Abstract
Local shape descriptors compactly characterize regions of a surface, and have been applied to tasks in visualization, shape matching, and analysis. Classically, curvature has be used as a shape descriptor; however, this differential property characterizes only an infinitesimal neighborhood. In this paper, we provide shape descriptors for surface meshes designed to be multi-scale, that is, capable of characterizing regions of varying size. These descriptors capture statistically the shape of a neighborhood around a central point by fitting a quadratic surface. They therefore mimic differential curvature, are efficient to compute, and encode anisotropy. We show how simple variants of mesh operations can be used to compute the descriptors without resorting to expensive parameterizations, and additionally provide a statistical approximation for reduced computational cost. We show how these descriptors apply to a number of uses in visualization, analysis, and matching of surfaces, particularly to tasks in protein surface analysis.
Index Terms: Curvature, descriptors, npr, stylized rendering, shape matching
1 Introduction
Local shape descriptors distill the shape of a region of a surface into a short vector of numbers, each corresponding to a property of the region. These descriptors have broad application when working with shapes: for example, they are used in visualizing and analyzing scientific data, shape matching, and in stylized rendering. While various local shape descriptors and methods for computing them exist, their inability to summarize the shape of larger regions limits their utility.
Our goal is a local surface shape descriptor that is applicable at different scales to summarize the shapes of differently sized neighborhoods. This allows it to be applied to smaller regions to capture small-scale detail, or to larger neighborhoods to summarize their overall shape. Regions of the surface may have one shape at a small scale, but a different shape at a larger scale (e.g. a small bump within a large bowl). This paper introduces an approach called multi-scale surface descriptors that meets this goal. We present a local shape descriptor that can be applied at multiple scales, along with techniques for computing them efficiently on a triangle mesh.
The shape of any finite region may contain arbitrary amounts of detail, therefore a shape descriptor can only provide a summary. For an infinitesimal region, the amount of detail is limited, so the shape can be completely described by its curvature. Curvature provides a compact local descriptor: three or four numbers are sufficient to characterize the shape for an infinitesimally small region. For finite-sized regions, however, the mathematics of curvature do not apply.
We provide a descriptor that captures the most significant features of the shape of a local surface region. The descriptor considers a local neighborhood around a central point with a roughly circular area specified by radial distance. It measures the degree and type of non-planarity of the region, for example encoding whether something is a steep bump or a shallow bowl. It also captures the degree and direction of anisotropy, identifying troughs and ridges. A key insight of our approach is that while these quantities are not sufficient to capture all details of the shape of a finite region, they do capture the most significant aspects of shape. We introduce robust and practical methods for computing these larger-scale surface descriptors, and show their usefulness in a number of applications.
One of our key motivating applications is the matching of molecular surface regions to identify potentially similar chemical functionality. An important aspect of this functionality is surface shape complementarity: a binding partner for a protein will often have locally complementary shape to its region of binding. Much as a key fits only its matching lock, complementarity implies that binding is highly stereospecific. Therefore, by characterizing the shape of a known binding pocket, and then using this information to identify similar regions in other proteins, we may find new targets for a given partner.
This application, which we discuss in more detail in §5.2, highlights many of the requirements for practical, effective local shape descriptors and the methods to compute them: they must operate over large enough neighborhoods to be chemically significant; they must be efficient, as we need to compute the descriptors for all points on each molecule in a database; they must be robust against poorly tesselated surfaces; and they must correspond to domain scientists’ intuition about shape and neighborhood. We provide the first shape descriptors that we feel are able to meet these needs.
1.1 Contribution
The contribution of this paper is an approach to local shape descriptors that provides a method for characterizing neighborhoods at multiple scales on the surface of a mesh. Our approach is the first to address all of the following goals for such descriptors:
It scales to describe larger neighborhoods. Curvature captures only infinitesimal regions and prior approaches to curvature computation focus on minimizing the size of regions to better approximate the differential case. In §3 we present methods that summarize a non-trivial region statistically.
It corresponds to intuitions about curvature. Like curvature, our descriptor captures the degree and type of non-planarity of a region, and the degree and direction of anisotropy.
It allows for control of scale in a simple way. Neighborhoods are specified by their center and a radius, so regions are controlled by physically relevant quantities of the surface. In contrast, methods such as mesh pre-filtering require a less direct specification of neighborhood size in terms of frequency (which can be difficult to explain to domain scientists), and filtering causes points to move, precluding localized assessment.
It compensates for issues in tessellation. The methods of §3.1 account for discretization, allowing our descriptor to be robust to poorly tesselated surfaces.
It affords efficient computation. Throughout §3 we present methods for computing the descriptors efficiently. In particular, our approach avoids expensive parameterizations and our approximations can provide good performance without resorting to expensive mesh operations such as exact geodesic computations.
It works in applications. In §5 we describe how our descriptor is effective in applications. By providing a larger-scale summarization of surface regions, it allows for simpler algorithms to make effective use of descriptors across a number of applications.
Our approach is closely related to prior work on using planar parameterizations and polynomial fitting for curvature estimation, which will be discussed in §2.1. However, unlike these methods that focus on the challenge of being robust with as small a region of surface as possible, our approach is designed to work with larger surface regions. This requires us to provide effective methods for finding the correct regions (§3.1) as well as to provide techniques for statistically summarizing the shape over these regions (§3.3, §3.3.1).
2 Related Work
2.1 Curvature
Many papers deal with the task of computing curvature for points on the surface: see [7] for an overview. Petitjean [25] surveys methods for estimating local surface quadratics, several of which inform our work. We draw inspiration for our point- and normal-weighting techniques from [24], who use a technique they describe as “normal vector voting” to compute curvature in the presence of noise.
Curvature estimation may break down in certain cases, as shown in [10]. Rusinkiewicz [28] shows how to avoid similar issues in per-vertex curvature computation by estimating the second fundamental tensor per-face, and incorporating that into a description of the 1-ring curvature about a point. In some cases, only a minimal description of the surface may be required, such as its radius of curvature. This can be found by fitting spheres to points on the surface [4]. Though more involved, ellipsoid fitting can indicate anisotropy [21].
Most discrete curvature fitting methods, such as Angle Deficit and Angle Excess, are very fast, but do not provide principle directions, and so cannot give anisotropy information [7, 32].
2.2 Applications
Local shape descriptors are useful for a number of applications. Many have been developed, in numerous forms, for the task of shape matching, and share a subset of our goals. Körtgen, et al. [19] and Gatzke, et al. [8] both use the statistics over the neighborhood surrounding a point to perform robust matching. Unlike ours, the former does not directly utilize the surface. The latter, while more similar to ours, differs in that they build up their descriptor using differential curvature estimates, rather than estimating curvature on the patch as a whole. Spin-images [16] represent such a neighborhood as a 2D texture, which can be quickly built, and can be used to perform rotationally-invariant matching. While fast, these are best suited for local feature comparison.
Gal [6] models the surface using local shape descriptors to construct partial matches, while [3] uses point signatures to detect rigid structures on a set of faces, which are then compared to one another. Goldman, et al. [11] describe a local quadratic shape descriptor that they use for molecular similarity searching. Though these methods each have similarities to ours, none satisfy all of our goals.
Many graphics and visualization techniques use curvature, and therefore may benefit from our improved descriptor. Gumhold [14] describes an algorithm to optimize the placement of lights in a scene, in order to emphasize high-curvature regions. Lee, et al. [20] have used curvature, along with globally discrepant lighting, to similarly emphasize the placement of specularities on the surface.
Toler-Franklin, et al. [34] describe how lighting derived from large-scale curvature can approximate local ambient occlusion, to better emphasize large concave features. Real-time rendering techniques also consider curvature: [35] show how extracting curvature information from image space can produce compelling lighting and shading in real-time, while [18] extracts curvature from volumetric data to enhance their visualizations.
Stylized rendering techniques benefit from the use of curvature as well. Principle curvatures can be used to depict the flow of curvature over the surface [9], and to then place textures along those flows, emphasizing important shape cues [13, 27, 2]. Line drawing techniques also utilize curvature [17, 5].
Curvature can directly inform another task, surface partitioning, by using high-water curvature as partition boundaries [22], or by placing seams in high-curvature regions to better hide them [31]. Mortara, et al. [23] uses the intersection of the surface and bubbles at multiple scales to estimate curvature for surface segmentation.
3 Multi-Scale Descriptors
Given a triangle mesh and a neighborhood defined by a center point i (a vertex in the mesh) and size d, our approach computes a local descriptor of this region as a statistical characterization of its shape. See Figure 1 for a depiction of this process. The radius, d, gives control over the scale of the neighborhood and has units of length with the same scale as the surface itself. The choice of radius depends on the application domain. For example, when considering molecular surfaces we choose d to be a size of biological relevance.
Fig. 1.
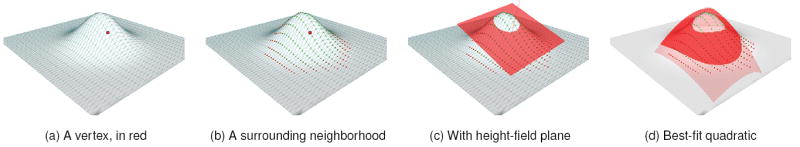
A demonstration of the steps involved in generating a descriptor for a single neighborhood. Our method starts with a vertex on the surface (a). It then generates a set of patches surrounding that vertex. One is shown in (b), with color given according to each vertex’s weight (greener vertices have high weight, and redder have low weight). A plane is constructed using a weighted average of the normals of that patch (c), onto which all points are projected. Finally, a quadratic surface is found to approximate that height field (d).
Our approach has the following steps:
The set of points, N(i), in the neighborhood around i are determined and a weight computed for each to compensate for issues in tessellation. In §3.1 we describe how the points can be determined efficiently and how the weights are chosen.
N(i) is represented as a height field on the surface tangent plane at i. This representation, motivated by methods in curvature estimation, simplifies the computation of the statistics in the next step. In §3.2 we describe how the tangent plane is determined and explain the height field representation.
The height field is characterized statistically to yield the descriptor. We provide two methods for this summarization. In §3.3 we describe an approach using robust least-squares fitting that directly applies the intuitions of curvature. In §3.4 we provide a method that is based on simpler intuitions of shape. In §4.2.1 we compare both approaches.
The result of these steps is: one number that describes the type and degree of non-planarity of the neighborhood, one number assessing the degree of anisotropy, and a vector giving the direction of anisotropy, assuming that there is sufficient anisotropy to determine the direction.
3.1 Neighborhood Construction
To characterize a given vertex, our method first finds the neighborhood of all vertices that are within a distance d from that vertex along the surface of the mesh, called a d-disc. Descriptors at multiple scales are constructed, then, by choosing a set of d values for a given surface. Small values characterize local regions, while larger values take more of the surface into account. In the latter case, small surface variations are deemphasized. Figure 4 shows the effect of varying patch sizes over the mesh.
Fig. 4.
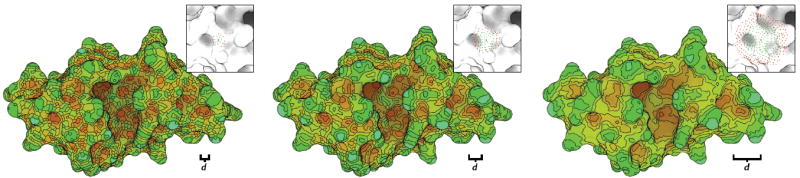
Shape description, represented at multiple scales, with peak-like regions in green and bowl-like regions in red. From left to right, the radius of the region, indicated by d, is increased. Inset, a single patch is shown for each scale, representing the neighborhood used for a single vertex.
To create uniformly-shaped patches on the surface, the geodesic distance should be used between points, as geodesics follow the shortest path on the surface. Computing geodesic distance on a mesh, however, is difficult because this path does not necessarily follow edges on the mesh. Even with efficient algorithms, such as [33], exact geodesic computation is prohibitively time-consuming and, as we describe below, unnecessary.
A common approximation to geodesic distance is simply to follow the edges between vertices. The distance between any two vertices, then, is the shortest patch along the edges connecting those vertices. This so-called Dijkstra path is an approximation to the true geodesic distance. Though fast to compute, Dijkstra distance can be an arbitrarily poor approximation to geodesic distance. This problem is significant in practice, as the use of Dijkstra distance in determining the neighborhood gives irregularly shaped regions, as shown in Figure 2, and is unacceptable for descriptor computation. The problem is not pathological, but rather, occurs frequently on real meshes.
Fig. 2.
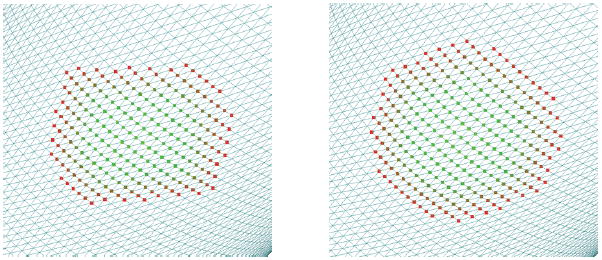
On the left is the result of performing a search along edges for those points that are less than a particular distance from a single vertex. Though, intuitively, this should form a circular patch because of the mesh’s tessellation, the patch has a non-circular shape. By adding virtual edges to 2-neighbors, we achieve a better result, at minimal incremental cost.
We introduce a modification to Dijkstra distance that improves its distance estimates. Our insight is that a large discrepancy in the distance computed between two vertices results from a large difference between the geodesic path and the edgewise path. Further, the majority of these cases arise in an edgewise path of length 2 (e.g., two hops). To close the gap, we simply add virtual edges between each point and all of its 2-neighbors, and perform a Dijkstra computation on the original mesh with these added neighbors. In theory, these “corner cutting paths” may still have lengths that are arbitrarily poor approximations of the geodesic distances, but we have not encountered such pathological cases in practice. Figures 2 and 3 show the results of this simple modification. We compare this method with true geodesics in §4.2.2.
Fig. 3.
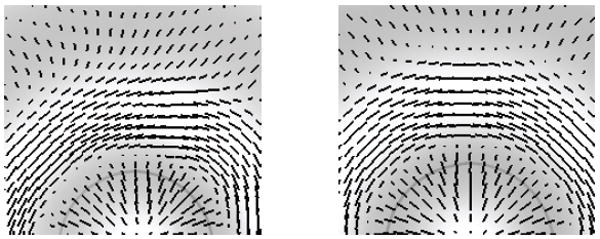
Here, directions of least curvature are found for a radially symmetric cosine wave. On the left, we show the result from computing distance along edges of the mesh. On the right, we add edges between 2-neighbors to better avoid tessellation issues. Note that the resulting directions are not symmetric on the left, but are on the right.
Values for d can be chosen in one of two ways. For surfaces with an inherent scale, such as proteins, d may be set according to the size of known features, to describe, for instance, atom- or residue-sized neighborhoods on the surface; for molecular examples in this paper, we set d to 4 and 8 Ångströms, respectively. For surfaces that have no inherent scale, we can simply let the user choose their own, or use a heuristic, such as a multiple of the median edge length, to automatically pick a reasonable size.
3.1.1 Weighting
Before using the constructed neighborhood, N(i), our algorithm first adjusts the weights of its individual vertices:
Points are weighted according to the area of the surface nearest to them. This reduces the effect of any variability of the size of triangles on a surface.
Points closer to the edge of the patch are weighted to contribute less to the overall shape description than those closer to the center. This compensates for the fact that, for any given angular wedge, outer regions will contain more surface area than inner regions.
Given these two factors, our equation for the weight of a vertex p ∈ N(i) is:
| (1) |
where A(p) refers to the area of the faces surrounding p and D(i, p) refers to the shortest distance from p to i. Note that dividing by D(i, p) ensures that concentric rings in the neighborhood have equal weight. Normalizing these weights produces the final values:
| (2) |
3.2 Height Field
As described in [7], a common method for estimating curvature on the surface of a mesh is to construct a height-field function, parameterized by two variables, u and υ:
| (3) |
There are several reasons one might want to do this. First, projection to a height field acts as a change of coordinate system. In this case, the height field approximates a tangent plane, and is therefore a more natural basis for computing differential curvature, as well as our curvature. Second, by projecting a set of points onto a height-field, a 3D problem is reduced to a 2D problem, reducing its degrees of freedom, and therefore, its complexity.
The coordinates for each point on the patch are found by projecting that point onto a plane. The choice of plane will affect how good the overall parameterization will be. As we will describe below in §3.2.1, one measure of ‘goodness’ is to minimize the difference between the normals on the surface, and the normal of the plane of projection. So our method finds the plane normal Ni(p) as the weighted average of all surrounding vertex normals:
| (4) |
This vector is normalized to produce the final height-field normal, . It should be noted that while [1] give a more accurate method for averaging normals than our linear approximation, we have not seen a case in practice that would benefit from their method.
As a final step, we ensure that the central vertex in a patch is placed at the origin (so u0 = υ0 = 0). This is accomplished by subtracting the projected u, υ for that vertex from the u and υ of all other vertices.
3.2.1 Avoiding Issues with Height Fields
Several problems can arise from the use of height functions: first, projection does not preserve relative distances in the (u, υ) parameter space. Second is the issue of foldover: F(u, υ) may have multiple, conflicting values at a given u and υ. This is generally the result of including triangles that face away from the plane of projection.
We have experimented with various parameterization techniques, including exponential maps [30], to directly address both of these issues. Though this is a viable alternative, our techniques do not require a parameterization of the surface. Also, because we are using an orthonormal projection, relative 3D distance is preserved. Thus we are still able to achieve a reliable fit and projection isn’t a large issue. Local parameterizations, on the other hand, besides being slower to compute than height-field projections, may break down with distance; since we fit relatively large patches, this presents a problem.
The second problem, foldover, is more of a concern, but it can be mitigated by simply throwing away those vertices whose normals point away from the plane of projection. While this doesn’t completely remove foldover, remaining conflicting vertices have very low weight, so they do not tend to affect the result, as described in §4.2.5. Usage of robust statistics to lower the weight of outliers, as described below in §3.3.1, further improves matters.
3.3 Fitting with Quadratics
After representing the surface region as a height field, our approach must compute a descriptor that summarizes its shape statistically. Because the region may consist of a large number of points, the height field may have an arbitrarily complex shape. To summarize this shape, we first approximate it by a simpler shape that is easier to characterize. We fit a quadratic function to the height field. We choose quadratic functions because they are the simplest form that can sufficiently express the shape variability we need to encode.
Note that if the region were infinitesimal, its shape would exactly fit the quadratic form, and the coefficients of this quadratic would yield the curvature at the center point of the region. This is the basis of several curvature estimation approaches [7]. However, rather than trying to find a minimal set of points to constrain the quadratic, we instead use a large collection of points over the neighborhood and find the best-fit quadratic.
The form of equation to fit is:
| (5) |
Each point of the height field (ui, υi, zi) provides a linear constraint on the 5 degrees of freedom of the quadratic. Note that we do not include a constant term, which forces our function through the central vertex in a patch.
The simplest fitting method is to solve the set of linear constraints according to a weighted least-squares metric, with the weights from §3.1.1. Because there is a small number of variables, our implementation solves these linear least-squares problems by forming the normal equations (multiplying the matrix by its transpose), and solving the resulting linear system using the Cholesky factorization. The use of linear least squares for polynomial fitting is discussed in [26].
Once parameters have been found for the quadratic, the principle directions and magnitude of curvature of the quadratic patch can be estimated by finding the eigenvectors and eigenvalues, respectively, of the second fundamental form:
| (6) |
For our descriptor, we use these principle curvatures of the quadratic. Specifically, we include the eigenvector corresponding to the smallest eigenvalue (i.e., with shallowest curvature), along with the degree of anisotropy, which is derived from the ratio of the largest absolute eigenvalue to the smallest.
3.3.1 Robust Statistics
When there are many points, even the best fit quadratic surface may be a poor approximation. In particular, the least squares metric used for fitting is sensitive to outliers: a small number of badly fit points can skew the results. In local descriptor computations, outliers can come from surface foldovers, sharp discontinuities, as well as noise on the surface itself.
We find that by applying robust statistical techniques [15] to our initial fit, we can lessen the impact of outliers. Using M-estimation, with the original quadratic fit as a prior, regions that have folded under are quickly identified as outliers. Their weights are then lowered, reducing their contribution, and a new surface is fit. This can be iteratively applied to reweight points, driving the least-squares solution toward a better fit with those points that remain. In practice, we find that a single iteration of this re-weighting is required to provide sufficient robustness.
3.4 Moment-Based Surface Description
A different description of the shape of the local region is based on a simpler intuition that less directly corresponds with curvature. In §4.2.3, we contrast the two descriptors and show that their results are similar.
Consider the height field patch from the region as a rigid object, with the points having mass proportional to their weights. If the center of mass of the object is above the plane, the center point is likely to be at the bottom of a bowlike shape (or the top of a peak if the mass center is below). The distance between the center and the plane gives an assessment of how peak/bowl-like the shape is. Similarly, we can consider the statistical trend of the mass distribution. If the moment of inertia is strongly directional, then the region is anisotropic.
These simple intuitions lead to a very efficient statistical characterization of the height field. The centroid of the neighborhood is found quickly by averaging, and its height value gives the assessment of non-planarity. By treating the height field as an image (with the height mapping to intensity), image statistic methods can be used to determine the distribution. Image moment computations [12] determine the degree and direction of anisotropy.
3.4.1 2D Moment Computation
Image moments can be used to statistically deconstruct an image to find, for instance, its area, center of mass, and orientation. It is this last property, described using the second central moment, that we use to interpret anisotropy in a height field.
The second central moment is used to find the direction of highest intensity in an image, which can be used to uncover its orientation. For our method, consider the projected distance (or height) of a point on the plane as ‘intensity’. Intuitively, ridges in the height field will have the highest such intensity. Ridges, by definition, have lowest curvature along their major axis. By finding this direction, we then find an approximation to the direction of least curvature of the neighborhood.
These major and minor axes of intensity can be found by first constructing the central moments [12] up to order two:
| (7) |
| (8) |
Axes of intensity are then found by taking solving for the eigenvectors and eigenvalues of the covariance matrix:
| (9) |
One minor wrinkle: as described, this technique works only for ridges. To properly characterize neighborhoods that represent valleys, we simply invert the heights, replacing F(u, υ) with −F(u, υ) in equation (7).
4 Results
In this section, we discuss the performance and correctness of our descriptors. For all results below, the symbol ‘±’ represents a range of one standard deviation from a given value.
In this paper, we have implemented our multi-scale surface descriptors in a visualization testbed that runs under Windows on PCs. All figures in this paper are generated from within this testbed. For all molecular examples in this paper, we use MSMS [29] to generate an initial molecular surface as a triangle mesh.
4.1 Performance
To assess performance of our method, we built a corpus of 30 meshes, ranging in size from 2,600 to 170,000 vertices. Descriptors were computed over the surface of each mesh. On our test set, the time needed to construct these descriptors using quadratics ranged from 3 seconds to 4 minutes per mesh on an Intel Core 2 Duo, E8500 with 3GB of RAM. Runtime was generally a function of the largest neighborhood size, as a larger neighborhood has more vertices to consider, as well as the size of the mesh itself, as a set of descriptors is built for each vertex. In our testing, construction of vertex neighborhoods takes about the same amount of time as fitting the quadratics over those neighborhoods. We found moment computation to run three times faster than quadratic fitting. Combining these timings, moments can be calculated, on average, 50% faster than quadratics.
Our system provides the ability to cache the results of computing our descriptors onto disk for future use, so in cases when meshes must be visited many times, such as in the matching scenario we describe in §5.2, we do not have to repeatedly reconstruct them.
4.2 Evaluation
Tradeoffs were made in the construction of our descriptor to achieve high performance. In the following sections we assess the impact of these tradeoffs, both in terms of accuracy, as well as in terms of our sensitivity to both noise and to differences in surface tessellation.
4.2.1 Descriptor Equivalence
We describe a set of moment-based descriptors which further reduce computational requirements. These may not necessarily agree with quadratic descriptors. In these cases, we consider the quadratic descriptors to be the the ‘correct’ result. To assess the degree to which moments get the wrong answer, we ran both algorithms against the same corpus of meshes. On each mesh, we found the average angular difference over each mesh between the principle direction produced by moments and the direction produced by quadratics. For all meshes, this value averaged 16° ± 12°. The curvature values produced agreed with each other with an R2 value ranging from .81 to .95. This suggests that moments, by and large, produce similar results to quadratics, a result reinforced by visual inspection. We found that the majority of the cases where moments fail to agree with quadratic fitting occurred in two problem areas: edges, where quadratics were better able to fit in the absence of data, and regions with significant foldover. We are investigating methods to compensate for these scenarios.
We also compared our method to Rusinkiewicz’s local curvature estimation [28]. To do so, we constructed descriptors with small neighborhoods (equivalent, approximately, to 2-rings). We found that the principle directions found by quadratic descriptors deviated, on average, 14° ± 11°. Our curvature values agreed with the mean of theirs with an average R2 value of .83. It is difficult, however, to compare our methods directly as their method aims to deal with instantaneous neighborhoods directly, while ours cannot.
4.2.2 Geodesics vs. 2-Ring Approximation
We described in §3.1 a simple method for improving Dijkstra searches by adding virtual edges between 2-ring neighbors. This method, while more tolerant of issues with tessellation, is still an approximation to true geodesic distance. To quantify the improvements our method produces, and to determine how close our approximation comes, we compare geodesic neighborhoods of varying size, generated using the accurate method in [33], on the surface of our corpus to both our method, as well as to standard Dijkstra distance.
We find that for all protein surfaces in our corpus, for a patch radius of 8 Ångströms, our 2-ring approximation finds between 97 – 100% of the vertices in a geodesic patch of the same radius, with an average distance for the missing vertices of .1 Ångströms from the patch boundary. Standard Dijkstra finds 84–90%, with an average distance within .5 Ångströms of the boundary.
We also tested against a torus model, containing a “grain” in its tessellation. Because, unlike proteins, this model does not have an intrinsic scale, we assign one according to its median edge length, ‘em ’ = .8. For patches of size d = 10 * em ≃ .6, our approximation finds 92–95% of the vertices, with an average distance of .02. Standard Dijkstra finds only 76 – 80%, with average distance of .05.
4.2.3 Noise Sensitivity
To gauge the ability of our surface descriptor to tolerate noise, we used the same models as above, and then perturbed each vertex a random distance, up to .5 Ångströms, along its normal. Figure 5 shows one example protein, before and after this process. For all other models, vertices were perturbed up to .75 * em. Curvatures were assessed for each vertex on the original model, then compared against the same vertex on the perturbed model.
Fig. 5.
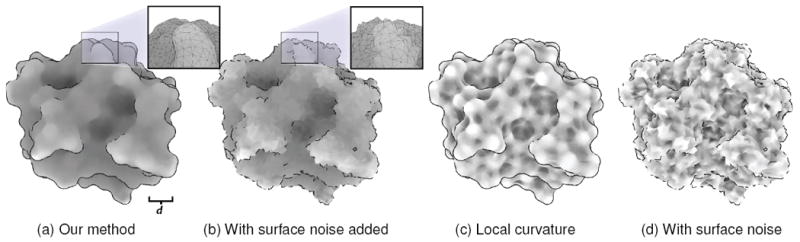
A depiction of the results from a test of our descriptors’ sensitivity to surface noise. (a) Shows our results on the surface of a protein, with lighter colors denoting areas of higher positive curvature. (b) Shows the same surface after noise is introduced. This test is repeated in (c) and (d) using local curvature estimation. Note the similarities between the results in (a) and (b), indicating a resilience to noisy surfaces, unlike (c) and (d).
For small patch sizes of around 2 Ångströms (or 2 * em), we found that this difference between respective vertices, on average, accounted for 19%±26% error in the reported curvature, versus the actual curvature. For patches 4 times larger, this dropped to 2%±6%. In contrast, local curvature had 25%±36% error. This is in accord with our visual observations that descriptors formed from larger neighborhoods seem more resilient to noise.
4.2.4 Tessellation Sensitivity
To test our method’s sensitivity to varying tessellation, we used models with uniform tessellation and known curvature. Tessellation “noise” was introduced by repeatedly selecting a triangle from the mesh, inserting a new vertex in the center of this triangle, and projecting this new vertex onto the surface of the model (for this reason, models with known analytic forms were used). This process creates meshes with high variability in the tessellation, and many examples of bad tessellations, such as high valence vertices and sliver triangles.
Two models were used in testing, a unit sphere and a unit torus; the sphere model began with 2,562 vertices, to which 5,000 more are added during subdivision. Figure 6 shows the sphere before and after subdivision. The torus begins with 4,800 vertices, and again 5,000 are added. One minor wrinkle in our testing: because the tessellated meshes had a much smaller em relative to the non-tesselated meshes, we set the tesselated model’s scale equal the original.
Fig. 6.
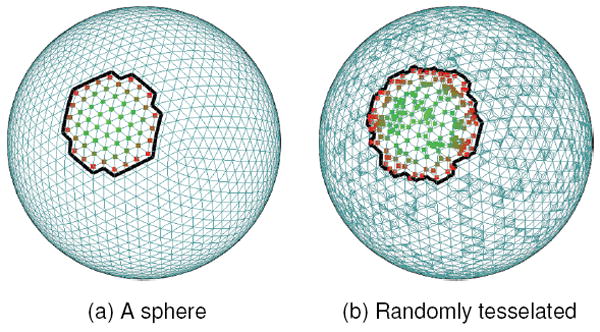
This figure illustrates one of our test cases for tessellation sensitivity. (a) Shows a sphere, with 5,120 faces. (b) Shows the same sphere after 5,000 rounds of subdivision, with replacement. Also shown is a sample neighborhood on each sphere. Note the uneven distribution of sample points arising from subdivision.
Tests were conducted in a similar manner as in §4.2.3: each vertex in the original mesh was compared against the same vertex in the tesselated mesh. In the sphere, average error ranged from .02% ± .01% for small patches to .07% ± .1% in large patches. In the torus, error ranged from .2% ± .1% to 1% ± .3%, respectively.
4.2.5 Foldover
As discussed in §3.2.1, we discard vertices that face away from the plane to avoid having foldover bias our results. In our protein corpus, we have found that, on average, 23% ± 12% of the vertices of the largest patches (8 Ångströms) are discarded. Though this sounds wasteful, on average the closest discarded vertex is 2.75 ± .73 Ångströms from the center of the patch, which means that, on average, only the largest descriptors have any foldover, and those have enough samples to be meaningful. Other meshes in our test set performed similarly, or better.
5 Applications
We have found a number of applications that benefit from our multi-scale, anisotropic descriptors. In the following sections, we describe several of these applications. This list is not intended to be comprehensive, but rather a sampling of the possible uses for our method.
5.1 Multi-scale Lighting
Rusinkiewicz, et al., [34] describe a method for multi-scale lighting which uses curvature information at large scales. They show that by darkening regions of the mesh that are concave at these scales, they produce results similar to those of local ambient occlusion.
Our system does not use curvature directly, but rather uses a descriptor built from large neighborhoods to identify concave regions. These are similarly darkened. Figure 7 demonstrates our results, and compares it against simply lighting using local curvature. Note that, especially with larger descriptors, lighting with a multi-scale descriptor produces similar results to ambient occlusion. Because, however, we only capture local phenomena, we are unable to correctly shade flat areas that are deep within a groove. Nevertheless, we achieve interesting lighting results, which can, for instance, darken features of a particular size for better emphasis.
Fig. 7.
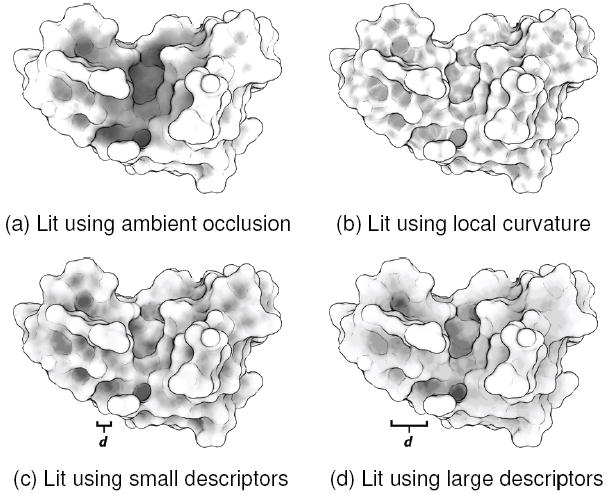
Depicted here are four lighting schemes applied to a ribonuclease molecule (PDB ID 1MO7). (a) Is lit using ambient occlusion, which darkens interior points. This is a global effect, which helps to emphasize the large active cleft. (b) Uses local curvature, darkening concave regions. Note that very small features are emphasized, but the cleft is hard to make out. (c) Uses our descriptor, with an atom-sized (4 Ångström) neighborhood radius. (d) With a residue-sized (8 Ångström) radius.
5.2 Multi-scale/Anisotropic Matching
Next, we look at the task of surface matching: given a point on the surface, find similar points, either on the same model, or on other models. We are interested in this task as a component of matching functional sites between proteins. Figure 8 demonstrates a preliminary result, produced by simply matching the curvature and degree of anisotropy for a source point against those of all other points.
Fig. 8.
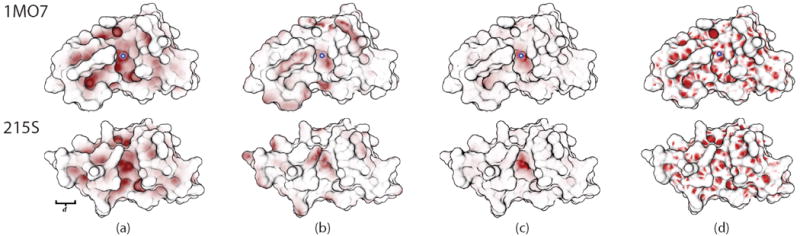
Matching two ribonuclease proteins (PDB IDs 1MO7 and 215S). Both have similar functional sites, occupying the center groove. (a) Shows large-scale curvature matching for the picked site (blue and white circle in the center of each top image). In (b), degree-of-anistropy is used instead. In (c), both metrics are combined, yielding a more accurate match in the bottom image to the picked point in the top image. Finally, (d) shows, for comparison, the results from matching using Rusinkiewicz’s local curvature estimation technique [34].
Each vertex is assigned a feature vector with 8 values, formed from the curvature and anisotropy of four differently-sized patches centered at that vertex. The match between two vertices is then given by the normalized dot product of the feature vector for each vertex.
5.3 Segmentation
We now look at the task of surface segmentation, which operates on the curvature of a surface to produce regions of similar curvature. An early surface segmentation technique, called the watershed method [22], segmented the surface according to curvature. Segments are demarcated by ridges of high curvature, such that all vertices within a segment have lesser or equal curvature than the boundary vertices. Segments can flow into neighbors if their height is lower than a threshold parameter. This parameter, defined by the difference between the curvature at the highest-curvature vertex in the segment and the lowest, has a large effect on the number of segments produced.
The watershed method, while fast and simple, is sensitive to noise in curvature. Tuning for the ‘minimum height’ of a segment can help by merging some small patches caused by noise. Unfortunately, this may also cause larger segments to merge, too, making precise segmentation difficult. In Figure 9, we show that by simply changing the segmentation to use larger patches, which in turn exhibit smoother variations in curvature, we see a significantly improved segmentation, without a need for excessive tuning.
Fig. 9.
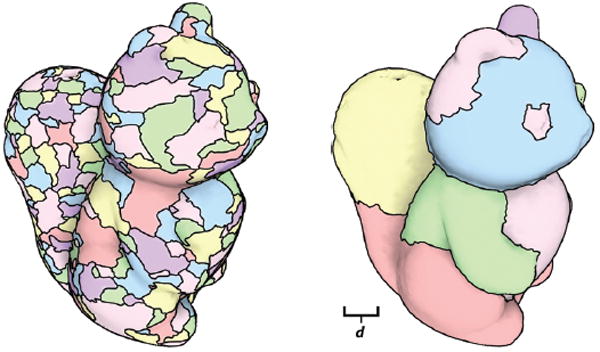
A demonstration of the usage of large-scale descriptors within the watershed algorithm [22]. On the left, the result of segmenting the surface using local curvature values. On the right, shape is estimated using our descriptor, and the same algorithm run. Note the improvement in patch boundaries, and the lack of small, isolated segments. The same overflow threshold of .9 is used in both cases.
5.4 Stylized Rendering
Finally, we demonstrate the effects of large-scale curvature on hatching. Hatching simulates the brush strokes an artist might make to convey the shape of a surface. Girshick, et al. [9] note that these strokes very often are made along the lines of principle curvature on a surface, and that their length and distribution is directly related to the degree to which a surface is curved.
Flat regions, containing little detail, need only a few strokes, or possibly none at all, to convey their shape. Highly curved regions, meanwhile, need far more to properly convey.
We show that depending on application, it may not be desirable to emphasize all small features, especially bumps and wiggles. To do so may require adding numerous additional strokes, cluttering up the image, reducing understandability and increasing rendering times. On the surface of a protein, for instance, atom-scale features abound. Representing all of these using hatching strokes makes for a less comprehensible image (see Figure 10).
Fig. 10.
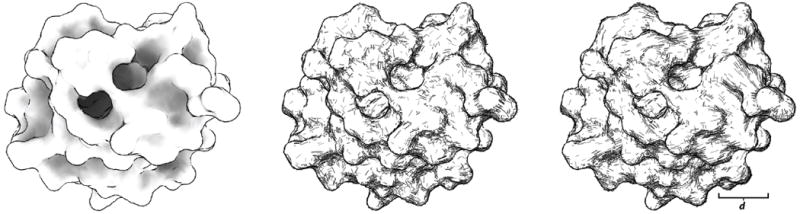
A demonstration of hatching using multi-scale descriptors, with lines drawn along directions of principle curvature. On the left, a Hydrolase molecule (PDB ID 6RNT). In the center, local curvature is used to place lines, using the method described in [28]. On the right, curvature is estimated using a much larger patch (with a radius of 8 Ångströms). Note that the stroke lines are more uniformly oriented, with fewer ‘stray’ lines.
6 Discussion
Our prototype shows that multi-scale surface descriptors can provide intuitive shape characterization of regions in a triangulated mesh. These descriptors resemble curvature, but capture the shape of larger neighborhoods. They are practical to compute on modest hardware, are robust to poor tessellation, and are useful in a range of applications, including surface segmentation, shading and matching.
While our method works well for our desired applications, it does have a few limitations. First, unless the neighborhood being characterized is quadratic, which for our large patches is almost never true, our descriptor is a crude approximation of that neighborhood. This is a fundamental issue with our method, one which makes it unsuitable for applications which require analytical precision. We might partially address this by including the quality-of-fit, or residual error, in the descriptor itself. This, at least, could allow applications to factor in quality when using our descriptors. We do not yet know how much this would help.
Our method, as it produces values comparable to mean curvature, does not discriminate between saddle and planar regions. This is a shortcoming, and though our descriptors meet the requirements of our applications without such discrimination, we would like to address this in future work.
In the future, we would also like to expand each mentioned application to utilize multiple scales in concert. As our initial experiments indicate that the values found for a given point at various scales are highly correlated, we believe that we can incorporate a large number of scales into a compact, robust descriptor.
Finally, we would like to improve the performance of our descriptors. To that end we are investigating techniques for using decimation to further reduce the number of descriptors required to fully describe a mesh. We are also looking into methods for directly aggregating descriptors, to achieve the same goals.
Acknowledgments
We thank the Center for Eukaryotic Structural Genomics for access to new structural results. Cipriano was supported by NIH training grant NLM-5T15LM007359.
Contributor Information
Gregory Cipriano, Email: gregc@cs.wisc.edu.
George N. Phillips, Jr., Email: phillips@biochem.wisc.edu.
Michael Gleicher, Email: gleicher@cs.wisc.edu.
References
- 1.Buss S, Fillmore J. Spherical averages and applications to spherical splines and interpolation. ACM Trans Graph. 2001;20(2):95–126. [Google Scholar]
- 2.Cai Y, Dong F. Surface hatching for medical volume data. Computer Graphics, Imaging and Vision: New Trends, 2005 International Conference on. 2005:232–238. [Google Scholar]
- 3.Chua C, Jarvis R. Point signatures: A new representation for 3D object recognition. International Journal of Computer Vision. 1997 [Google Scholar]
- 4.Coleman RG, Burr MA, Souvaine DL, Cheng AC. An intuitive approach to measuring protein surface curvature. Proteins: Structure, Function, and Bioinformatics. 2005;61(4):1068–1074. doi: 10.1002/prot.20680. [DOI] [PubMed] [Google Scholar]
- 5.DeCarlo D, Finkelstein A, Rusinkiewicz S, Santella A. Suggestive contours for conveying shape. ACM Trans Graph. 2003;22(3):848–855. [Google Scholar]
- 6.Gal R, Cohen-Or D. Salient geometric features for partial shape matching and similarity. ACM Trans Graph. 2006;25(1):130–150. [Google Scholar]
- 7.Gatzke T, Grimm C. Estimating curvature on triangular meshes. International Journal of Shape Modeling. 2006 June;12(1):1–29. [Google Scholar]
- 8.Gatzke T, Grimm C, Garland M, Zelinka S. Curvature maps for local shape comparison. Shape Modeling and Applications, 2005 International Conference. 2005:244–253. [Google Scholar]
- 9.Girshick A, Interrante V, Haker S, Lemoine T. Proceedings of the 1st international symposium on Non-photorealistic animation and rendering. Annecy, France: ACM; 2000. Line direction matters: an argument for the use of principal directions in 3D line drawings; pp. 43–52. [Google Scholar]
- 10.Goldfeather J, Interrante V. A novel cubic-order algorithm for approximating principal direction vectors. ACM Trans Graph. 2004 [Google Scholar]
- 11.Goldman B, Wipke WT. Quadratic shape descriptors. 1. rapid superposition of dissimilar molecules using geometrically invariant surface descriptors. Journal of Chemical Information and Computer Sciences. 2000 May;40(3):644–658. doi: 10.1021/ci980213w. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez R, Woods R. Digital Image Processing. Prentice-Hall; 2002. [Google Scholar]
- 13.Gorla G, Interrante V, Sapiro G. Texture synthesis for 3D shape representation. Visualization and Computer Graphics, IEEE Transactions on. 2003;9(4):512–524. [Google Scholar]
- 14.Gumhold S. Proceedings of the conference on Visualization ’02. Boston, Massachusetts: IEEE Computer Society; 2002. Maximum entropy light source placement; pp. 275–282. [Google Scholar]
- 15.Huber P. Robust Statistics. Wiley-Interscience; Feb, 1981. [Google Scholar]
- 16.Johnson A. Spin-images: a representation for 3-D surface matching. Carnegie Mellon University, The Robotics Institute; 1997. [Google Scholar]
- 17.Kalogerakis E, Nowrouzezahrai D, Simari P, Mccrae J, Hertzmann A, Singh K. Data-driven curvature for real-time line drawing of dynamic scenes. ACM Trans Graph. 2009;28(1):1–13. [Google Scholar]
- 18.Kindlmann G, Whitaker R, Tasdizen T, Moller T. Proceedings of the 14th IEEE Visualization 2003 (VIS’03) IEEE Computer Society; 2003. Curvature-Based transfer functions for direct volume rendering: Methods and applications; p. 67. [Google Scholar]
- 19.Kortgen M, Park G, Novotni M, Klein R. 3D shape matching with 3D shape contexts. 7th Central European Seminar on Computer Graphics; Apr. 2003. [Google Scholar]
- 20.Lee C, Hao X, Varshney A. Geometry-dependent lighting. Visualization and Computer Graphics, IEEE Transactions on. 2006 doi: 10.1109/TVCG.2006.30. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Griffiths J. Least squares ellipsoid specific fitting. Geometric Modeling and Processing, 2004 Proceedings. 2004:335–340. [Google Scholar]
- 22.Mangan A, Whitaker R. Partitioning 3D surface meshes using watershed segmentation. Visualization and Computer Graphics, IEEE Transactions on. 1999;5(4):308–321. [Google Scholar]
- 23.Mortara M, Patan G, Spagnuolo M, Falcidieno B, Rossignac J. Blowing bubbles for Multi-Scale analysis and decomposition of triangle meshes. Algorithmica. 2003;38(1):227–248. [Google Scholar]
- 24.Page D, Sun Y, Koschan A, Paik J, Abidi M. Normal vector voting: crease detection and curvature estimation on large, noisy meshes. Graph Models. 2002;64(3/4):199–229. [Google Scholar]
- 25.Petitjean S. A survey of methods for recovering quadrics in triangle meshes. ACM Computing Surveys. 2002;2:161. [Google Scholar]
- 26.Pratt V. Direct least-squares fitting of algebraic surfaces. SIGGRAPH Comput Graph. 1987;21(4):145–152. [Google Scholar]
- 27.Praun E, Webb M, Finkelstein A. Real-time hatching. PROCEEDINGS OF SIGGRAPH 2001. 2001:579–584. [Google Scholar]
- 28.Rusinkiewicz S. Proceedings of the 3D Data Processing, Visualization, and Transmission, 2nd International Symposium. IEEE Computer Society; 2004. Estimating curvatures and their derivatives on triangle meshes; pp. 486–493. [Google Scholar]
- 29.Sanner M, Olson A, Spehner J. Proceedings of the eleventh annual symposium on Computational geometry. Vancouver, British Columbia, Canada: ACM; 1995. Fast and robust computation of molecular surfaces; pp. 406–407. [Google Scholar]
- 30.Schmidt R, Grimm C, Wyvill B. Interactive decal compositing with discrete exponential maps. ACM Transactions on Graphics. 2006;25(3):603–613. [Google Scholar]
- 31.Sheffer A, Hart JC. Proceedings of the conference on Visualization ’02. Boston, Massachusetts: IEEE Computer Society; 2002. Seamster: inconspicuous low-distortion texture seam layout; pp. 291–298. [Google Scholar]
- 32.Stokely E, Wu S. Surface parametrization and curvature measurement of arbitrary 3-D objects: five practical methods. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1992;14(8):833–840. [Google Scholar]
- 33.Surazhsky V, Surazhsky T, Kirsanov D, Gortler S, Hoppe H. Fast exact and approximate geodesics on meshes. ACM Trans Graph. 2005;24(3):553–560. [Google Scholar]
- 34.Toler-Franklin C, Finkelstein A, Rusinkiewicz S. Proceedings of the 5th international symposium on Non-photorealistic animation and rendering. San Diego, California: ACM; 2007. Illustration of complex real-world objects using images with normals; pp. 111–119. [Google Scholar]
- 35.Vergne R, Barla P, Granier X, Schlick C. Proceedings of the 6th international symposium on Non-photorealistic animation and rendering. Annecy, France: ACM; 2008. Apparent relief: a shape descriptor for stylized shading; pp. 23–29. [Google Scholar]


