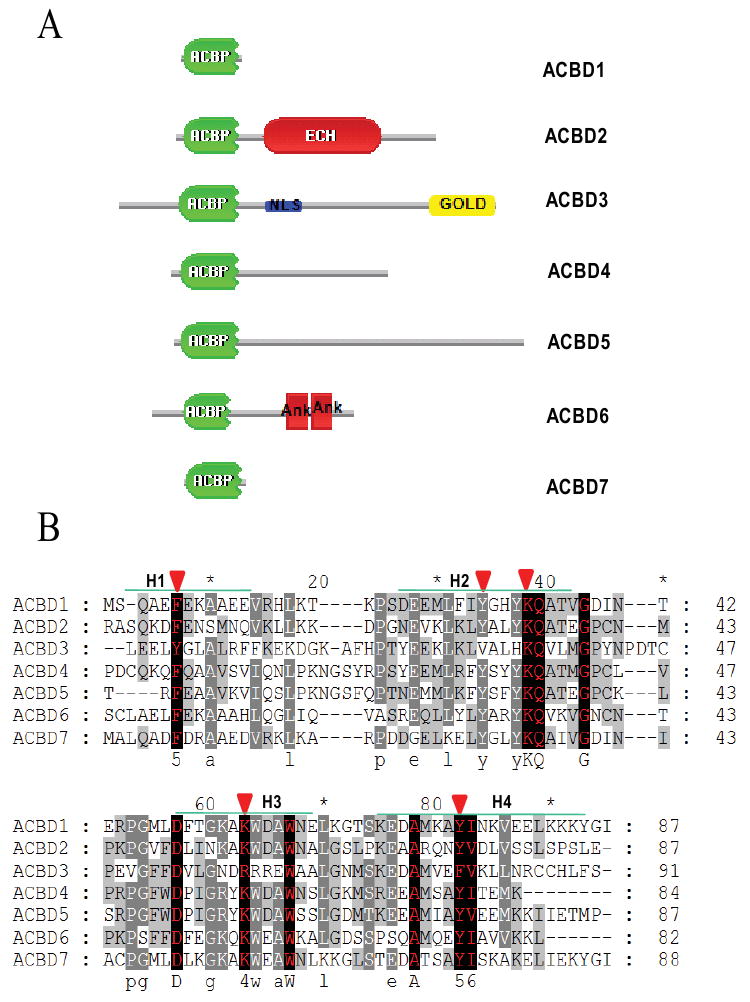
Schematic domain architecture and sequence alignment of ACBD3 and other ACBP domain-containing proteins in humans. A. Organization of ACBP domain-containing proteins. ACBD3 contains an ACBP domain (shown in green) at the N-terminus, a GOLD (for for Golgi dynamics) domain at the C-terminus, and a putative nuclear localization signal (NLS) located between the ACBP- and GOLD- domains. ACBD2 has one enoyl-CoA hydratase/isomerase (ECH) domain closer to the C-terminus. ACBD6 possesses two Ankyrin repeats (Ank) at the C-terminus. B. The positions of the four alpha-helices are indicated (H1-4). Red arrowheads indicate the important binding sites for acyl-CoA type ligands [62]. The conserved sites are in red with black background. C. A neighbor-joining (NJ) tree calculated from the amino acid sequences of human and mouse ACBP domain-containing proteins. The numbers at each node indicate the percentage of bootstrap values from 1000 resamplings. Bar indicates that the estimated genetic distance is proportional to the horizontal length of each branch.
3.1. Acyl-Coenzyme A binding domain containing 1 (ACBD1)
The ACBD1 (previously known as ACBP or liver ACBP, L-ACBP) gene encodes a cytosolic polypeptide of approximately 10-kDa, that binds long-chain (C12-C22) acyl-CoA esters with high affinity [20-25]. ACBD1 is recognized as a protein important for intracellular transport of acyl-CoAs, a function that can be traced back to very early evolutionary history [26]. Previous studies have suggested that ACBD1 also functions as pool former and transporter of cytosolic acyl-CoA either protected against acyl-CoA hydrolases or protecting fundamental cellular processes from being affected by the long-chain acyl-CoA esters [27,28]. Additionally, ACBD1 was also found in the nucleus of rat liver cells, where it involved in regulation of acyl-CoA-dependent processes [29]. Recently, ACBD1 has been proposed to play a role in vesicular trafficking by targeting to the endoplasmic reticulum (ER) and Golgi in a ligand-binding-dependent manner, i.e. either targeting to ER after binding with acyl-CoA or targeting to Golgi induced by fatty acid [30].
Studies examining DNA microarray using yeast acyl-CoA-binding protein (Acb1p, the ortholog of ACBD1) deletion and expression of Acb1p mutant protein, which is unable to bind acyl-CoA, revealed an Acb1p-dependent connection between fatty acid metabolism and transcriptional regulation of phospholipid biosynthesis [31]. This connection was due either to the formation of the Acb1p–acyl-CoA ester complex directly, or to the ability of Acb1p to donate acyl-CoA esters to utilizing systems [31]. Although both ACBD1 and liver-type fatty acid binding protein (L-FABP) play a similar role in microsomal phosphatidic acid biosynthesis via shuttling acyl-CoA between the mitochondria and ER (microsomes), only ACBD1 protects the long chain fatty acyl-CoAs from microsomal acyl-CoA hydrolase activity and therefore ACBD1 may affect the fatty acid-mediated gene regulation through the availability of acyl-CoA [32,33]. Furthermore, Acb1p has been shown to be involved in ceramide synthesis and normal vacuole function [34].
ACBD1 was also previously known as diazepam binding inhibitor (DBI) [20] and endozepine [21], a peptide cleaved by endo- and exo-peptidases to form bioactive processing products [35]. DBI and some of its processing products were reported to displace benzodiazepines from the plasma membrane GABAA receptor and the mitochondrial translocator protein (18 kDa; TSPO), a protein previously known as the peripheral-type benzodiazepine receptor, and induce cholesterol transfer into the mitochondria of steroidogenic cells, the rate-determining step in steroid biosynthesis [36,37]. Thus, the interaction between ACBD1 and TSPO is thought to be critical in the acute stimulation of adrenal steroidogenesis by adrenocorticotropic hormone (ACTH) [36-38].
3.2. Acyl-Coenzyme A binding domain containing 2 (ACBD2)
ACBD2 (MW: 40.18 kDa) is also known as peroxisomal D3, D2-enoyl-CoA isomerase (PECI) [39] or diazepam-binding inhibitor-related protein 1 (DRS-1) [40]. ACBD2 contains an N-terminal ACBP-like domain and a C-terminal enoyl-CoA isomerase/hydratase (ECH) domain [41]. ACBD2 acts to isomerize 3-cis and 3-trans double bonds into the 2-trans form in a range of enoyl-CoA species. Human ACBD2 has been found to be localized to peroxisomes by subcellular fractionation and immunofluorescence microscopy [39]. Since ACBD2 is essential for the metabolism of unsaturated fatty acids, it may be a candidate gene involved in defects of the peroxisomal fatty acid metabolism. Actually, ACBD2 has linked with acquired aplastic anemia via serving as an autoantigen eliciting immune attack against hematopoietic stem cells [40,42].
Additionally, an alternatively spliced, partially processed human ACBD2 gene has been linked to human antigens recognized by autologous antibodies in patients with renal cell carcinoma [43]. ACBD2 has also been linked to human hepatocellular carcinoma, where it is associated with hepatocellular carcinoma-associated antigen 64 [44]. Therefore, it would be interesting to examine whether defects in peroxisomal fatty acid metabolism are related to cancer development.
Furthermore, certain intermediates of extramitochondrial fatty acid oxidation (e.g., the long-chain dicarboxylic fatty acids) impair mitochondrial function. Some of these intermediates are implicated as modulators of gene expression through their interaction with the peroxisome proliferator-activated receptor [45]. Moreover, increased peroxisomal branched chain fatty acid oxidation during fatty acid metabolism has been proposed to be a marker for prostate malignant transformation [46].
3.3. Acyl-Coenzyme A binding domain containing 3 (ACBD3)
ACBD3 (MW: 60.59 kDa) was first identified by yeast 2-hybrid screening and mammalian cell co-precipitation studies as a TSPO (PBR) and Protein Kinase-A Regulatory Subunit Iα (PKARIA)-binding protein. ACBD3 was named as the PBR-associated protein 7 (PAP7) and as the Golgi complex-associated protein GCP60 [3,5]. Subsequently, several cellular signaling pathways were linked with this protein. After ACBD3 is released during the fragmentation of the Golgi, ACBD3 can interact in a tissue and cell-specific manner with (i) TSPO and PKARIA to mediate steroidogenesis, (ii) divalent metal transporter 1 (DMT1) and a 30 kDa brain-enriched member of the Ras family of small monomeric G proteins (Dexras1) to regulate iron homeostasis, (iii) a golgin-160 fragment to regulate cell apoptosis, and (iv) a membrane associated protein, numb homolog (Drosophila) to regulate neuronal differentiation. These pathways will be discussed in detail (see below).
3.4. Acyl-Coenzyme A binding domain containing 4 (ACBD4)
ACBD4 (MW: 34.76 kDa) is one of the nineteen most up-regulated genes known to be induced by the histone deacetylase inhibitor valproic acid (VPA) in a panel of cancer cell lines [47]. VPA is used clinically as an anticonvulsant and mood-stabilizing drug to treat many neurological conditions such as epilepsy, bipolar disorder, migraine headaches, and schizophrenia. However, the biological function of ACBD4 is still not well understood.
3.5. Acyl-Coenzyme A binding domain containing 5 (ACBD5)
Previous studies have shown that ACBD5 (MW: 54.79 kDa) is up-regulated in wild-type neurospheres, a free-floating structure generated by neural stem cells (NSCs) in vitro, compared with PTEN null neurosphere cultures [48]. Genes up-regulated in PTEN null neuroshperes include doublecortin, glutamate receptors, GABA receptors, and glutamate dehydrogenase. Considering the roles of these proteins together with ACBD5 in brain growth and development, we speculate that ACBD5 may be linked to cell differentiation and metabolism.
3.6. Acyl-Coenzyme A binding domain containing 6 (ACBD6)
Human ACBD6 (MW: 31.15 kDa) possesses two Ankyrin repeats, each approximately 33 amino acids long. These repeats function in protein-protein interactions through a core helix-loop-helix structure, with a beta-hairpin/loop region projecting out from the helices at a 90° angle [49]. ACBD6 prefers to bind unsaturated long-chain acyl-CoAs (C18:1-CoA and C20:4-CoA), rather than saturated, C16:0-CoA, acyl species. Since ACBD6 expression is restricted to bone marrow, spleen, placenta, cord blood, circulating CD34(+) progenitors, and embryonic-like stem cells derived from placenta, it is assumed that ACBD6 functions in hematopoiesis and blood vessel development [50].
Moreover, ACBD6 is one of seven genes that are responsive to dihydrotestosterone (5α-DHT) treatment [51]. Since DHT is a major bioactive androgen, these results indicate that ACBD6 is an androgen receptor target gene.
3.7. Acyl-Coenzyme A binding domain containing 7 (ACBD7)
ACBD7 (MW: 9.79 kDa) is expressed in spleen, thymus and brain, and is very similar to ACBD1 in predicted amino acid sequence. Due to its expression in brain, it was previously called brain ACBP (B-ACBP) [26]. However, there have been no reports solely concerning ACBD7, likely because ACBD7 is not as ubiquitous as ACBD1.
3.8. Other ACBP domain-containing proteins
Endozepine-like peptide (ELP; MW: 9.86 kDa), also known as ACBD1-isoform 5 or testes ACBP (T-ACBP), is a testis-specific isoform of the ubiquitous ACBD1 [26]. ELP is highly expressed at both the mRNA and protein levels in the late haploid stages of spermatogenesis [52]. Although it is highly conserved, along with ACBD1, in diverse mammals and shares a similar ability to bind mid–long chain acyl–CoA, it is considered an essential component of haploid sperm. Only two pseudogenes corresponding to ACBD1-isoform 5 have been found in the primate testis, including humans. Thus, it has been proposed that a slow process of the gene silencing occurs during evolution in humans. ELP may be related to the subfertility of human males, compared with non-human primates [53].
4. Characteristic features of domain architecture and evolutionary history
4.1. ACBP domain
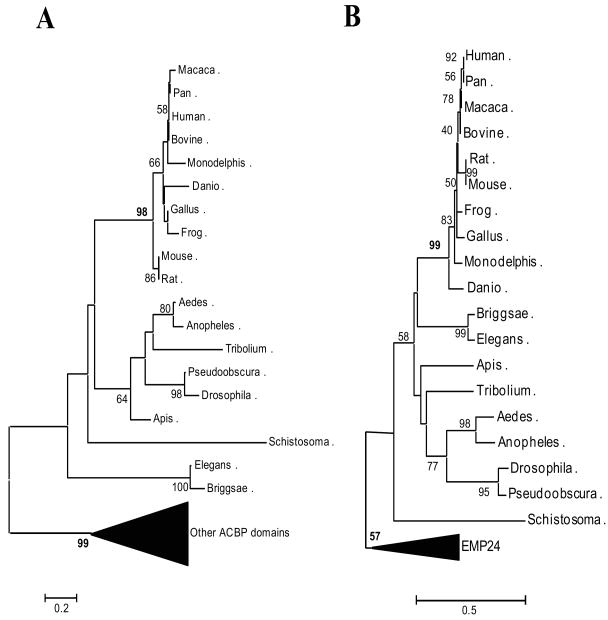
The prototypical and archetypal member of the family of ACBP domain-containing proteins is Acyl-CoA-binding protein (ACBD1, ACBP or DBI). ACBD1 binds medium- and long-chain acyl-CoA esters with high affinity. ACBD1 likely acts as an intra-cellular carrier of acyl-CoA esters with multiple functions [24,54]. Additionally, numerous physiological and biochemical functions have been reported for ACBD1 under the name DBI, because of the ability of DBI to displace diazepam from GABAA receptor, thus acting as a putative neurotransmitter [35,55] and a regulator of insulin release from pancreatic cells [56,57]. ACBD1 also acts as a mediator in corticotropin-dependent adrenal steroidogenesis by interacting with the mitochondrial TSPO protein. TSPO is a high affinity drug and cholesterol binding protein, mediating cholesterol import into mitochondria, the rate-determining step in steroid biosynthesis [58]. ACBD1 is widely distributed in the tissues of vertebrates, insects, plants and yeast. Six other proteins have been found to possess an ACBP-domain.
The 3-D structures of the ACBP domains of ACBD1, ACBD2 and ACBD6 have been deduced by NMR spectroscopy and X-ray diffraction. Although there are conformational differences in loops and turns among the proteins, the ACBD domains are largely conserved, consisting of four short alpha-helices and three connecting beta-strands [23,59,60]. The predicted 3D structures of the rest ACBP domain-containing proteins were compared with their homologous models generated through Swiss Model Server under project (optimize) mode (http://swissmodel.expasy.org/SWISS-MODEL.html) (Fig. 3). All these ACBP domains have a similar electrostatic surface potential, which may suggest that ACBDs possess a positive binding cavity of Acyl-CoA. ACBP binds acyl-CoA through not only the hydrophobic interactions, but also the electrostatic interactions between the protein and its substrates [61,62].
Fig. 3.
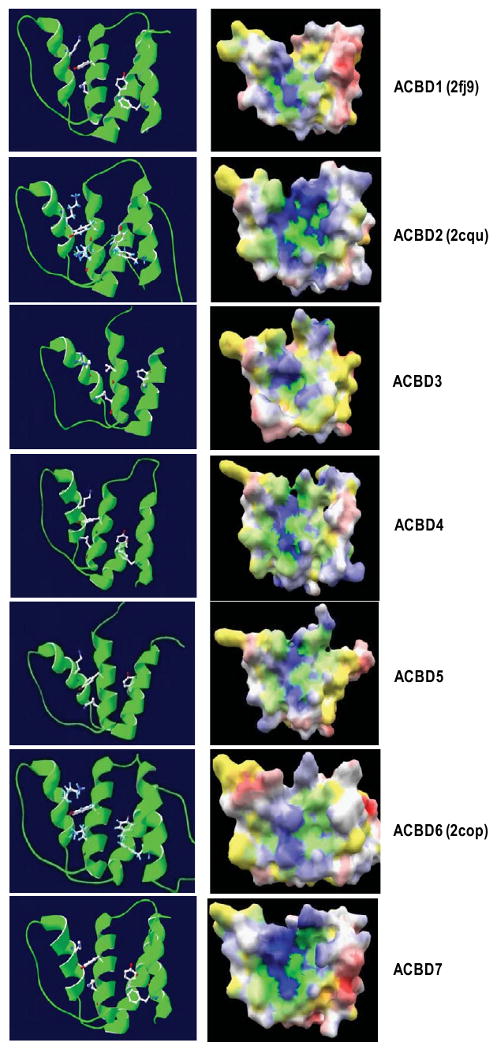
Comparison of the 3D structures of ACBP domains from each ACBP domain-containing protein. Left panel: Ribbon presentation of 3D models of each ACBP domain. The five conserved amino acids comprising the acyl-CoA binding sites are presented in a ball-and stick model with CPK colors. Right panel: Electrostatic potentials are mapped on the protein surface colored from red (negative) to blue (positive) and yellow (hydrophobic) using eF-Surf service (http://ef-site.hgc.jp/eF-surf) and jV Version 3.6 [109]. The positive binding cavity is shown. Crystal structures are from human ACBD1 (2FJ9), ACBD2 (2CQU) and ACBD6 (2COP). Homology models are deduced via the Swiss-model services using NMR- and/or crystal structures of human ACBD1 and ACBD6 (2FJ9 and 2COP) as templates.
Further analysis of the molecular surfaces revealed some differences in electrostatic potentials between the ACBP domains of ACBD3 and ACBD1 (Fig. 3). The binding surface of the palmitoyl-coenzyme A ligand in ACBD3 is slightly more negatively charged than ACBD1. However, the four to five residues responsible for ligand binding are similar [63-65]. These residues provide positive patches, which are assumed to be important for substrate binding via the electrostatic interactions between the adenosine 3′-phosphate of the CoA and ACBP [63,66] (Fig, 4). It is important to note that substitutions among the conserved residues important for substrate binding may be responsible for conferring binding properties for acyl-CoA with variable chain lengths; for instance, trypanosome acyl-CoA-binding protein, which has two amino acid substitutions among the five important residues, possesses the greatest affinity for shorter chain acyl-CoAs, such as lauroyl-CoA [67]. Generally, the acyl-CoA binding sites, which are illustrated by positively charged residues, are located in a hydrophobic pocket on the surface of each ACBP domain.
Fig. 4.
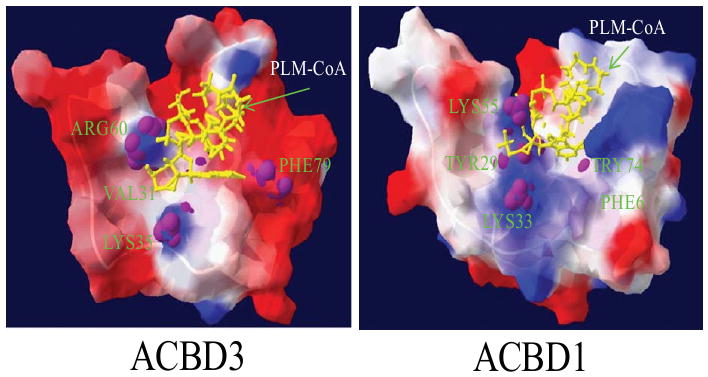
Electrostatic potential of the ACBP domains from human ACBD3 (left) and ACBD1 (right), superimposed with a stick representation of the palmitoyl-coenzyme A ligand (in yellow) and backbone of bovine ACBP as a template (PDB: 1ACA) [62]. The electrostatic molecular surfaces are colored by electrostatic potential: red, negatively charged regions, blue, positively charged areas, and white, hydrophobic surface. The four/five residues, which are necessary for ligand binding, are indicated in the figure [63].
4.2. GOLD domain
The beta-strand rich GOLD domain has been found in several eukaryotic Golgi and lipid-trafficking proteins. The typical GOLD domain is between 90 and 150 amino acids in length. Two categories of GOLD domain-containing proteins have been proposed: the p24-like category which has a single GOLD-domain (category 1) and those with one GOLD domain at the extreme amino or carboxyl terminus plus an additional lipid-binding related domain (category 2) [68]. The GOLD domain-containing proteins encompass the P24 families of proteins, which include endosome membrane proteins (emp24s) and a calnexin-associated integral membrane glycoprotein of the ER (GP25L) in eukaryotes, as well as other category 2 members, such as GCP60 (ACBD3).
GOLD domain-containing proteins are involved in diverse functions, such as protein-protein interactions [68], but they are all related to a basic cellular process, such as protein-lipid binding [69]. The main function of the GOLD domain is to provide a binding site for membrane lipids and proteins. The GOLD domain coexists with other conserved globular domains, such as the pleckstrin homolog (PH) domain involved in protein-protein interactions, the RUN domain associated with Ras-like GTPase signaling [70], the CRAL/TRIO domain named after cellular retinal and the TRIO guanine exchange factor, which binds small lipophilic molecules such as retinol, vitamin E, squalene, and inositol [71], the FYVE zinc finger domain, which binds lipids, the oxysterol-binding domain responsible for sterol binding, and the ACBP domain binding medium- and long-chain acyl-CoA esters. On the other hand, p24 proteins, which contains a single GOLD domain, are involved in COPI (referring to specific coat protein complex I)- and COPII-mediated vesicular transport as major membrane components of coated vesicles [72]. Therefore, GOLD-domain containing proteins may function by bringing cargo forward from the ER and binding to coat proteins by their cytoplasmic domains [68,72].
ACBD3, a GOLD-domain containing protein, possesses one ACBP domain at its N-terminus and one GOLD domain at its C-terminus (Fig. 2). Overexpression of a portion of ACBD3, including the GOLD domain, causes abnormality of the Golgi structure as well as disruption of protein transport from the ER to the Golgi [3]. The GOLD domain seems to be responsible for the Golgi localization of ACBD3, whereas the ACBP domain functions to presumably bind acyl-CoA derivatives. Thus, the GOLD domain of ACBD3 appears to confer the ability in determining its intracellular (Golgi) localization and trafficking. Although the ACBP domain of ACBD3 remains functionally elusive, it likely reflects specific biochemical features of this protein.
4.3. Vertebrate ACBD3
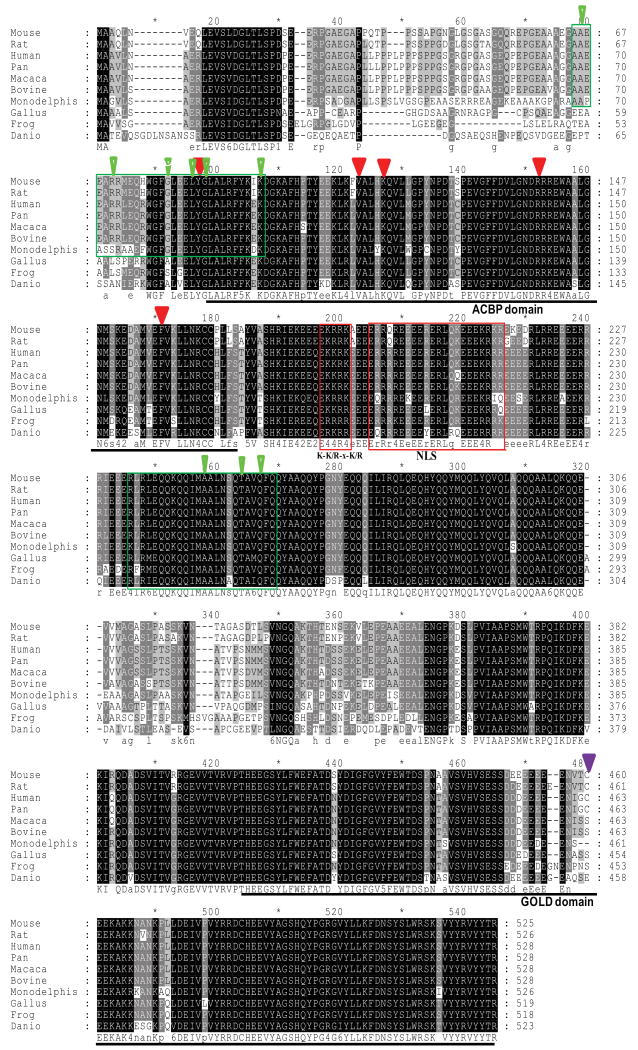
Sequence analysis showed that vertebrate ACBD3 proteins form a highly conserved protein family (Fig. 5). All members of vertebrate ACBD3 proteins have one extra conserved coil-coil domain and one or two nuclear localization signals (NLS) between the two major ACBD and GOLD domains. Additionally, portions of the human ACBD3 protein from residue 68–97 have some similarities to one isoform (designed as N0 splice) of dual specificity protein kinase A anchoring protein 1 (D-AKAP1) targeting domain 1–30, which binds to both the regulatory subunits (RI and the RII) of cAMP-dependent protein kinase (PKA) [73,74]. ACBD3 sequences are highly conserved in this region among mammals, but not in non-mammalian vertebrates. In contrast, the portion of the protein with similarity to the dual type I and type II domain (317–338) of D-AKAP1 and hPAP7 238–259 are identical for all vertebrates (Fig. 5) [19,75]. This observation may suggest that there is one identical mechanism existing in mammals, but with some variations in cAMP-ACBD3-mediated signaling in non-mammal vertebrates. Since the partial sequence of the AKAP disrupting peptide overlaps with the N-terminal ACBP domain, it is possible that the AKAP activity of ACBD3 depends on this domain.
Fig. 5.
Multiple alignment of ACBD3 proteins from vertebrates. Sequences were aligned using the ClusterW algorithm. Sequences are designated by species and were obtained from Genbank: human (NP_073572), mouse (NP_573488), rat (AAH83877), pan (gorrila; XP_001140648), bovine (XP_001251579), macaca (monkey; XP_001091471), Monodelphis (opossum; XP_001368267), Gallus (chicken; NP_001026214), frog (AAI35954), and danio (zebrafish; CAI21107). Shaded areas represent conserved amino acids. The numbers indicate amino acid positions. Conserved amino acids are indicated by upper case letters, while less conserved amino acids are represented by lower case letters at the foot of the alignment. The two main domains, ACBP and GOLD, are indicated with underlines. Two putative nuclear localization signals are boxed in red. The five putative acyl-CoA ligand-binding residues are marked with arrowheads. The green boxes indicate the N0 isoform of D-AKAP1 targeting domain 1–30, human PAP7 peptide residue 68–97, the dual type I and type II domain (317–338) of D-AKAP1, and hPAP7 238–259. Green arrows indicate conserved sites, as shown previously. The purple arrow indicates the location of cys463 in human ACBD3 that interacts with golgin-160-(140–311) in a redox dependent reaction.
In contrast to vertebrate ACBD3, a database sequence search indicated that invertebrate ACBD3 members are more divergent in the sequence. None of the invertebrate ACBD3s contains an NLS between the ACBD and GOLD domains (data not shown). So far the only experimental report mentioning ACBD3 from invertebrates is the ACBD3 from Drosophila [7]. This vertebrate-specific sequence feature may attribute some more advanced functions in this highly evolved group of organisms. Nevertheless, both vertebrate and invertebrate ACBD3s share a conserved function in the numb signaling in asymmetric cell division [7].
Rodent and human ACBD3 proteins were extensively studied by our group. We found that ACBD3 is a mediator of the cAMP-induced protein-protein interactions driving cholesterol transport into mitochondria. Thus, ACBD3 proteins increased steroid formation by steroid synthesizing cells, such as the Leydig cells of the testis [5,19]. Reduction of ACBD3 gene expression in Leydig cells attenuated hormone-induced steroid formation [5]. Expression of ACBD3 proteins lacking the ACBP domain in Leydig cells reduced the hormone–stimulated steroid production. Deletion of the mouse ACBD3 gene was embryonic lethal as early as at 8.5 days [7] (Wang, Liu, Papadopoulos, unpublished).
Nevertheless, knockout of the mouse ACBD3 gene revealed a potential link between ACBD3 and numb signaling in asymmetric cell division [7]. Rat ACBD3 was shown to be involved in the N-methyl-D-aspartate (NMDA) receptor-nitric oxide (NO)-Dexras1-PAP7-DMT1-iron uptake signaling cascade [6]. Human ACBD3 was found to regulate nuclear translocation and golgin-160 fragments (GOLGA3, Golgi autoantigen, golgin subfamily a, 3) [3,76]. Nuclear translocation of golgin-160-(140–311) is a highly coordinated event regulated not only by the cleavage of the golgin-160 head but also by the oxidation state of GCP60 [16,17]. Human ACBD3 is highly expressed in steroidogenic tissues, where it follows the pattern of PRKAR1A expression, suggesting that it participates in PRKAR1A-mediated tumorigenesis and hypercortisolism [77].
Protein functions can be attributed to specific protein domains within their molecular architecture. Protein domains serve as the basic functional units of the proteins as well as the basic evolutionary units that drive new protein formation through domain fusion, recombination and shuffling, thus promoting proteomic complexity [78-80]. Although two main domains are present in the ACBD3, it is clear that there is no domain shuffling in ACBD3 proteins from worms to humans (Fig. 6; Table 2). Phylogenetic analysis of the ACBD3 domains shows that ACBD3 has unique features distinct from other ACBP proteins. The estimated genetic distance between ACBD3 and other ACBP domain containing proteins is remarkable. Even among the important conserved binding sites for acyl-CoA type ligands, four of the five amino acids have been replaced by an alternative amino acid with various biochemical properties: one hydrophilic polar/hydrophobic nonpolar amino acid replacement (Tyr87Phe), two hydrophobic/hydrophilic amino acid replacements (Tyr87Phe and Phe161Tyr) and one similar amino acid replacement (Arg142Lys) (Fig. 2B). Interestingly, another domain of ACBD3, the GOLD domain, is also highly conserved from worm to humans (Fig. 5 and 6), suggesting that ACBD3 is unique as a tethering protein in the assembly of Golgi-membrane-associated protein complexes.
Fig. 6.
Neighbor-joining (NJ) trees of the ACBP domain, the GOLD domain from ACBD3 proteins, and other domain-containing proteins. A. ACBP domain-containing proteins. ACBP domain-containing proteins (other than ACBD3) are in a compressed branch with a strong bootstrap support (99% from 1000 resamplings). B. GOLD domain-containing proteins. The EMP24 family (integral membrane components of endoplasmic reticulum-derived COPII-coated vesicles) is clearly in a separated compressed clade in this NJ tree, suggesting that the two main branches are present in the GOLD/EMP24 family. The number at each node represents the percentage of NJ bootstrap values from 1000 replicates. The estimated genetic distance is proportional to the horizontal length of each branch. The GenBank accession numbers and other abbreviations used here are listed in Table 2.
Table 2.
Accession numbers of the genes used in this study
| Abbreviationa | Species | Accession No. |
|---|---|---|
| Vertebrates | ||
| ACBD3 | Homo sapiens | NP_073572 |
| ACBD3 | Mus musculus | NP_573488 |
| ACBD3 | Rattus norvegicus | AAH83877 |
| ACBD3 | Pan troglodytes | XP_001140648 |
| ACBD3 | Bos Taurus | XP_001251579 |
| ACBD3 | Macaca mulatta | XP_001091471 |
| ACBD3 | Monodelphis domestica | XP_001368267 |
| ACBD3 | Gallus gallus | NP_001026214 |
| ACBD3 | Xenopus laevis | AAI35954 |
| ACBD3 | Danio rerio | CAI21107 |
| ACBD1-iso1 | Homo sapiens | NP_065438 |
| ACBD1-iso2 | Homo sapiens | NP_001073332 |
| ACBD1-iso3 | Homo sapiens | NP_001073331 |
| ACBD2 | Homo sapiens | NP_006108 |
| ACBD4 | Homo sapiens | NP_078998 |
| ACBD5 | Homo sapiens | NP_001035938 |
| ACBD6 | Homo sapiens | NP_115736 |
| ACBD7 | Homo sapiens | NP_001034933 |
| ACBD1-iso5 | Homo sapiens | EAW90652 |
| ACBD1-iso1-mouse | Mus musculus | NP_001033088 |
| ACBD1-iso2-mouse | Mus musculus | NP_031856 |
| ACBD2-mouse | Mus musculus | NP_035998 |
| ACBD4-mouse | Mus musculus | NP_080264 |
| ACBD5-mouse | Mus musculus | NP_001095907 |
| ACBD6-mouse | Mus musculus | NP_082526 |
| ACBD7-mouse | Mus musculus | NP_084339 |
| ACBD1-iso5-mouse | Mus musculus | NP_067269 |
| EMP24_human | Homo sapien | NP_998766 |
| EMP24_Pan | Pan troglodytes | XP_001164357 |
| EMP24_Pongo | Pongo pygmaeus | Q5R9S0 |
| EMP24_Bos | Bos Taurus | XP_870562 |
| EMP24_Canis | Canis familiaris | XP_547929 |
| EMP24_Rattus | Rattus norvegicus | XP_001061720 |
| EMP24_Mus | Mus musculus | NP_001028647 |
| EMP24_Monodelphis | Monodelphis domestica | XP_001374103 |
| EMP24_Xenopus | Xenopus laevis | AAI06450 |
| EMP24_Tetraodon | Tetraodon nigroviridis | CAG10605 |
| EMP24_Danio | Danio rerio | NP_001018152 |
| Invertebrates | ||
| ACBD3_Aedes | Aedes aegypti | EAT42117 |
| ACBD3_Anopheles | Anopheles gambiae | XP_312883 |
| ACBD3_Tribolium | Tribolium castaneum | XP_972065 |
| ACBD3_pseudoobscura | Drosophila pseudoobscura | XP_001354572 |
| ACBD3_Drosophila | Drosophila melanogaster | NP_608348 |
| ACBD3_Apis | Apis mellifera | XP_394773 |
| ACBD3_briggsae | Caenorhabditis briggsae | CAE68127 |
| ACBD3_elegans | Caenorhabditis elegans | NP_001041025 |
| ACBD3_Schistosoma | Schistosoma japonicum | AAW25632 |
In the remaining sections of the review, we will focus on the distribution and function of ABCD3, a protein that we initially described in 2001 to play a critical role in the regulation of steroid hormone biosynthesis [5].
5. Localization and function
5.1. Tissue distribution of ACBD3
ACBD3 is highly expressed in several different types of cells and tissues, including Leydig and germ cells of the testis, theca and granulosa cells of the ovary, fasciculata reticularis and glomerulosa cells of the adrenal gland, the hippocampus, and specific neuronal and glial cells of the cortex [5]. ACBD3 immunoreactivty has also been detected in the paraventricular and superoptic nuclei regions of the hypothalamus, as well as in liver and kidney [5,77].
5.2. Intracellular localization of ACBD3
ACBD3 was first observed to be intracellularly localized in the Golgi body, as well as in mitochondria [3,5]. However, during cell mitosis, ACBD3 is released from the Golgi body into the cytosol, where it binds to the mitochondrial TSPO protein [5,19], PRKARIA (protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) to target it to mitochondria [5], to golgin-160 caspase cleavage fragments (amino acids 140–311)[16], or to Numb-like protein, driving neuronal differentiation [7]. In the later case, ACBD3 plays a role in cellular asymmetric division in neural progenitor cell-fate specification. Since asymmetric cell division occurs widely from embryonic stem cells to other mammalian cultured cell lines, it would be of interest to further investigate this aspect of ACBD3 function [81].
As noted earlier in this review, the vertebrate ACBD3s can have one or two nuclear localization signals (NLS) of the form ER4E4RERLQKE3KR3. NLS have been found in 193 proteins, of which 98% are verified nuclear proteins (http://cubic.bioc.columbia.edu/cgi/var/nair/resonline.pl). Moreover, ACBD3 contains nine predicted DNA binding sites with the sequence motifs E/DRnED (with n>4); within a helix-loop-helix domain between the ACBP- and the GOLD- domains. However, immunolocalization of ACBD3 and the expression of exogenous ACBD3 in cells failed to show nuclear localization of the protein (Fig. 7) [19]. Nevertheless, it cannot be excluded that ACBD3 might move to the nucleus following the Golgi disassembly under certain circumstances, such as apoptosis.
Fig. 7.
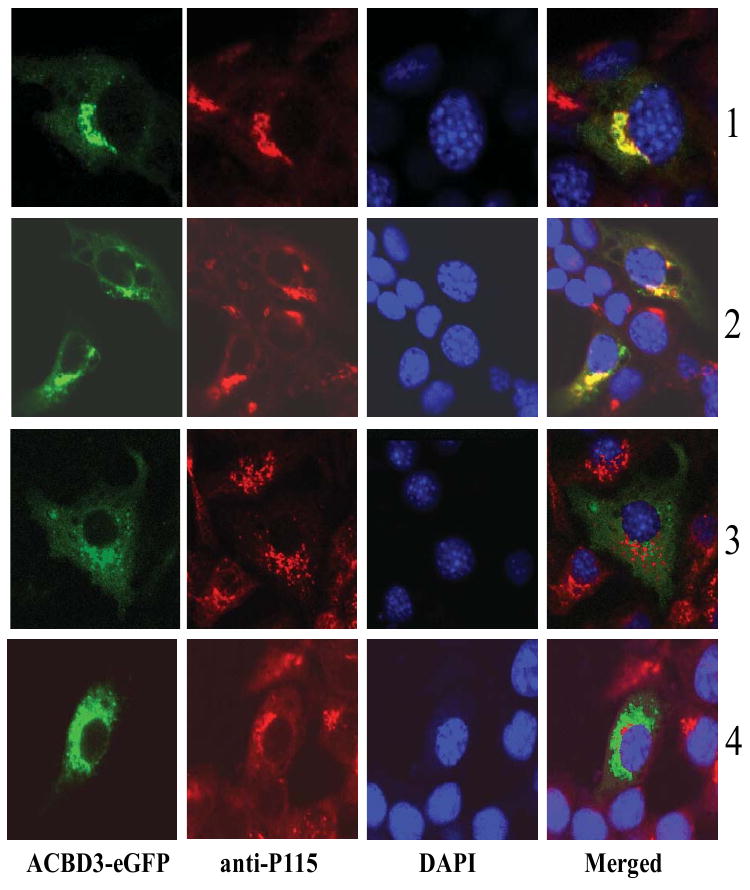
Redistribution of ACBD3 after MA-10 cells were treated with hCG (2), H-89 (3), and hCG/H-89 (4). The MA-10 cells transfected with plasmid ACBD3-eGFP without any treatment was used as control (1). The vesicle docking protein p115 was used as a marker of Golgi body, while DAPI staining marked the nucleus.
6. ACBD3 in steroidogenesis
The mechanism by which hormones, such as the gonadotropin luteinizing hormone (LH), and the corticotrophin ACTH, bind to G-protein coupled plasma membrane receptors, inducing cAMP synthesis and stimulating steroid formation has been an area of interesting research [82-84]. Although many details are still unknown, it is clear that cAMP rapidly activates cholesterol transport from intracellular stores into mitochondria, where steroidogenesis begins. The rate-limiting step in the hormonal regulation of steroidogenesis is the transfer of cholesterol into mitochondria, where TSPO [85], ACBD1 (DBI) [86] and ACBD3 (PAP7) [84] play critical roles.
ACBD3 is critical for the formation of a macromolecular signaling complex (scaffold) through its interactions with TSPO and PRKARIA, acting as an A-kinase anchoring protein (AKAP) [5,19,75,77,87,88]. This interaction allows for the targeting of a few molecules of cAMP to the mitochondria to locally activate (phosphorylate) substrates. Such activation is involved in the steroidogenic acute regulatory protein (StAR) initiation of cholesterol transfer, via TSPO, to the inner mitochondrial membrane cytochrome P450 side-chain cleavage (CYP11A1) enzyme, which catalyzes the metabolism of cholesterol to pregnenolone [19,84,89].
A series of ACBD3 suppression and overexpression studies, along with expression of dominant negative mutants and in vitro reconstitution experiments in steroid synthesizing and control cells, support the idea that upon hormonal stimulation, the mitochondrial proteins TSPO and voltage-dependent anion-selective channel (VDAC) form a complex that allows for the recruitment and anchoring of ACBD3, PRKAR1A, ACBD1 and StAR on the surface of the mitochondria and subsequent cholesterol transfer [5,19,75,77,84,88]. Similar conclusions were reached concerning the orthologous human gene, hACBD3, which is highly expressed in steroidogenic tissues and follows the pattern of PRKARIA expression. In this regard, hACBD3 appears to participate in PKARIA-mediated hormone-independent hypercortisolism [75,77].
ACBD3 is also mechanistically involved in the Golgi fragmentation elicited by human chorionic gonadotropin (hCG) in the steroidogenic testicular Leydig cells. ACBD3 is released from the Golgi after hCG treatment, but not released after treatment with a protein kinase A inhibitor, H-89 (Fig. 7). ACBD3 is distributed in the cytosol after treatment with both hCG and H-89 (Fig. 7), while hCG-stimulated steroid hormone production is blocked (Liu J and Papadopoulos V, unpublished). Since H-89 functions to block Golgi fragmentation and dispersal in mitosis, and also blocks ER export [90,91], it is clear that ACBD3-mediated hCG-induced cholesterol transport and steroid formation is closely related to proper Golgi fragmentation as well as normal ER-Golgi trafficking.
A gene interaction network centered on ACBD3, and constructed through the IntNetDB server (http://hanlab.genetics.ac.cn/sys/) [92], reveals that ACBD3 is related to steroid and cholesterol synthesis-related genes, including STAR (steroidogenic acute regulatory protein), SCP2 (sterol carrier protein 2), hormone nuclear receptor (NR0B1, nuclear receptor subfamily), Acyl coenzyme-related ACBD1 and BACH (acyl-CoA thioesterase 7), Golgi proteins (BLZF1, basic leucine zipper nuclear factor 1), lipid degradation protein (AZGP1, alpha-2-glycoprotein 1, zinc-binding), and several p24 family members (Fig. 8). These data suggest that ACBD3 functions as an AKAP in mitochondrial cholesterol transport. Cyclic AMP-dependent steroid hormone production serves as a central node in the steroidogenesis.
Fig. 8.
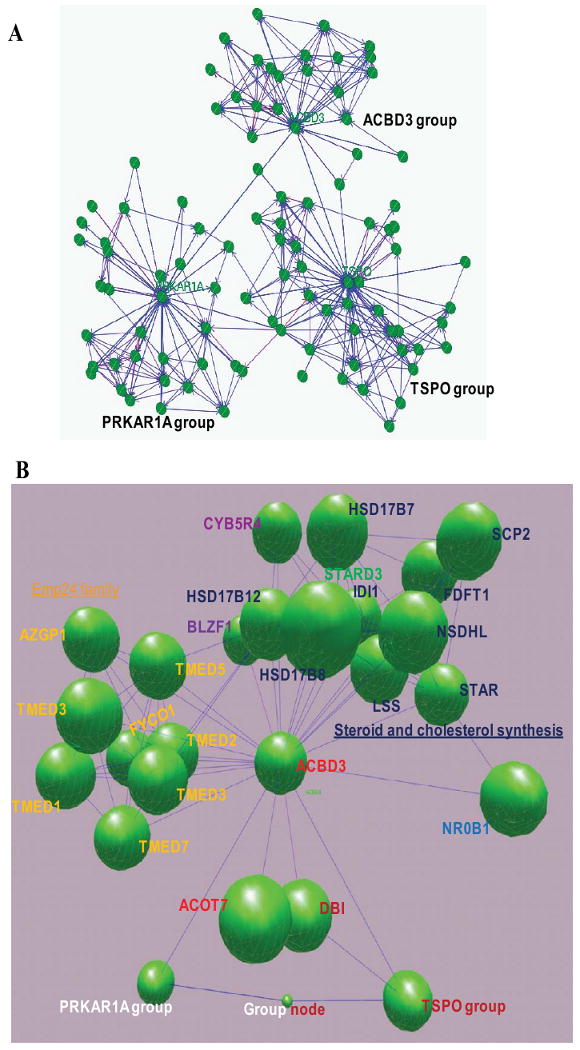
ACBD3, a central node in steroidogenesis. A. Gene networks were constructed through the IntNetDB server (http://hanlab.genetics.ac.cn/IntNetDB.htm) and visualized by BioLayout (http://www.biolayout.org) using human ACBD3 (Gene ID: 64746), TSPO (Gene ID: 706), and PRKAR1A (Gene ID: 5573) genes as probes, respectively. B. A 3D graph of the ACBD3 gene network, with collapsed TSPO- and PRKAR1A- groups. Steroid and cholesterol synthesis-related genes (in green) in the ACBD3 gene network: STAR, steroidogenic acute regulatory protein; SCP2, sterol carrier protein 2; FDFT1, farnesyl-diphosphate farnesyltransferase 1 (squalene synthase); NSDHL, NAD(P) dependent steroid dehydrogenase-like; LSS, lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase); IDI1, isopentenyl-diphosphate delta isomerase 1; STARD3, StAR-related lipid transfer (START) domain containing 3; HSD17B7, hydroxysteroid (17-beta) dehydrogenase 7; HSD17B12, hydroxysteroid (17-beta) dehydrogenase 12; HSD17B8, hydroxysteroid (17-beta) dehydrogenase 8; CYB5R4, cytochrome b5 reductase 4 (N-terminal cytochrome b5 and cytochrome b5 oxidoreductase). Hormone nuclear receptor (in dark blue): NR0B1, nuclear receptor subfamily 0, group B, member 1. Acyl coenzyme-related: DBI, diazepam binding inhibitor (GABA receptor modulator, acyl-Coenzyme A binding protein); BACH (ACOT7), acyl-CoA thioesterase 7 (brain acyl-CoA hydrolase). Golgi proteins: BLZF1, basic leucine zipper nuclear factor 1 (JEM-1). Lipid degradation: AZGP1, alpha-2-glycoprotein 1, zinc-binding. Emp24 family (in yellow): TMED1, transmembrane emp24 protein transport domain containing 1; TMED9, transmembrane emp24 protein transport domain containing 9; TMED5, transmembrane emp24 protein transport domain containing 5; TMED3, transmembrane emp24 protein transport domain containing 3; RNP24, transmembrane emp24 domain trafficking protein 2 (transmembrane emp24 domain trafficking protein 2); TMED7, transmembrane emp24 protein transport domain containing 7; FYCO1, FYVE and coiled-coil domain containing 1.
7. ACBD3 in apoptosis
In addition to the maintenance of the structure of the Golgi apparatus by interaction with giantin, ACBD3 is also involved in apoptosis through interaction with golgin-160 caspase cleavage fragments requiring a single, critical, oxidized cysteine residue in ACBD3 (Cyc-363) [3,16,17].
Golgin-160 is a ubiquitous protein in vertebrates, which localizes to the cytoplasmic side of the Golgi complex. The protein is composed of an N-terminal domain, containing a Golgi targeting sequence, a cryptic NLS, and three aspartates as caspase cleavage sites. The C-terminus contains a large coiled-coil domain. During the initiation of apoptosis, caspase cleavage of the head domain leads to the accumulation of cleavage products in the nucleus [93].
The significance of the presence of the golgin-160 fragment in the nucleus is not clear. ACBD3 demonstrates a strong interaction with caspase-generated golgin-160 fragments containing amino acids 140-311. ACBD3 holds the fragment in the cytoplasm through a single oxidized cysteine residue, when the fragment and/or ACBD3 are overexpressed. Thus it has been suggested that the nucleus localized golgin-160 fragments (amino acids 140-311) might promote cell survival after apoptotic insults, since overexpression of ACBD3 make cells more sensitive to apoptosis induced by staurosporine [17]. However, overexpression of the golgin-160 fragment (amino acids 140-311) in the cells with a caspase-resistant golgin cannot overcome the sensitivity of the apoptosis induced by staurosporine [94], suggesting that the cleavage of golgin in vivo seems to play a central role in the transduction of apoptotic signals. Also, to induce apoptosis, both Golgi apparatus fragmentation and disruption of actin filament organization induced by staurosporine are necessary steps. Therefore, the exact role of the Golgi protein ACBD3 in this process remains unclear. Moreover, the caspase-cleaved fragment of an additional Golgi protein (p115) also caused Golgi fragmentation and induced apoptosis in a similar fashion to golgin-160 [14,95]. Whether ACBD3 is also related to the p115 caspase-cleaved fragment awaits further investigation.
8. ACBD3 in iron-homeostasis
Iron-homeostasis is essential for neuronal function. Excess iron induces neurodegeneration. ACBD3 has been reported to be involved in iron homeostasis via its interaction with the divalent metal transporter 1 [DMT1, or SLC11A2 (solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2)] and Dexras1, a 30 kDa brain-enriched member of the Ras family of small monomeric G proteins [6]. Dexras1 has been identified as a target of neuronal nitric oxide synthase (nNOS) via the nNOS adaptor protein CAPON, enhancing NO –mediated signaling [96]. Dexras1 was found to bind to ACBD3; Then ACBD3 in turn binds to the divalent metal transporter (DMT1), an iron import channel. Finally, the NMDA (N-methyl-D-aspartic acid)-NO-Dexras1-ACBD3-DMT1 cascade was shown to mediate physiologic influences of NMDA on iron uptake [6].
9. ACBD3 in neurogenesis
Neurogenesis is one of the mechanisms underlying neuronal plasticity, enabling organisms to adapt to environmental changes and influencing learning and memory throughout life. During neurogenesis, neural progenitor cells divide asymmetrically to self-renew and produce a neuron by segregating cytosolic Numb proteins to one daughter cell. Numb proteins play two evolutionarily conserved roles in the maintenance of stem cell in mammalian neurogenesis: (i) determination of the fate of the progenitor neuronal cell and (ii) neuronal differentiation [97].
ACBD3 interacts physically with Numb through an essential Numb domain during mitotic Golgi fragmentation. ACBD3 is thought to diversify cell fates in asymmetric cell division. ACBD3 determines self-renewal and differentiation of neural progenitor cell via inhibition of neuron formation, due to its continued presence during the progenitor cell cycle [7]. Therefore, released ACBD3 during Golgi fragmentation in mitosis regulates Numb signaling through the subcellular asymmetrical distribution of ACBD3, which may provide a general mechanism to couple cell fate specification within the cell cycle. Taken together with other functions presented here, it is clear that ACBD3 plays a central role in several cellular signaling cascades after it is released from the Golgi apparatus in response to the second messengers cAMP and nitric oxide, either from hormone treatment of the cell or from synthesis by the enzyme nitric oxide synthase (NOS), in steroidogenesis and iron homeostasis. ACBD3 may also act via direct physical interaction with other signaling molecules, such as Golgi-160 large fragment and the Numb protein during mitosis in apoptosis and neurogenesis (Fig. 9).
Fig. 9.
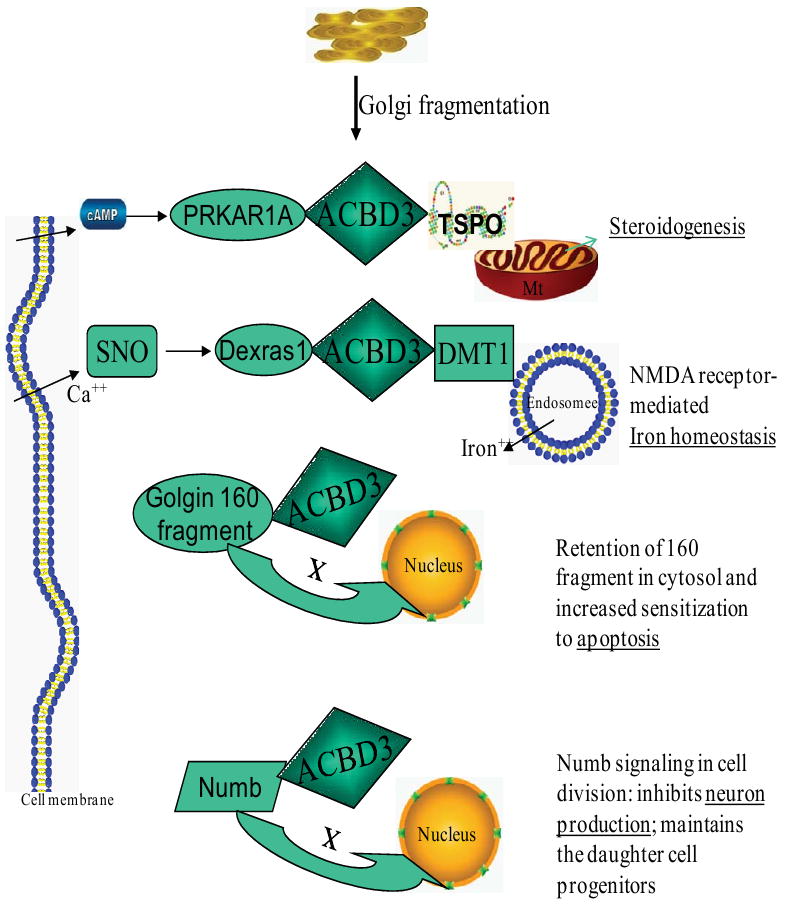
ACBD3-mediated signaling pathway and cellular regulation. Following release after Golgi fragmentation, ACBD3 interacts with several other signaling molecules. ACBD3 then regulates diverse cellular processes, including steroidogenesis, iron homeostasis, apoptosis, and neurogenesis. ACBD3 serves as an intermediate, and may be involved in the signaling protein complex related to steroidogenesis and NMDA receptor-mediated iron homeostasis. ACBD3 holds a Golgi 160 fragment and numb protein within the cytoplasm. ACBD3 then regulates cell apoptosis and neurogenesis. Mt, mitochondria; TSPO, translocator protein, 18 kDa; PRKAT1A, protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1); cAMP, cyclic adenosine monophosphate; DMT1, divalent metal transporter 1; Dexras1, a 30 kDa brain-enriched member of the Ras family of small monomeric G proteins; SNO, S-nitrosothiol; Golgin-160 fragment, caspase cleavaged fragment of the golgin family of Golgi-localized proteins; Numb, numb homolog (Drosophila).
10. ACBD3 in embryonic development
Generation of null alleles of the ACBD3 gene are embryonically lethal in mice [7] (Wang H, Liu J and Papadopoulos V, unpublished data). Although ACBD3 heterozygous mice are viable, fertile and similar to their wild-type littermates, no homozygous mutants survive postnatally. Embryos are dying between 8.5 to 10.5 days of age, suggesting that ACBD3 is essential for embryogenesis. Homozygotes are much smaller than the wild-type littermates at E8.5, with defects in axial turning and neural tube closure at E9.5. At E10.5, embryos become necrotic and are absorbed [7] (Wang H, Liu J and Papadopoulos V, unpublished data). In most PAP7 mutants, the most dramatic change seen was reduction of the thickness of the neuroepithelium, indicating reductions in progenitor cell numbers, since the vast majority of cells in the wild-type neuroepithelium are progenitor cells. Due to premature progenitor cell depletion the phenotype of ACBD3 mutants was deemed similar to the loss of both m-numb and Numbl [7,98,99].
11. ACBD3 in cancer
The expression profile of ACBD3 has been tightly linked with PKARIA in primary pigmented nodular adrenocortical disease (PPNAD) tissues. This indicates that ACBD3 may participate in PKARIA-mediated tumorigenesis and hormone-independent hypercortisolism [75]. Moreover, microarray data from published cancer-related databases has shown that ACBD3 is involved in cell cycle control and is regulated by retinoblastoma protein (pRB), a critical tumor suppressor [100].
A recent clinical trial examined the safety and efficacy of celecoxib, a selective COX-2 inhibitor, approved by the Food and Drug Administration for prevention of colon cancer in patients with familial adenomatous polyposis (FAP) syndrome. ACBD3 was found to be one of 173 genes (out of 9,128 screened genes) that showed a statistically significant (P < 0.001) difference in expression, upon pretreatment versus post-treatment with celecoxib, but not placebo [101]. Additionally, ACBD3 has been one of 51 candidate genes for discriminating between responders (PR) and non-responders (PD) to gefitinib (Iressa, ZD1839), an inhibitor of epidermal growth factor receptor-tyrosine kinase. Gefitinib has shown potent anti-tumor effects and improved symptoms and quality-of-life in a subset of patients with advanced non-small cell lung cancer (NSCLC). The expression of ACBD3 is 3.8 fold higher in non-responders (PD) than in responders (PR), suggesting that ACBD3 may play a role in the underlying mechanism of the response of lung cancer cells to gefitinib [102].
Finally, ACBD3 is upregulated by photodynamic therapy (PDT). PDT involves the use a photosensitizer, visible light, and tissue oxygen to generate cytotoxic reactive oxygen species. The reactive oxygen species cause tumor ablation. The study provides evidence that a subset of genes induced by PDT may be related to cell survival and the cell death response [103]. Interestingly, it is believed that PDT therapy is mediated at least in part via the mitochondrial TSPO [104], which directly interacts with ACBD3, as described above.
It has been widely accepted that asymmetric division occurs in stem cells. Asymmetric division results in one daughter cell that resembles the parent stem and a second cell that differentiates into a tissue-specific progenitor or a postmitotic cell. Disturbance of this delicately balanced asymmetric process has long been speculated to produce cancerous growth[105,106]. Cancer stem cells, which drive growth and metastasis, have been identified in leukemia and in solid tumors of the breast and brain. Moreover, inappropriate regulation of the self-renewal of somatic stem cells is believed to cause neoplastic proliferation and cancer formation [107]. Thus, the regulation of stem cell self-renewal and tumor suppression via asymmetric cell division is critical and delicate [105,108]. ACBD3 plays at least a partial role in the regulation of this balanced process in neurogenesis. Further investigations of the role of ACBD3 in cancer stem cell differentiation may help increase the understanding of the onset of cancer in other stem cell systems, such as germ-line disorders.
12. Conclusion
The data presented in this review suggest that ACBD3 is a critical component in Golgi-body platform-based cell signaling. As ACBD3 becomes further understood through genome-wide investigations, we believe that new information will clarify its biological functions. Because of the various names assigned to this protein based on its function in distinct cell signaling pathways, there may continue to be confusion regarding communication about this protein. Therefore, we propose to adopt the name ACBD3 assigned by the human genome organization (HUGO).
The most detailed research about ACBD3 focuses on its functions as a TSPO-associated protein and AKAP in the cAMP-dependent steroidogenesis, as well as its role in the Dexras1-ACBD3-DMT1-mediated signaling pathway that connects neurotransmission with iron uptake. Although the detailed mechanisms of ACBD3 action, beyond protein targeting and protein-protein interactions, remains unclear, it is likely linked to hormones and other factors that elicit fragmentation of the Golgi apparatus. ACBD3 likely leads to a series of cellular processes mediating steroid hormone formation, iron homeostasis and other functions. From the available information, ACBD3 emerges as a multifunctional signaling molecule that plays a role in the stem cell differentiation in neurogenesis via an asymmetric cell division. Thus, ACBD3 is likely to be important in cancer stem cell differentiation and carcinogenesis, hematopoetic stem cell differentiation, as well as spermatogenesis.
Acknowledgments
This work was supported by National Institutes of Health grant HD37031 (to V.P.). V.P. was also supported by a Canada Research Chair in Biochemical Pharmacology and M.C. by a Royal Victoria Hospital Centenary award. The Research Institute of MUHC is supported by a Center grant from Le Fonds de la recherche en santé du Québec.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barr FA. Golgi inheritance: shaken but not stirred. J Cell Biol. 2004;164:955–958. doi: 10.1083/jcb.200402011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misumi Y, Sohda M, Ikehara Y. GCP60, a novel Golgi phosphoprotein, interact with giantin and involved in the maintenance of the Golgi structure. Cell Struct Funct. 2000;25:410. [Google Scholar]
- 3.Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J Biol Chem. 2001;276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- 4.Barr FA, Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr Opin Cell Biol. 2003;15:405–413. doi: 10.1016/s0955-0674(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- 6.Cheah JH, Kim SF, Hester LD, Clancy KW, Patterson SE, III, Papadopoulos V, Snyder SH. NMDA receptor-nitric oxide transmission mediates neuronal iron homeostasis via the GTPase Dexras1. Neuron. 2006;51:431–440. doi: 10.1016/j.neuron.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 8.Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: how and why? Curr Opin Cell Biol. 2003;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- 9.Nozawa K, Casiano CA, Hamel JC, Molinaro C, Fritzler MJ, Chan EKL. Fragmentation of Golgi complex and Golgi autoantigens during apoptosis and necrosis. Arthritis Res. 2002;4:R3. doi: 10.1186/ar422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nardini M, Span S, Cericola C, Pesce A, Massaro A, Millo E, Luini A, Corda D, Bolognesi M. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carcedo CH, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- 12.Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol. 2008;10:1146–1153. doi: 10.1038/ncb1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancini M, Machamer CE, Roy S, Nicholson DW, Thornberry NA, Casciola-Rosen LA, Rosen A. Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J Cell Biol. 2000;149:603–612. doi: 10.1083/jcb.149.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu R, Novikov L, Mukherjee S, Shields D. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J Cell Biol. 2002;159:637–648. doi: 10.1083/jcb.200208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe M, Lane JD, Woodman PG, Allan VJ. Caspase-mediated cleavage of syntaxin 5 and giantin accompanies inhibition of secretory traffic during apoptosis. J Cell Sci. 2004;117:1139–1150. doi: 10.1242/jcs.00950. [DOI] [PubMed] [Google Scholar]
- 16.Sbodio JI, Hicks SW, Simon D, Machamer CE. GCP60 preferentially interacts with a caspase-generated golgin-160 fragment. J Biol Chem. 2006;281:27924–27931. doi: 10.1074/jbc.M603276200. [DOI] [PubMed] [Google Scholar]
- 17.Sbodio JI, Machamer CE. Identification of a redox-sensitive cysteine in GCP60 that regulates its interaction with golgin-160. J Biol Chem. 2007;282:29874–29881. doi: 10.1074/jbc.M705794200. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson JG, Lippincott-Schwartz J. Sorting and signaling at the Golgi complex. Cell. 2000;101:693–696. doi: 10.1016/s0092-8674(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Rone MB, Papadopoulos V. Protein-protein interactions mediate mitochondrial cholesterol transport and steroid biosynthesis. J Biol Chem. 2006;281:38879–38893. doi: 10.1074/jbc.M608820200. [DOI] [PubMed] [Google Scholar]
- 20.Guidotti A, Forchetti CM, Corda MG, Konkel D, Bennett CD, Costa E. Isolation, characterization, and purification to homogeneity of an endogenous polypeptide with agonistic action on benzodiazepine receptors. Proc Natl Acad Sci U S A. 1983;80:3531–3535. doi: 10.1073/pnas.80.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoyab M, Gentry LE, Marquardt H, Todaro GJ. Isolation and characterization of a putative endogenous benzodiazepineoid (endozepine) from bovine and human brain. J Biol Chem. 1986;261:11968–11973. [PubMed] [Google Scholar]
- 22.Marquardt H, Todaro GJ, Shoyab M. Complete amino acid sequences of bovine and human endozepines. Homology with rat diazepam binding inhibitor. J Biol Chem. 1986;261:9727–9731. [PubMed] [Google Scholar]
- 23.Andersen KV, Poulsen FM. Three-dimensional structure in solution of acyl-coenzyme A binding protein from bovine liver. J Mol Biol. 1992;226:1131–1141. doi: 10.1016/0022-2836(92)91057-v. [DOI] [PubMed] [Google Scholar]
- 24.Knudsen J, Burton M, Faergeman NJ. Long chain acyl-CoA esters and acyl-CoA binding protein (ACBP) in cell function. In: van der Vusse Ger., editor. Advances in molecular and cell biology. Vol. 33. Lipobiology. Elsevier; 2004. [Google Scholar]
- 25.Faergeman NJ, Wadum M, Feddersen S, Burton M, Kragelund BB, Knudsen J. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol Cell Biochem. 2007;299:55–65. doi: 10.1007/s11010-005-9040-3. [DOI] [PubMed] [Google Scholar]
- 26.Burton M, Rose TM, Faergeman NJ, Knudsen J. Evolution of the acyl-CoA binding protein (ACBP) Biochem J. 2005;392:299. doi: 10.1042/BJ20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen JT, Rosendal J, Knudsen J. Interaction of acyl-CoA binding protein (ACBP) on processes for which acyl-CoA is a substrate, product or inhibitor. Biochem J. 1993;292:907. doi: 10.1042/bj2920907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elholm M, Garras A, Neve S, Tornehave D, Lund TB, Skorve J, Flatmark T, Kristiansen K, Berge RK. Long-chain acyl-CoA esters and acyl-CoA binding protein are present in the nucleus of rat liver cells. J Lipid Res. 2000;41:538–545. [PubMed] [Google Scholar]
- 30.Hansen JS, Faergeman NJ, Kragelund BB, Knudsen J. Acyl-CoA-binding protein (ACBP) localizes to the endoplasmic reticulum and Golgi in a ligand-dependent manner in mammalian cells. Biochem J. 2008;410:463–472. doi: 10.1042/BJ20070559. [DOI] [PubMed] [Google Scholar]
- 31.Feddersen S, Neergaard TBF, Knudsen J, Fμrgeman NJ. Transcriptional regulation of phospholipid biosynthesis is linked to fatty acid metabolism by an acyl-CoA-binding-protein-dependent mechanism in Saccharomyces cerevisiae. Biochem J. 2007;407:219. doi: 10.1042/BJ20070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolly CA, Wilton DC, Schroeder F. Microsomal fatty acyl-CoA transacylation and hydrolysis: fatty acyl-CoA species dependent modulation by liver fatty acyl-CoA binding proteins. BBA-Mol Cell Biol L. 2000;1483:185–197. doi: 10.1016/s1388-1981(99)00170-5. [DOI] [PubMed] [Google Scholar]
- 33.Knudsen J, Neergaard TB, Gaigg B, Jensen MV, Hansen JK. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J Nutr. 2000;130:294S–298S. doi: 10.1093/jn/130.2.294S. [DOI] [PubMed] [Google Scholar]
- 34.Faergeman NJ, Feddersen S, Christiansen JK, Larsen MK, Schneiter R, Ungermann C, Mutenda K, Roepstorff P, Knudsen J. Acyl-CoA-binding protein, Acb1p, is required for normal vacuole function and ceramide synthesis in Saccharomyces cerevisiae. Biochem J. 2004;380:907–918. doi: 10.1042/BJ20031949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa E, Guidotti A. Diazepam binding inhibitor(DBI): a peptide with multiple biological actions. Life Sci. 1991;49:325–344. doi: 10.1016/0024-3205(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 36.Papadopoulos V, Guarneri P, Kreuger KE, Guidotti A, Costa E. Pregnenolone biosynthesis in C6-2B glioma cell mitochondria: regulation by a mitochondrial diazepam binding inhibitor receptor. Proc Natl Acad Sci U S A. 1992;89:5113–5117. doi: 10.1073/pnas.89.11.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadopoulos V. Peripheral-type benzodiazepine/diazepam binding inhibitor receptor: biological role in steroidogenic cell function. Endocr Rev. 1993;14:222–240. doi: 10.1210/edrv-14-2-222. [DOI] [PubMed] [Google Scholar]
- 38.Cavallaro S, Pani L, Guidotti A, Costa E. ACTH-induced mitochondrial DBI receptor(MDR) and diazepam binding inhibitor (DBI) expression in adrenals of hypophysectomized rats is not cause-effect related to its immedite steroidogenic action. Life Sci. 1993;53:1137–1147. doi: 10.1016/0024-3205(93)90550-m. [DOI] [PubMed] [Google Scholar]
- 39.Geisbrecht BV, Zhang D, Schulz H, Gould SJ. Characterization of PECI, a novel monofunctional dealta(3), delta(2)-enoyl-CoA isomerase of mammalian peroxisomes. J Biol Chem. 1999;274:21797–21803. doi: 10.1074/jbc.274.31.21797. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, Chuhjo T, Sugimori C, Kotani T, Lu X, Takami A, Takamatsu H, Yamazaki H, Nakao S. Diazepam-binding inhibitor-related protein 1: a candidate autoantigen in acquired aplastic anemia patients harboring a minor population of paroxysmal nocturnal hemoglobinuria-type cells. Blood. 2004;104:2425–2431. doi: 10.1182/blood-2004-05-1839. [DOI] [PubMed] [Google Scholar]
- 41.Suk K, Kim YH, Hwang DY, Ihm SH, Yoo HJ, Lee MS. Molecular cloning and expression of a novel human cDNA related to the diazepam binding inhibitor. Biochim Biophys Acta. 1999;1454:126–131. doi: 10.1016/s0925-4439(99)00033-2. [DOI] [PubMed] [Google Scholar]
- 42.Nakao S, Feng X, Sugimori C. Immune pathophysiology of aplastic anemia. Int J Hematol. 2005;82:196–200. doi: 10.1532/IJH97.05116. [DOI] [PubMed] [Google Scholar]
- 43.Scanlan MJ, Gordan JD, Williamson B, Stockert E, Bander NH, Jongeneel V, Gure AO, Jaeger D, Jaeger E, Knuth A. Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, Zhao HT, Rui JA, Leng XS. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies 1. J Immunol. 2002;169:1102–1109. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- 45.Ockner RK, Kaikaus RM, Bass NM. Fatty-acid metabolism and the pathogenesis of hepatocellular carcinoma: review and hypothesis. Hepatology. 1993;18:669–676. [PubMed] [Google Scholar]
- 46.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer P D. 2006;9:230–234. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 47.Files A, Chavez-Blanco A, Perez-Plasencia C, Perez-Cardenas E, Carrasco-Legleu C, Rangel-Lopez E, Segura-Pacheco B, Taja-Chayeb L, Trejo-Becerril C, Gonzalez-Fierro A. Antineoplastic effects of the DNA methylation inhibitor hydralazine and the histone deacetylase inhibitor valproic acid in cancer cell lines. Cancer Cell Int. 2006;6:2. doi: 10.1186/1475-2867-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le BJ, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc Natl Acad Sci U S A. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foord R, Taylor IA, Sedgwick SG, Smerdon SJ. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat Struct Biol. 1999;6:157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 50.Soupene E, Serikov V, Kuypers FA. Characterization of an acyl-coenzyme A binding protein predominantly expressed in human primitive progenitor cells. J Lipid Res. 2008;49:1103–1112. doi: 10.1194/jlr.M800007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, Cogan JP, Arasheben A, Tilley WD, Scher HI, Gerald WL, Buchanan G, Coetzee GA, Frenkel B. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer. 2007;6:39. doi: 10.1186/1476-4598-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valentin M, Balvers M, Pusch W, Weinbauer GF, Knudsen J, Ivell R. Structure and expression of the mouse gene encoding the endozepine-like peptide from haploid male germ cells. Eur J Biochem. 2000;267:5438–5449. doi: 10.1046/j.1432-1327.2000.01603.x. [DOI] [PubMed] [Google Scholar]
- 53.Ivell R, Pusch W, Balvers M, Valentin M, Walther N, Weinbauer G. Progressive inactivation of the haploid expressed gene for the sperm-specific endozepine-like peptide (ELP) through primate evolution. Gene. 2000;255:335–345. doi: 10.1016/s0378-1119(00)00317-6. [DOI] [PubMed] [Google Scholar]
- 54.Mandrup S, Faergeman NJ, Knudsen J. Structure, function, and phylogeny of acyl-CoA binding protein. Vch Verlagsgesellschaft Mbh; 2004. pp. 151–171. [Google Scholar]
- 55.Alho H, Costa E, Ferrero P, Fujimoto M, Cosenza-Murphy D, Guidotti A. Diazepam-binding inhibitor: a neuropeptide located in selected neuronal populations of rat brain. Science. 1985;229:179–182. doi: 10.1126/science.3892688. [DOI] [PubMed] [Google Scholar]
- 56.Borboni P, Condorelli L, De Stefanis P, Sesti G, Lauro R. Modulation of insulin secretion by diazepam binding inhibitor and its processing products. Neuropharmacology. 1991;30:1399–1403. doi: 10.1016/s0028-3908(11)80008-0. [DOI] [PubMed] [Google Scholar]
- 57.Chen Z, Agerberth B, Gell K, Andersson M, Mutt V, Ostenson CG, Efendic S, Barros-Soderling J, Persson B, Jornvall H. Isolation and characterization of porcine diazepam-binding inhibitor, a polypeptide not only of cerebral occurrence but also common in intestinal tissues and with effects on regulation of insulin release. Eur J Biochem. 1988;174:239–244. doi: 10.1111/j.1432-1033.1988.tb14088.x. [DOI] [PubMed] [Google Scholar]
- 58.Papadopoulos V, Berkovich A, Krueger KE, Costa E, Guidotti A. Diazepam binding inhibitor and its processing products stimulate mitochondrial steroid biosynthesis via an interaction with mitochondrial benzodiazepine receptors. Endocrinology. 1991;129:1481–1488. doi: 10.1210/endo-129-3-1481. [DOI] [PubMed] [Google Scholar]
- 59.Kragelund BB, Knudsen J, Poulsen FM. Acyl-coenzyme A binding protein (ACBP) BBA-Mol Cell Biol L. 1999;1441:150–161. doi: 10.1016/s1388-1981(99)00151-1. [DOI] [PubMed] [Google Scholar]
- 60.Taskinen JP, van Aalten DM, Knudsen J, Wierenga RK. High resolution crystal structures of unliganded and liganded human liver ACBP reveal a new mode of binding for the acyl-CoA ligand. Proteins. 2007;66:229–238. doi: 10.1002/prot.21124. [DOI] [PubMed] [Google Scholar]
- 61.Abo-Hashema KAH, Cake MH, Lukas MA, Knudsen J. The interaction of acyl-CoA with acyl-CoA binding protein and carnitine palmitoyltransferase I. Int J Biochem Cell B. 2001;33:807–815. doi: 10.1016/s1357-2725(01)00049-8. [DOI] [PubMed] [Google Scholar]
- 62.Kragelund BB, Andersen KV, Madsen JC, Knudsen J, Poulsen FM. Three-dimensional structure of the complex between acyl-coenzyme A binding protein and palmitoyl-coenzyme A. J Mol Biol. 1993;230:1260–1277. doi: 10.1006/jmbi.1993.1240. [DOI] [PubMed] [Google Scholar]
- 63.Kragelund BB, Poulsen K, Andersen KV, Baldursson T, Kroll JB, Neergard TB, Jepsen J, Roepstorff P, Kristiansen K, Poulsen FM. Conserved Residues and Their Role in the Structure, Function, and Stability of Acyl-Coenzyme A Binding Proteinå. Biochemistry. 1999;38:2386–2394. doi: 10.1021/bi982427c. [DOI] [PubMed] [Google Scholar]
- 64.Larsen MK, Tuck S, Faergeman NJ, Knudsen J. MAA-1, a novel acyl-CoA-binding protein involved in endosomal vesicle transport in Caenorhabditis elegans. Molecular biology of the cell. 2006;17:4318. doi: 10.1091/mbc.E06-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng B, Cai X, Zhu G. Functional characterization of a fatty acyl-CoA-binding protein (ACBP) from the apicomplexan Cryptosporidium parvum. Microbiology. 2006;152:2355. doi: 10.1099/mic.0.28944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faergeman NJ, Sigurskjold BW, Kragelund BB, Andersen KV, Knudsen J. Thermodynamics of ligand binding to acyl-coenzyme a binding protein studies by titration calorimetry. Biochemistry(Easton) 1996;35:14118–14126. doi: 10.1021/bi960545z. [DOI] [PubMed] [Google Scholar]
- 67.Milne KG, Ferguson MAJ. Cloning, expression, and characterization of the acyl-CoA-binding protein in African trypanosomes. J Biol Chem. 2000;275:12503–12508. doi: 10.1074/jbc.275.17.12503. [DOI] [PubMed] [Google Scholar]
- 68.Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3:research0023. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito K, Tautz L, Mustelin T. The lipid-binding SEC14 domain. BBA-Mol Cell Biol L. 2007;1771:719–726. doi: 10.1016/j.bbalip.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 70.Callebaut I, Goud B, Mornon JP. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- 71.Stocker A, Tomizaki T, Schulze-Briese C, Baumann U. Crystal structure of the human supernatant protein factor. Structure. 2002;10:1533–1540. doi: 10.1016/s0969-2126(02)00884-5. [DOI] [PubMed] [Google Scholar]
- 72.Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud JP, Thomas DY, Bergeron JJ, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J Biol Chem. 1997;272:8057. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 74.Jarnaess E, Ruppelt A, Stokka AJ, Lygren B, Scott JD, Tasken K. Dual specificity A-kinase anchoring proteins (AKAPs) contain an additional binding region that enhances targeting of protein kinase A type I. J Biol Chem. 2008;283:33708. doi: 10.1074/jbc.M804807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu J, Li H, Papadopoulos V. PAP7, a PBR/PKA-RIalpha-associated protein: a new element in the relay of the hormonal induction of steroidogenesis. J Steroid Biochem Mol Biol. 2003;85:275–283. doi: 10.1016/s0960-0760(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 76.Fritzler MJ. Molecular characterization of two human autoantigens: unique cDNAs encoding 95-and 160-kD proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, Matyakhina L, Han Z, Sandrini F, Bei T, Stratakis CA, Papadopoulos V. Molecular cloning, chromosomal localization of human peripheral-type benzodiazepine receptor and PKA regulatory subunit type 1A (PRKAR1A)-associated protein PAP7, and studies in PRKAR1A mutant cells and tissues. FASEB J. 2003;17:1189–1191. doi: 10.1096/fj.02-1066fje. [DOI] [PubMed] [Google Scholar]
- 78.Rajalingam R, Parham P, bi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors 1. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 79.Vogel C, Teichmann SA, Pereira-Leal J. The Relationship between domain duplication and recombination. J Mol Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 80.Kaessmann H, Zollner S, Nekrutenko A, Li WH. Signatures of domain shuffling in the human genome. Genome Res. 2002;12:1642–1650. doi: 10.1101/gr.520702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duan H, Jefcoate CR. The predominant cAMP-stimulated 3 × 5 kb StAR mRNA contains specific sequence elements in the extended 3′UTR that confer high basal instability. J Mol Endocrinol. 2007;38:159–179. doi: 10.1677/jme.1.02153. [DOI] [PubMed] [Google Scholar]
- 83.Miller WL. Mechanism of StAR's regulation of mitochondrial cholesterol import. Mol Cell Endocrinol. 2007;265:46–50. doi: 10.1016/j.mce.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 84.Papadopoulos V, Liu J, Culty M. Is there a mitochondrial signaling complex facilitating cholesterol import? Mol Cell Endocrinol. 2007;265-266:59–64. doi: 10.1016/j.mce.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 85.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Boujrad N, Hudson JR, Papadopoulos V. Inhibition of hormone-stimulated steroidogenesis in cultured Leydig tumor cells by a cholesterol-linked phosphorothioate oligodeoxynucleotide antisense to diazepam-binding inhibitor. Proc Natl Acad Sci U S A. 1993;90:5728–5731. doi: 10.1073/pnas.90.12.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hauet T, Liu J, Li H, Gazouli M, Culty M, Papadopoulos V. PBR, StAR, and PKA: partners in cholesterol transport in steroidogenic cells. Endocr Res. 2002;28:395–401. doi: 10.1081/erc-120016814. [DOI] [PubMed] [Google Scholar]
- 88.Liu J, Cavalli LR, Haddad BR, Papadopoulos V. Molecular cloning, genomic organization, chromosomal mapping and subcellular localization of mouse PAP7: a PBR and PKA-RIalpha associated protein. Gene. 2003;308:1–10. doi: 10.1016/s0378-1119(03)00453-0. [DOI] [PubMed] [Google Scholar]
- 89.Culty M, Luo L, Yao ZX, Chen H, Papadopoulos V, Zirkin BR. Cholesterol transport, peripheral benzodiazepine receptor, and steroidogenesis in aging Leydig cells. J Androl. 2002;23:439–447. [PubMed] [Google Scholar]
- 90.Altan-Bonnet N, Phair RD, Polishchuk RS, Weigert R, Lippincott-Schwartz J. A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc Natl Acad Sci U S A. 2003;100:13314–13319. doi: 10.1073/pnas.2234055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Puri S, Linstedt AD. Capacity of the Golgi Apparatus for Biogenesis from the Endoplasmic Reticulum. Mol Biol Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xia K, Dong D, Han JD. IntNetDB v1.0: an integrated protein-protein interaction network database generated by a probabilistic model. BMC Bioinformatics. 2006;7:508. doi: 10.1186/1471-2105-7-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J Biol Chem. 2002;277:35833–35839. doi: 10.1074/jbc.M206280200. [DOI] [PubMed] [Google Scholar]
- 94.Maag RS, Mancini M, Rosen A, Machamer CE. Caspase-resistant Golgin-160 disrupts apoptosis induced by secretory pathway stress and ligation of death receptors. Mol Biol Cell. 2005;16:3019–3027. doi: 10.1091/mbc.E04-11-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukherjee S, Chiu R, Leung SM, Shields D. Fragmentation of the Golgi apparatus: an early apoptotic event independent of the cytoskeleton. Traffic. 2007;8:369–378. doi: 10.1111/j.1600-0854.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 96.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 97.Petersen PH, Tang H, Zou K, Zhong W. The enigma of the numb-Notch relationship during mammalian embryogenesis. Dev Neurosci. 2006;28:156–168. doi: 10.1159/000090761. [DOI] [PubMed] [Google Scholar]
- 98.Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- 99.Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci. 2004;7:803–811. doi: 10.1038/nn1289. [DOI] [PubMed] [Google Scholar]
- 100.Finocchiaro G, Mancuso F, Muller H. Mining published lists of cancer related microarray experiments: identification of a gene expression signature having a critical role in cell-cycle control. BMC Bioinformatics. 2005;6 4:S14. doi: 10.1186/1471-2105-6-S4-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glebov OK, Rodriguez LM, Lynch P, Patterson S, Lynch H, Nakahara K, Jenkins J, Cliatt J, Humbyrd CJ, Denobile J, Soballe P, Gallinger S, Buchbinder A, Gordon G, Hawk E, Kirsch IR. Celecoxib treatment alters the gene expression profile of normal colonic mucosa. Cancer Epidemiol Biomarkers Prev. 2006;15:1382–1391. doi: 10.1158/1055-9965.EPI-04-0866. [DOI] [PubMed] [Google Scholar]
- 102.Kakiuchi S, Daigo Y, Ishikawa N, Furukawa C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, Sone S, Nakamura Y. Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839) Hum Mol Genet. 2004;13:3029–3043. doi: 10.1093/hmg/ddh331. [DOI] [PubMed] [Google Scholar]
- 103.Buytaert E, Matroule JY, Durinck S, Close P, Kocanova S, Vandenheede JR, de Witte PA, Piette J, Agostinis P. Molecular effectors and modulators of hypericin-mediated cell death in bladder cancer cells. Oncogene. 2008;27:1916–1929. doi: 10.1038/sj.onc.1210825. [DOI] [PubMed] [Google Scholar]
- 104.Chen Y, Zheng X, Dobhal MP, Gryshuk A, Morgan J, Dougherty TJ, Oseroff A, Pandey RK. Methyl pyropheophorbideùa analogues: potential fluorescent probes for the peripheral-type benzodiazepine receptor. Effect of central metal in photosensitizing efficacy. J Med Chem. 2005;48:3692–3695. doi: 10.1021/jm050039k. [DOI] [PubMed] [Google Scholar]
- 105.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 106.Clevers H. Stem cells, asymmetric division and cancer. Nat Genet. 2005;37:1027–1028. doi: 10.1038/ng1005-1027. [DOI] [PubMed] [Google Scholar]
- 107.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 108.Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Prog Mol Subcell Biol. 2007;45:205–225. doi: 10.1007/978-3-540-69161-7_9. [DOI] [PubMed] [Google Scholar]
- 109.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]