Abstract
We report the discovery of small molecules that target the Rho pathway, a central regulator of cytokinesis, the final step in cell division. We have developed a method to target a small molecule screen towards a specific pathway, which should be widely applicable to study any signaling pathway. In a chemical genetic variant of a classical modifier screen, we used RNA interference (RNAi) to sensitize cells and identified small molecules that suppressed or enhanced the RNAi phenotype. We have discovered promising candidate molecules, which we named Rhodblock 1–8, and we identified the target of Rhodblock 6 as Rho kinase. Several Rhodblocks inhibit a function of the Rho pathway in cells: the correct localization of phosphorylated myosin light chain during cytokinesis. Rhodblocks differentially perturb Rho pathway proteins in cells and can be used to dissect the mechanism of the Rho pathway during cytokinesis.
INTRODUCTION
Rho GTPases are key regulators of cell division and control other processes involving the cytoskeleton, such as cell migration, contraction and adhesion1. With Rho GTPases at the center of complicated signaling cascades that are only partially understood, different branches of these pathways cooperate to coordinate these processes. Small GTPases regulate their downstream effectors by switching between two states, active (GTP-bound) and inactive (GDP-bound)1. This cycling is controlled by a number of regulatory proteins such as guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Improper regulation of the Rho pathway has been implicated in cardiovascular diseases and cancer2,3. For example, RhoA is highly overexpressed in breast tumors and malignancy is correlated with high RhoA expression4,5. Increasing evidence suggests that altered Rho signaling contributes to cancer onset, invasion and metastasis, but little is known about the underlying mechanisms6,7. In this study, we focus on the role of the Rho pathway in cytokinesis, the final step of cell division, where cells physically separate8. As a key regulator, the Rho pathway participates in all steps of cytokinesis, from the initial specification of the location of the cleavage furrow, to constriction and final abscission. Small molecules targeting the Rho pathway would be very useful, both as biological probes and as therapeutic leads9,10.
Our options to identify small molecules that affect pathways have been limited. Pure protein screens target single proteins while phenotypic screens target entire processes, irrespective of a specific pathway. Despite serious efforts, especially with the oncogenic GTPase Ras, small molecules that target the GTP-binding pocket in small GTPases have been elusive because GTP affinity in GTPases is much higher than ATP affinity in kinases11. This is why we decided to develop a strategy to target the GTPase signaling pathway rather than the GTPase’s enzymatic activity. Rho associates with many regulatory and downstream effector proteins, which are potential small molecule targets. It is difficult to target these proteins using conventional biochemical assays because inhibition of their enzyme activity is often not readily detectable. Here, we report the development of a phenotypic screening approach that allows us to target a pathway independent of specific enzyme activities. We indentified pathway-specific small molecules and show that they perturb the Rho pathway in cells.
RESULTS
Screen concept and design
Our goal was the identification of small molecules that specifically target the Rho pathway. Inspired by classical genetic experiments, we designed a phenotypic screening strategy analogous to a genetic modifier screen, but perturbed cells by small molecules and RNA interference (RNAi), instead of genetic mutations. By using RNAi to impair signaling through the Rho pathway, we decreased the amount of compound needed to detect a phenotype. To ensure specificity, we prioritized compounds that exhibit stronger defects in RNAi-sensitized cells vs. wild-type cells.
We used success or failure of cytokinesis as a measure of Rho activity. Failed cytokinesis leads to the formation of binucleated cells, which was the readout in the screen. We chose to deplete Rho itself because it is tractable, ideally positioned within the signaling cascade and biologically and clinically relevant. We modestly impaired cytokinesis using partial RNAi depletion of Rho, added small molecules, and identified compounds that suppressed or aggravated RNAi-induced cytokinesis defects (Fig. 1a). We expected to find enhancers and suppressors because the pathway is both positively and negatively regulated.
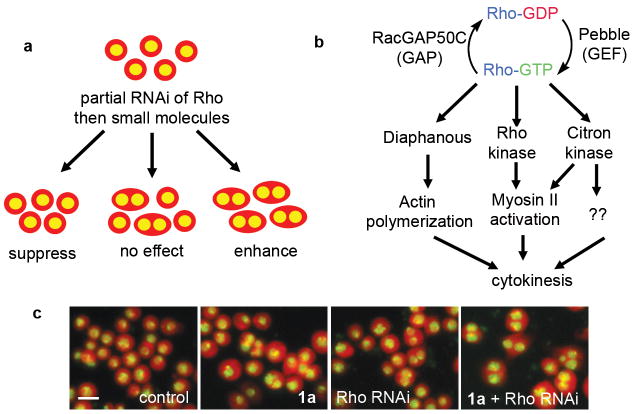
Figure 1.
(a) A small molecule/RNAi modifier screen. (Whole cells are cartooned in red, nuclei in yellow). (b) Simplified diagram of Rho signaling during cytokinesis. (c) Example of synergy between Rho RNAi and a hit compound (Rhodblock 1a). Note how the percentage of binucleate cells is relatively low in cells that are treated with RNAi or compound, but increases with both RNAi and compound treatment (Drosophila Kc167 cells are shown in red, nuclei in yellow). The scale bar shown in the control image represents 10μm.
A key feature of our strategy is to achieve an intermediate RNAi phenotype. During RNAi in Drosophila cells, which we used in this screen, double-stranded (ds)RNA corresponding in sequence to mRNA encoding the target protein is added to cells. The mRNA is destroyed and no new protein can be synthesized, resulting in the depletion of the target protein over time. We used the gradual decrease in Rho protein during RNAi treatment to obtain our intermediate phenotype. We optimized the assay to reproducibly yield intermediate depletion of Rho by varying the sequence and dose of the dsRNA and the length of the RNAi experiment (Supplementary Fig. 1).
Synergy between Rho pathway proteins
We first confirmed that the concept of the screen was feasible, i.e. that we could observe synergy, measured as a substantial increase in binucleate cells that is larger than the effect of two independent treatments, if we blocked different branches of the Rho pathway at the same time. We performed double RNAi experiments where two different pathway proteins (see Fig. 1b for examples) were depleted simultaneously and observed synergy between these protein pairs (Supplementary Fig. 2). Conversely, we did not observe synergy between Rho RNAi and RNAi of proteins or small molecule inhibitors that target cytokinesis, but not the Rho pathway (Supplementary Methods). It is important to include such Rho-independent controls because the screen could potentially result in other outcomes such as the identification of small molecules that modulate the process of RNAi itself.
We also confirmed that it was possible to observe synergy between Rho RNAi and pathway-specific small molecule treatment. When we tested GSK269962A12, a Rho kinase inhibitor, we observed strong synergy with Rho RNAi. Our screening concept predicts that a compound that is specific for the Rho pathway should inhibit cytokinesis in non-RNAi treated cells at higher concentrations than in cells sensitized by RNAi treatment, which is what we observed with GSK269962A (Supplementary Fig. 3).
Automated image analysis
Although the human eye can readily detect changes in the ratio of mononucleate to binucleate cells (Fig. 1c), a major challenge in our screening protocol was to automate the image analysis, both to allow high throughput and to quantify our screening output. We used the CellProfiler software package13 and its recently developed machine learning capability14 to differentiate between cells with one nucleus or two nuclei using machine learning guided by visual inspection (Supplementary Methods and Supplementary Fig. 4).
Screen results in 9 small molecule enhancers of Rho RNAi
We screened ~38,000 compounds from commercial sources and natural product extracts. During the screen, we arrayed RNAi-treated (or wild-type) Drosophila Kc167 cells into 384-well plates, treated with small molecules (nominal concentration ~ 25 μM), fixed and stained cells and DNA with fluorescent markers. We collected images by automated fluorescence microscopy and performed automated image analysis to identify wells containing active small molecules (see Supplementary Table 1 for screen summary). To calibrate our screen, we first screened a collection of compounds with known biological activities. In addition to enhancers, we found several compounds that suppressed the RNAi phenotype, i.e. they inhibited the formation of binucleate cells (Supplementary Table 2). Most of these compounds were cell cycle inhibitors that arrest cells before they get to the division stage. To ensure that the suppressors we indentified in the full screen target the Rho pathway rather than a different step earlier in the cell cycle, a careful cell cycle analysis is needed for this class of compounds. We therefore initially focused on enhancers and selected the 9 most active compounds, which we named Rhodblock 1–8, for further evaluation (Fig. 3). Each compound caused a substantial increase in the proportion of binucleate cells in a partial Rho RNAi background (Fig. 1c).
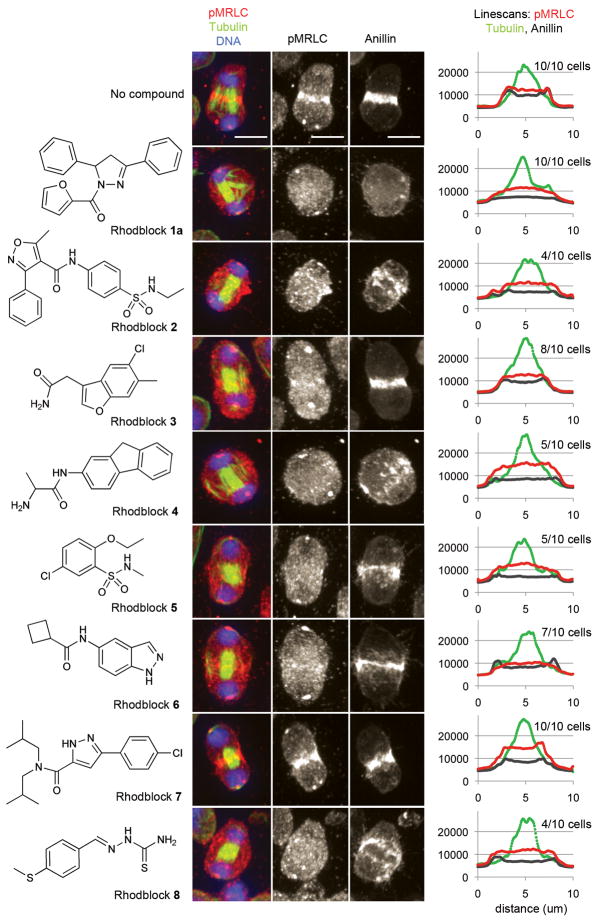
Figure 3. Several Rhodblocks prevent the accumulation of phospho-myosin regulatory light chain and/or Anillin at the cleavage furrow.
The chemical structures of Rhodblocks 1a–8 are shown on the left. Immunofluorescence images of representative phenotypes for each Rhodblock are shown in the middle. Phospho-MRLC (red), tubulin (green) and DNA (blue) have been visualized in Drosophila Kc167 cells. For greater clarity, the middle panel shows gray-scale images of phospho-MRLC staining only. The right panel shows gray-scale images of Anillin staining in the same cell. The images were taken under identical conditions and were processed identically (see Supplementary Methods). The scale bar shown in the control images represents 5μm. We analyzed images of 10 cells for each condition (4 h treatment with compound at the minimally synergistic concentration after overnight Rho RNAi sensitization). For each cell we analyzed, we placed a line across the cleavage furrow and quantitated the fluorescence intensities for phospho-MRLC, Anillin and tubulin staining (see Supplementary Methods). We then averaged the line scans for cells exhibiting the phospho-MRLC phenotype shown in this figure (shown on the right). The number of cells represented by the image is shown in parentheses above the linescans for each compound. In the linescans, the x axis represents fluorescence intensity (AU). Bumps in fluorescence intensities at the edge of the cell are characteristic of an intact furrow (e.g. control cell), uniform fluorescence intensities across the entire cell are characteristic of a missing furrow (e.g. Rhodblock 1a). In Rhodblock 6-treated cells, Anillin forms a furrow while phospho-MRLC does not.
We purchased the nine Rhodblocks, confirmed their identity by analytical chemistry and tested them at different concentrations to determine the minimal concentration at which we could observe robust synergy with Rho RNAi (Table 1). To rule out possible effects on RNAi rather than synergy with the Rho pathway, we tested the active compounds in cells treated with a protein inhibitor of Rho, C3 transferase (CT04)15. All compounds synergized with CT04 as well as with Rho RNAi. As predicted by our screening concept and the GSK269962A experiment discussed above, we would expect the Rhodblocks to be active in the absence of Rho RNAi at higher concentrations. Rhodblock 1a, our most active compound, is active at 100 μM and synergizes with Rho RNAi at 10 μM. Our collection of active compounds included a small molecule, Rhodblock 1b, that was structurally related to Rhodblock 1a, allowing for a rudimentary structure-activity analysis. To evaluate the importance of the substituents on Rhodblock 1, we obtained compound 1c, which varies from 1a only in the furan substituent (Supplementary Fig. 5). The activity of 1c was similar to the less active 1b, suggesting that the furan is an important determinant of activity.
Table 1.
Synergy ratios for Rhodblocks 1–8 in cells sensitized by partial RNAi of Rho pathway proteins (RhoA, Pebble, RacGAP50C, Diaphanous, Citron kinase, Rho kinase). Strong interactions (ratio>4) are highlighted in red, medium interactions (2–4) in orange and weaker interactions (1.3–2) in yellow. Insignificant changes (0.7–1.2) are pale yellow and antagonistic interactions pale green. Please see methods and supporting information for data and details on the statistical analysis.
| Rho | Pbl | RacGAP | Dia | CK | Rok | |
|---|---|---|---|---|---|---|
| 1a (10 μM) | 14.7 | 2.3 | 2.1 | 1.3 | 0.7 | 0.9 |
| 1b (30 μM) | 8.1 | 0.9 | 1.1 | 0.8 | 0.2 | 0.7 |
| 2 (30 μM) | 4.0 | 4.2 | 1.1 | 2.7 | 0.7 | 1.7 |
| 3 (100 μM) | 9.0 | 1.9 | 0.4 | 1.4 | 0.8 | 0.7 |
| 4 (30 μM) | 2.9 | 2.1 | 1.7 | 2.3 | 0.8 | 1.5 |
| 5 (100 μM) | 5.9 | 1.8 | 0.7 | 1.1 | 0.7 | 0.8 |
| 6 (100 μM) | 3.5 | 2.6 | 0.6 | 1.3 | 0.8 | 1.9 |
| 7 (30 μM) | 3.0 | 1.4 | 1.5 | 1.2 | 0.8 | 0.5 |
| 8 (30 μM) | 4.0 | 2.0 | 1.1 | 0.8 | 0.7 | 1.2 |
Quantitation of Synergy
To gain a more quantitative understanding of the strength of the synergistic interactions between our compounds and Rho RNAi, we calculated a “synergy ratio” (Table 1). We define the synergy ratio as the ratio of the observed phenotype over the expected phenotype. If the effects of small molecule treatment and partial Rho RNAi are independent of each other, they can be represented in a multiplicative model (i.e. the “expected phenotype”). All compounds selected for this study have a synergy ratio of ~3 or higher for Rho RNAi (Table 1), meaning that the observed synergistic phenotype shows at least a three-fold increase relative to the background.
Rhodblocks synergize with other Rho pathway proteins
After establishing synergy with Rho RNAi, we explored the Rhodblocks’ interactions with other Rho pathway proteins. As our screen targets a pathway rather than a single protein, we expected to obtain compounds that target different proteins within the Rho pathway. Because each protein within the pathway has different functions and interaction partners, compounds that target different proteins should show differential levels of synergy with other pathway proteins. We therefore analyzed the effect of the Rhodblocks on cells where other Rho pathway proteins had been partially depleted by RNAi (Table 1). We chose the regulatory GAP (called RacGAP50C in Drosophila 16 , MgcRacGAP in mammals17 and cyk-4 in C. elegans 18) and GEF (called pebble in Drosophila 19 , Ect2 in mammals) as well as effector proteins Diaphanous20, Rho kinase and Citron kinase (Fig. 1b). We quantified synergistic interactions for each small molecule/RNAi pair and showed that each compound has a unique synergy pattern (Table 1, Supplementary Tables 3 and 4).
We performed the entire panel in parallel to reduce error due to experimental variations and used conditions optimized for Rho RNAi, which could explain why the highest synergy ratios were observed in cells sensitized by Rho RNAi. For example, we observed relatively low synergy ratios with Rho kinase RNAi. Rho kinase is a stable protein and it has been reported that extended RNAi treatments are needed to observe a robust phenotype21. When we sensitized cells by longer Rho kinase RNAi, we observed an increase in synergy at lower concentrations of Rhodblock 6 (see next section). Interestingly, some compounds showed both synergy and antagonism (i.e. some suppress the RNAi phenotype rather than enhance it), especially in RacGAP-sensitized cells. RacGAP is thought to have opposing roles as a Rho deactivator and as a scaffold required for correct Rho localization and activation22, which could explain positive and negative interactions with our small molecules. We conclude from these experiments that our compounds are likely to have diverse targets within the Rho pathway.
Rhodblock 6 inhibits Rho kinase
To further support our initial strong, but circumstantial, evidence that we have discovered compounds that target the Rho pathway, we wanted to directly measure inhibition of Rho pathway activity, both in vitro and in cells. There are no specific biochemical assays for many proteins in the pathway. Rho kinase (Rok in Drosophila), however, can be readily assayed in a kinase assay. We purified FLAG-tagged Rok and tested our compounds at the concentrations at which we observed synergy in cells (Table 1, Supplementary Fig. 6). Rhodblock 6 inhibited Rok activity robustly and in a dose-dependent manner (Fig. 2). It also inhibited its human ortholog, ROCK I (Supplementary Fig. 7), and, similarly to other ROCK inhibitors, it causes the disruption of stress fibers in human HeLa cells (Fig. 2). The formation of stress fibers in human cells is one of the functions of the Rho pathway mediated by ROCK23. Rok’s functions during cytokinesis include the phosphorylation of myosin regulatory light chain (see next section) as well as inhibition of myosin phosphatase (Drosophila Mbs). Mbs and Rok have antagonistic functions and Mbs RNAi can partially rescue Rok RNAi phenotypes21. We observed a significant reduction in binucleate cells when Mbs was depleted in Rhodblock 6-treated cells (Supplementary Table 5). These data suggest that Rok is a significant cellular target of Rhodblock 6 and provide further evidence that our screen is a means to identify compounds that target the Rho pathway.
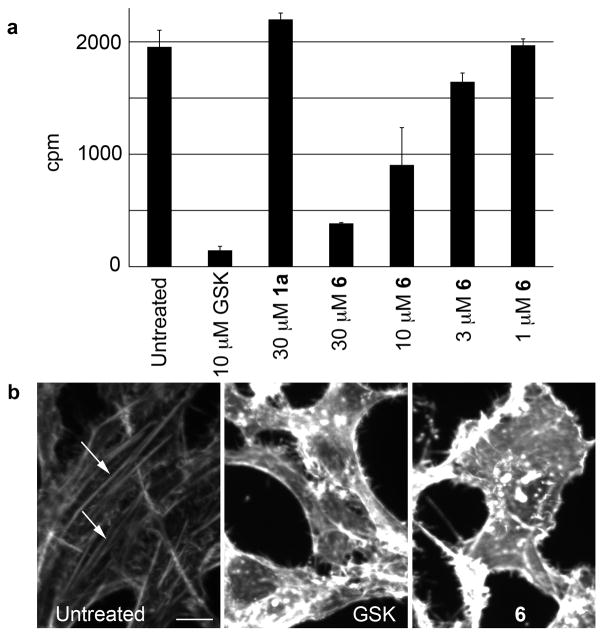
Figure 2.
(a) Drosophila Rho kinase assay data for Rhodblock 6. A dose-response curve is shown as well as control data for the Rho kinase inhibitor GSK269962A and Rhodblock 1a. For a full panel, see Supplementary Figure 6. Error bars indicate standard deviation (n=2). (b) Rhodblock 6 (100μM) and GSK269962A (10 μM) cause disappearance of actin stress fibers in HeLa cells (white arrows in control image. HeLa cells were treated for 20 h and fixed. Actin was visualized using phalloidin staining. The scale bar shown in the control image represents 10μm.
Rhodblocks inhibit a cellular function of the Rho pathway
We next investigated whether our Rhodblock compounds affected a specific function of the Rho pathway in cells, which would be the most conclusive validation of our approach and is a key feature in our goal to use these compounds as small molecule probes. Several proteins in the pathway, including Rok, cooperate to localize myosin II at the cleavage furrow and activate it by phosphorylating Serine-21 on myosin regulatory light chain (MRLC) in Drosophila cells24. By using a phospho-specific antibody, we can evaluate if this branch of the Rho pathway has been perturbed. Treatment of cells with eight (Rhodblocks 1–6, 8) of the nine Rhodblocks at the lowest synergistic concentration caused a variably penetrant decrease in phosphorylated MRLC and its mislocalization from the cleavage furrow (Fig. 3). We expected to observe this phenotype for Rhodblock 6 because it inhibits Rok, a protein known to be involved in myosin phosphorylation. The other Rhodblocks, however, do not inhibit Rok in vitro, suggesting that they target different proteins within the pathway. Since myosin phosphorylation is just one of the cellular functions of the Rho pathway, we expect the compound that did not inhibit myosin phosphorylation to target a different branch of the Rho pathway.
Rhodblocks perturb key cytokinesis proteins
After establishing that most of our compounds inhibit a function of the Rho pathway, our next goal was to study the role of the pathway during cytokinesis. We did this initially by evaluating the localization of Rho pathway proteins in the presence of small molecules. We focused on Rhodblocks 1a, 3, and 6 because they show potent synergy with Rho RNAi and are the most penetrant inhibitors of MRLC phosphorylation, i.e. we know that they target the Rho pathway. To minimize possible off-target effects, we performed all detailed cellular studies using the Rhodblocks at their minimal synergistic concentrations and amplified the effects of our small molecules with an overnight Rho RNAi treatment before small molecule addition. We did not observe a significant reduction in the number of cytokinetic cells showing decreased Rho staining at the cleavage furrow after overnight treatment (Supplementary Fig. 8). Unlike in the screen, where we treated sensitized cells with small molecules for 24 h to allow most cells to complete a cell cycle and to enter (and fail) cytokinesis, we used a shorter (4 h) treatment in the detailed studies. Although fewer cells will be in the process of failing cytokinesis, failure will be acute, allowing an analysis of the localization of cytokinesis proteins at the cleavage site before cells adapt to the new conditions. The ability to use acute short-term treatments is a major advantage of small molecules over genetic approaches such as RNAi.
Efforts to dissect signaling cascades within the Rho pathway have primarily focused on evaluating localization patterns of pathway proteins. Many Rho pathway proteins that are involved in cytokinesis are co-dependent for localization and the requirement of one protein for correct localization of a second suggests that the first protein is upstream of the second in the signaling cascade. Correct localization does not mean that a particular protein is active, but in the absence of assays for protein activity for many Rho pathway proteins, it gives some indication as to their function. We therefore analyzed the effects of our compounds on proteins that localize to the ingressing cleavage furrow such as actin, phospho-MRLC (as described above), Anillin, a septin (Drosophila Peanut) and Rho itself. We also analyzed microtubule structures as well as RacGAP and the kinesin-6 Pavarotti (called MKLP1 in mammals), which form the microtubule-bound Centralspindlin complex25. For a discussion of these proteins within the context of their role during cytokinesis, please see the Discussion section. The three Rhodblocks we chose for further analysis showed different localization patterns for different proteins, further supporting our hypothesis that they target different proteins within the pathway (Fig. 3). None of the compounds disrupted Centralspindlin localization, and only Rhodblock 1a had an effect on microtubule structures. Instead of forming a single midzone microtubule bundle, midzone microtubules in Rhodblock 1a-treated cells often bundled into two or more structures (Figs. 3 and 4). Rhodblock 6, the Rok inhibitor, inhibited phospho-MRLC localization, but did not have an effect on other proteins. Rhodblock 3 inhibited phospho-MRLC localization as well as furrow localization of the septin Peanut and increased Peanut’s localization on microtubules, but did not affect any of the other proteins we tested. Relocalization of Peanut to midzone microtubules has been reported for Anillin RNAi26. Rhodblock 1a, in contrast, inhibited furrow localization of the cortical proteins we tested (Actin, Anillin, Peanut), with Peanut strongly associating with microtubules. Rho was also mislocalized in about 50% of cells (Supplementary Fig. 9).
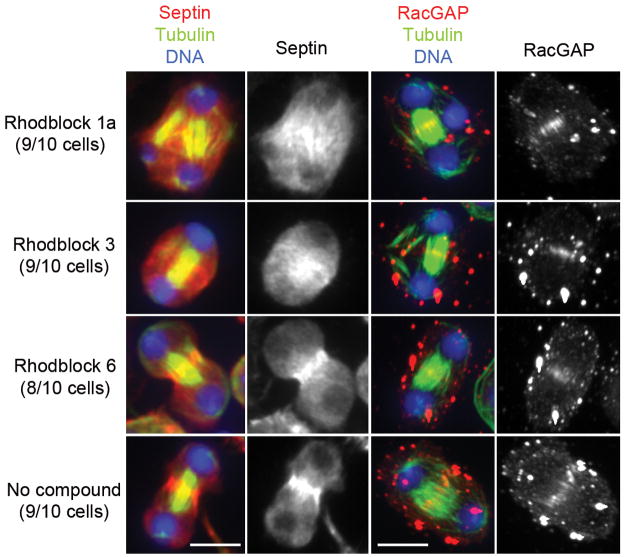
Figure 4. Effect of Rhodblocks 1a, 3 and 6 on cytokinesis protein localization.
The septin Peanut and RacGAP are shown in red in the color-combined figure and again for greater clarity in grey in the neighboring image. Microtubules are shown in green and DNA in blue. Cells were treated with compound for 4 h at the minimally synergistic concentration after overnight Rho RNAi sensitization. The septin Peanut localizes to midzone microtubules in Rhodblock 1a-treated cells and is partially microtubule-bound and partially diffuse in Rhodblock 3-treated cells. We analyzed images of 10 cells for each condition for Peanut staining. The number of cells represented by the image is shown in parentheses for each compound. RacGAP localization is not perturbed by any Rhodblocks. See Supplementary Figure 9 for Actin, Rho and Pavarotti staining. The images for each set of markers were taken under identical conditions and were processed identically (see Supplementary Methods). The scale bar represents 5μm.
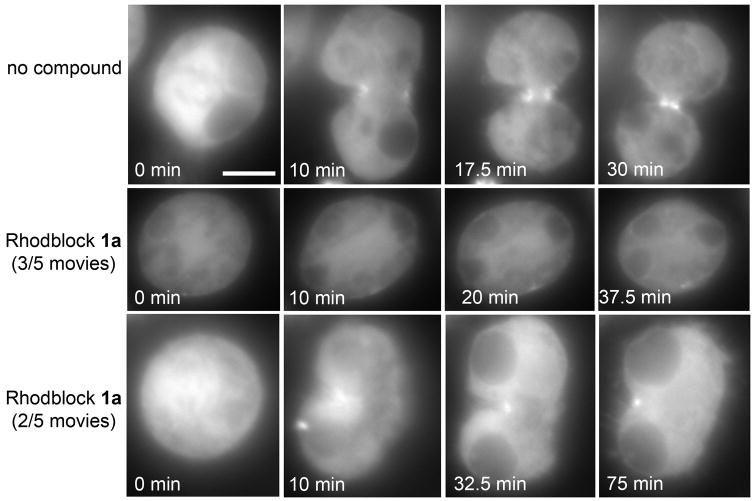
One of the major advantages of small molecule probes is that they are ideal for live imaging. We assessed the effect of Rhodblocks 1a, 3 and 6 on Drosophila S2 cells labeled with GFP-MRLC (Fig. 5 and Supplementary Fig. 10). We were able to observe cytokinesis inhibition in real time with Rhodblocks 1a and 6, but not with Rhodblock 3, even at 200 μM in Rho RNAi sensitized cells. Since the overall level of MRLC is higher in cells expressing GFP-MRLC, it is possible that the target of Rhodblock 3 is more directly connected to MRLC and therefore sensitive to myosin concentrations. For Rhodblocks 1a and 6, we used the faint localization of GFP-MRLC to the mitotic spindle identify cells in metaphase, added compound and watched the cells undergo (or fail) cytokinesis. We did not observe furrow ingression in any of the five cells we evaluated in the presence of Rhodblock 6. In two movies, cells failed to elongate after metaphase (Supplementary Fig. 10), similarly to phenotypes reported for rok RNAi21. We expected to see some variations in cellular responses because, given the speed of cell division, it is difficult to add compound at exactly the same stage in each replicate movie and small changes in the time of addition can have big effects because of the tight temporal regulation of cytokinesis. We also found that some cells treated with Rhodblock 1a briefly attempted to form a partial furrow, which then fell apart, resulting in a binucleate cell (Fig. 5). Some cells did not assemble a furrow and failed cytokinesis without attempting to ingress.
Figure 5. Movie stills of GFP-MRLC S2 cells treated with 100μM Rhodblock 1a after overnight Rho RNAi sensitization.
In 3/5 movies (middle panel), no furrows formed and cells failed to ingress. In 2/5 movies (lower panel) a partial furrow formed, briefly ingressed and broke apart. Movie timing was started at the beginning of anaphase. The scale bar shown in the first frame of the control movie represents 5μm.
Focusing in more detail on Rhodblocks 1a, 3 and 6, we have shown that these compounds have different effects on cytokinesis proteins. We conclude from these data that our compounds are useful probes to dissect the role of the Rho pathway during cytokinesis.
DISCUSSION
Combining different types of perturbations, for example genetic and small molecule treatments, can expand our understanding of complex biological processes27. Here, we report a strategy to discover small molecules that target signaling pathways, combining small molecule treatments with RNAi. Cells are sensitized to small molecules by lowering the levels of Rho, a key protein within the signaling pathway. Reducing the amount of target protein in a cell to identify specific small molecule ligands has been used successfully in the discovery of antibiotics28. Despite the appeal of this strategy, it has been difficult to adapt it to higher organisms because many proteins display functional redundancies, obscuring possible synergistic relationships. We overcame this limitation by performing our screen in Drosophila cells, a genetically less complex model system that is useful because many small molecules active in Drosophila are also active in human cells29. For example, RNAi depletion of one of the three human Rho isoforms does not cause cytokinesis failure,30 whereas depletion of the single isoform of Drosophila Rho inhibits cytokinesis. Our combination pathway screen has resulted in several small molecules that target the Rho pathway in Drosophila cells. We are currently investigating the effects of these small molecules on human cells.
Previous to this study, three classes of compounds were known to affect the Rho pathway: Rho kinase inhibitors are used in the clinic to treat cardiovascular diseases have been used as probe compounds to study aspects of the Rho pathway3,31. Statins inhibit HMG-CoA reductase (and ultimately cholesterol biosynthesis) and therefore also prevent isoprenylation required for active Rho31,32. While hugely successful in the clinic as cholesterol lowering agents, the statins are of limited use to study the Rho pathway because they affect multiple pathways. Recently, inhibitors of the human formin mDia have been reported33,34. In addition to discovering an inhibitor of Rho kinase (Rhodblock 6), we report several compounds that affect the Rho pathway. These compounds have different phenotypes suggesting that they have different mechanisms of action and therefore target pathway proteins that have not previously been targeted by a small molecule.
In this paper, we used myosin phosphorylation as a measure of Rho pathway activity. Interestingly, only one of the eight compounds (Rhodblock 6) that inhibited myosin phosphorylation inhibited Rok, the protein that is thought to be mainly responsible for MRLC phosphorylation21,35. It is likely that some of the Rhodblocks act upstream of Rok, resulting in the down-regulation of Rok and therefore eventual inhibition of myosin phosphorylation. Rhodblock 1a is a candidate for upstream action because it causes the mislocalization of several Rho pathway proteins. Rhodblock 3 inhibits recruitment of phospho-MRLC and Peanut, but not Anillin or other proteins, while Rhodblock 6 does not significantly inhibit cleavage furrow recruitment of any Rho pathway proteins other than phospho-MRLC. The factors that control recruitment of myosin to the cleavage furrow and its activation have been the subject of several recent studies35–37. There seems to be a consensus that some Rho pathway proteins that are required for active myosin are delivered along interzonal microtubule structures38,39. However, it is less clear how these proteins interact to achieve myosin activation. Our compounds prevent accumulation of phosphorylated myosin at the cleavage furrow while differentially affecting other Rho pathway proteins. We anticipate that the Rhodblocks will be useful in understanding this important aspect of cytokinesis regulation.
Investigations into the role of Rho pathway proteins during cytokinesis and determinants of their localization are active areas of research30,40–42. Successful cytokinesis requires that the components of the cytokinetic machinery be properly assembled, organized and maintained at the cleavage furrow. Recent studies indicate that Anillin functions as a key scaffolding protein that brings together other Rho pathway proteins including Rho, RacGAP and pebble as well as actin, myosin and the septin Peanut26,43–46. RacGAP interacts with the kinesin-6 protein Pavarotti to make up the Centralspindlin complex, which is critical for microtubule bundling, central spindle assembly and cytokinesis completion 25. Therefore, Anillin functions as a molecular bridge that links the actomyosin contractile ring with the Centralspindlin complex and spindle microtubules at the cleavage furrow. A combination of different Rhodblock treatments (Figs. 3, 4 and Supplementary Fig. 9) confirmed a sequential requirement of protein localizations during cytokinesis, i.e. myosin localizes independently of actin and Anillin47 and properly localized RacGAP is needed to localize Anillin, which is needed to localize Septin26,46. Rhodblock 3 gives us some insights into the organization of cortical Rho pathway proteins. It mislocalizes phospho-MRLC and septin, but not Anillin, suggesting that Anillin localization is independent of these proteins and that Anillin is an important early component of the furrow.
Anillin and RacGAP have been shown to interact directly in pulldown and yeast two hybrid experiments26,48. In Drosophila embryos, these two proteins are mutually required for localization, while in cultured cells RNAi of RacGAP disrupted Anillin, but not vice versa. Since Rhodblock 1a disrupts Anillin, but not RacGAP, it is likely to target the interaction between RacGAP and Anillin, either directly or by interfering with the regulation of this interaction. Because Rhodblock 1a is a small molecule that affects its target while it is still present in cells (unlike RNAi, see below) we can use it as a more direct means to dissect the mechanisms regulating the interaction between Anillin and RacGAP50C.
More generally, Rhodblock 1a perturbed the cortical proteins we tested (actin, Anillin, phospho-MRLC, Peanut and partially Rho), while leaving the Centralspindlin complex unaffected. This means that Rhodblock 1a disrupts the connection between the cortical and microtubule-bound activities of the Rho pathway, suggesting that microtubule-bound proteins are indeed responsible for the correct delivery of cortical Rho pathway proteins and function upstream within the signaling cascade. This notion is further supported by our live imaging data in GFP-MRLC labeled cells. In some cells, cytokinesis failed without assembly of a furrow, while in other cells a partial furrow briefly formed. As can be seen in figures 3 and 4, Rhodblock 1a induces additional midzone microtubule bundles that often point sharply towards the edge of the cells where the furrow would normally form. It is possible that cortical Rho pathway proteins are delivered to the furrow along these aberrant bundles, briefly attempt to form a furrow and then dissociate because they are asymmetric or because other ring assembly signals are lacking.
Most of the work from other labs discussed in the previous paragraphs has used RNAi to perturb Rho pathway signaling because few active small molecule inhibitors of the Rho pathway existed. Although there was generally good agreement between reported RNAi experiments and our compound treatments, it is important to keep in mind that small molecule treatments and RNAi can have different effects on cells. RNAi leads to the removal of the target protein, while a small molecule disrupts or inhibits a protein that is still present in the cell49. Since many of the Rho-regulated proteins like Anillin and the septins have important scaffolding functions, small molecules that affect these proteins, such as Rhodblocks 1a or 3, can be particularly useful because they allow manipulation of protein function without removing the protein. Therefore, as we identify more cellular targets of the Rhodblocks, we anticipate that we will gain further insight into the role of the Rho pathway in cytokinesis and other processes.
In addition to providing interesting and potentially valuable tools to study and manipulate the Rho pathway, our pathway screen based upon RNAi sensitization is a proof-of-principle study that should be widely applicable to many signaling pathways.
METHODS
Cell Culture
Drosophila Kc167 cells were grown at 25°C in Schneider’s medium (GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) and penicillin/streptomycin (Cellgro) in T25 and T75 flasks (BD Biosciences).
RNAi-sensitized small molecule screen
Details regarding preparation of double-stranded (ds)RNAs for RNAi treatments are described in the Supplementary Methods. On Day 1, 6 ml of serum-free Schneider’s medium containing 4 μg/ml Rho dsRNA was added to Drosophila Kc167 cells grown in T75 flasks at 25°C. After 1 h, 18 ml of Schneider’s medium supplemented with 10% heat-inactivated fetal bovine serum and penicillin/streptomycin was added and cells were incubated for 24 h. On Day 2, cells were arrayed in 384-well plates (Costar 3712) at 15,000 cells/well in 40μl complete growth medium. On Day 3, 100 nl of library compounds in DMSO was pin transferred into wells with Rho RNAi-sensitized cells and incubated 24 h. Cells were then fixed, stained, imaged and analyzed (see below). The screen was performed at ICCB-Longwood at Harvard Medical School. We screened the known bioactives collection from Biomol, fungal extracts (ICBG 2 and 4), the Starr foundation library, ChemBridge3, ChemDiv5 and ChemDiv4 libraries (http://iccb.med.harvard.edu/screening/compound_libraries/index.htm). For further screening details, see Supplementary Table 1. We ordered Rhodblocks 1–9, confirmed their identity and purity (Supplementary Methods and Supplementary Table 6) and used 10 mM or 50 mM stock solutions in DMSO in subsequent experiments.
Imaging
For screening, cells were fixed and permeabilized in 100 mM Pipes/KOH (pH 6.8), 10 mM EGTA, 1 mM MgCl2, 3.7% formaldehyde, and 0.2% TritonX-100 for 15 min and then washed in PBS. Whole cells were stained with 0.5 μg/ml NHS-tetramethylrhodamine (TMR, 5-[and-6]-carboxytetramethylrhodamine, succinimidyl ester C, Molecular Probes) in PBS. DNA was stained with 5 μg/ml Hoechst 33342 in TBST (TBS with 1% TritonX-100) for 15 min. Cells were then washed twice with TBST and sealed with aluminum seals (Costar) for image acquisition50. Cells were imaged using the ImageXpress Micro (Molecular Devices) at ICCB-Longwood using the 20X objective.
For spinning disk confocal microscopy, cells were grown on glass coverslips and were fixed and permeabilized as above. Cells were blocked in AbDil (TBST with 2% BSA) for 30 min and stained at 4°C overnight with one of the following antibodies diluted in AbDil: anti-phospho myosin light chain 2 (#3671S, Cell Signaling), anti-Anillin, anti-Peanut (gifts from Christine Field, Harvard Medical School), anti-Rho (p1D9; Iowa Hybridoma Bank), anti-RacGAP50C (a gift from Robert Saint, Australian National University) and anti-pavarotti. The anti-pavarotti antibody was raised in rabbits against the C-terminal peptide CNLGIEGHSSKKSKI. Actin was stained with TRITC-phalloidin (P1951, Sigma). Cells were washed with TBST and stained for 2 h with secondary antibodies (e.g. 1:1000 goat anti-rabbit or anti-rat Alexa Fluor 594 (Invitrogen)) followed by 2 h with 1:2000 fluorescein isothiocyanate-labeled anti-tubulin (DM1alpha, Sigma) in AbDil. The DNA was stained with 5 μg/ml Hoechst 33342 in TBST for 15 min followed by two TBST washes. Coverslips were mounted on glass slides using ProLong Gold (Invitrogen). Details regarding confocal image acquisition and linescan analysis are described in the Supplementary Methods.
For live-cell imaging, cells were allowed to attach on 25mm Round No. 1.5 glass coverslips (64-0715, Warner Instruments) for 1h prior to imaging. S2 cells expressing GFP fused to myosin regulatory light chain were a gift from E. Griffis, UCSF39. Myosin-GFP images were collected at 2.5 minute time intervals with a Nikon TE2000E microscope equipped with a 100X Plan Apo NA 1.4 objective lens.
RNAi of Rho pathway proteins
For table 1, cells were sensitized with a 1 day RNAi treatment (as described above) of Rho pathway proteins (RhoA, Pebble, RacGAP50C, Diaphanous, Citron kinase, Rho kinase at 4, 1, 5, 16, 1 and 16μg/ml dsRNA respectively), followed by a 24 h small molecule treatment (2 day total RNAi treatment). In the localization experiments in Figures 3 and 4 and Supplementary Figure 9, control and RNAi sensitized cells were treated for 4 h with compound. For double RNAi treatments, 10 μg/ml Dia dsRNA was used.
Synergy Ratios
The synergy ratio in Table 1 is defined by the following equation (Equation 1) where the observed phenotype refers to the percentage of binucleate cells experimentally measured after combined small molecule and RNAi treatment (ab), while the expected phenotype refers to the expected percentage of binucleate cells after combined treatment assuming a multiplicative model where small molecule (a) and RNAi treatments (b) are statistically independent. The background level of binucleate cells present in the untreated controls has been subtracted from all treatment values (indicated by the prime symbol).
A synergy ratio equal to 1 indicates no synergy between the small molecule and RNAi treatments. If there is synergy, i.e. cooperative effects between the two treatments, the level of binucleate cells should be higher than expected in a multiplicative model of individual treatments’ background levels, reflected in a synergy ratio greater than 1. A synergy ratio less than 1 indicates that one treatment suppresses the effects of the other. The maximum possible synergy ratio depends on the level of individual action. To allow a large range of synergy ratios, we set the experimental conditions reflected in Table 1 such that the individual actions were small.
Rho kinase assay
To obtain Drosophila Rho kinase, subconfluent Drosophila Kc167 cells in T75 flasks were transiently transfected with 40μg full length Rok-pAFW (FLAG plasmid pAFW was obtained from the Drosophila Gateway Collection) and expressed for 3 days. Rok-FLAG was then purified by pull-down from cell lysates using Anti-FLAG M2 agarose (Sigma). Aliquots of kinase were snap frozen and stored at −20°C. For the kinase assay, compounds were incubated with Rok-FLAG and myelin basic protein (MBP) substrate in kinase buffer (20mM Tris, 1mM MgCl2, 25mM KCl, 1mM DTT, 0.04mg/ml BSA). After 15 min, the kinase reaction was initiated by the addition of ATP (100μM final) including approximately 0.3μCi/μl [γ-32P]ATP. Reactions were performed in a total volume of 20μl. After 10 min, the reaction was terminated by spotting 17.5μl of the reaction mixture on P81-phosphocellulose paper (diameter 2.1cm, Whatman). P81 circles were then washed four times (5min each) with 0.75% phosphoric acid, once with acetone and dried. CPM values were then determined by liquid scintillation counting.
Supplementary Material
Acknowledgments
We thank the staff at ICCB-Longwood, the Nikon Imaging Center at HMS and the Broad Institute’s Imaging Platform for their assistance and Dr. Fritz Roth for helpful discussions. Funding for M.S.V., T.R.J. and A.E.C. was from the Life Sciences Research Foundation (Novartis), L’Oreal for Women in Science, the Society for Biomolecular Screening and NIH 5 RL1 CA133834-03. A.B.C., Y.S., A.D.T. and U.E. were supported by NIH grant R01 GM082834, the Claudia Adams Barr Program and the Dana-Farber Cancer Institute.
Footnotes
Author contributions
A.B.C. and U.S.E. designed the study. A.B.C., Y.S., A.D.T. and U.S.E. designed and conducted experiments and analyzed data. A.B.C., Y.S., M.S.V., T.R.J. and A.E.C. designed and performed automated image analysis. U.S.E. wrote the manuscript, with input from the other authors.
The authors declare no competing financial interests.
References
- 1.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 3.Budzyn K, Marley PD, Sobey CG. Targeting Rho and Rho-kinase in the treatment of cardiovascular disease. Trends Pharmacol Sci. 2006;27:97–104. doi: 10.1016/j.tips.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999;81:682–687. doi: 10.1002/(sici)1097-0215(19990531)81:5<682::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 7.Benitah SA, Valeron PF, van Aelst L, Marshall CJ, Lacal JC. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim Biophys Acta. 2004;1705:121–132. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Eggert US, Mitchison TJ, Field CM. ANIMAL CYTOKINESIS: From Parts List to Mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 9.Walker K, Olson MF. Targeting Ras and Rho GTPases as opportunities for cancer therapeutics. Curr Opin Genet Dev. 2005;15:62–68. doi: 10.1016/j.gde.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Aznar S, Fernandez-Valeron P, Espina C, Lacal JC. Rho GTPases: potential candidates for anticancer therapy. Cancer Lett. 2004;206:181–191. doi: 10.1016/j.canlet.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 12.Doe C, et al. Novel Rho kinase inhibitors with anti-inflammatory and vasodilatory activities. J Pharmacol Exp Ther. 2007;320:89–98. doi: 10.1124/jpet.106.110635. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones TR, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genth H, et al. Entrapment of Rho ADP-ribosylated by Clostridium botulinum C3 exoenzyme in the Rho-guanine nucleotide dissociation inhibitor-1 complex. J Biol Chem. 2003;278:28523–28527. doi: 10.1074/jbc.M301915200. [DOI] [PubMed] [Google Scholar]
- 16.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 17.Hirose K, Kawashima T, Iwamoto I, Nosaka T, Kitamura T. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol Chem. 2001;276:5821–5828. doi: 10.1074/jbc.M007252200. [DOI] [PubMed] [Google Scholar]
- 18.Maddox AS, Oegema K. Closing the GAP: a role for a RhoA GAP in cytokinesis. Mol Cell. 2003;11:846–848. doi: 10.1016/s1097-2765(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 19.Prokopenko SN, et al. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe N, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. Embo J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickson GR, Echard A, O’Farrell PH. Rho-kinase controls cell shape changes during cytokinesis. Curr Biol. 2006;16:359–370. doi: 10.1016/j.cub.2005.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zavortink M, Contreras N, Addy T, Bejsovec A, Saint R. Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J Cell Sci. 2005;118:5381–5392. doi: 10.1242/jcs.02652. [DOI] [PubMed] [Google Scholar]
- 23.Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 24.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 26.D’Avino PP, et al. Interaction between Anillin and RacGAP50C connects the actomyosin contractile ring with spindle microtubules at the cell division site. J Cell Sci. 2008;121:1151–1158. doi: 10.1242/jcs.026716. [DOI] [PubMed] [Google Scholar]
- 27.Lehar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 29.Perrimon N, Friedman A, Mathey-Prevot B, Eggert US. Drug-target identification in Drosophila cells: combining high-throughout RNAi and small-molecule screens. Drug Discov Today. 2007;12:28–33. doi: 10.1016/j.drudis.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Kamijo K, et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. [Google Scholar]
- 31.Ishizaki T, et al. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- 32.Liao JK, Laufs U. Pleiotropic effects of statins. Annu Rev Pharmacol Toxicol. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizvi SA, et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. pii. doi: 10.1016/j.chembiol.2009.10.006. S1074-5521(09)00333-0. 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauvin TJ, Fukui J, Peterson JR, Higgs HN. Isoform-selective chemical inhibition of mDia-mediated actin assembly. Biochemistry. 2009;48:9327–9329. doi: 10.1021/bi901354z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dean SO, Spudich JA. Rho kinase’s role in myosin recruitment to the equatorial cortex of mitotic Drosophila S2 cells is for myosin regulatory light chain phosphorylation. PLoS One. 2006;1:e131. doi: 10.1371/journal.pone.0000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci U S A. 2005;102:13473–13478. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 38.Foe VE, von Dassow G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J Cell Biol. 2008;183:457–470. doi: 10.1083/jcb.200807128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vale RD, Spudich JA, Griffis ER. Dynamics of myosin, microtubules, and Kinesin-6 at the cortex during cytokinesis in Drosophila S2 cells. J Cell Biol. 2009;186:727–738. doi: 10.1083/jcb.200902083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishima M, Glotzer M. Cytokinesis: a logical GAP. Curr Biol. 2003;13:R589–591. doi: 10.1016/s0960-9822(03)00521-9. [DOI] [PubMed] [Google Scholar]
- 42.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 44.D’Avino PP. How to scaffold the contractile ring for a safe cytokinesis - lessons from Anillin-related proteins. J Cell Sci. 2009;122:1071–1079. doi: 10.1242/jcs.034785. [DOI] [PubMed] [Google Scholar]
- 45.Hickson GR, O’Farrell PH. Anillin: a pivotal organizer of the cytokinetic machinery. Biochem Soc Trans. 2008;36:439–441. doi: 10.1042/BST0360439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hickson GR, O’Farrell PH. Rho-dependent control of anillin behavior during cytokinesis. J Cell Biol. 2008;180:285–294. doi: 10.1083/jcb.200709005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory SL, et al. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr Biol. 2008;18:25–29. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 49.Eggert US, Field CM, Mitchison TJ. Small molecules in an RNAi world. Mol BioSyst. 2006;2:93–96. doi: 10.1039/b515335b. [DOI] [PubMed] [Google Scholar]
- 50.Eggert US, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.