Abstract
The progression of prostate cancer from an organ-confined, androgen-sensitive disease to a metastatic one is associated with dysregulation of androgen receptor (AR)-regulated target genes and with a decrease in insulin-like growth factor-I receptor (IGF1R) expression. DNA methylation of CpG islands is an epigenetic mechanism associated with gene silencing. Recent studies have demonstrated that methylation occurs early in prostate carcinogenesis and, furthermore, may contribute to androgen independence. The methylation status of the AR and IGF1R genes was evaluated in a series of prostate cancer cell lines corresponding to early (benign) and advanced (metastatic) stages of the disease. Results of 5-Aza-2'-deoxycytidine (5-Aza) experiments, methylation specific PCR, and sodium bisulfite-direct DNA sequencing revealed that the AR promoter is hypermethylated in metastatic M12, but not in benign P69, cells. On the other hand, no methylation was seen in the IGF1R promoter at any stage of the disease. We show, however, that 5-Aza treatment, which caused demethylation of the AR promoter, led to a significant increase in IGF1R mRNA levels, whereas addition of the AR inhibitor flutamide decreased the IGF1R mRNA levels to basal values measured prior to the 5-Aza treatment. Given that the IGF1R gene has been identified as a downstream target for AR action, our data is consistent with a model in which the AR gene undergoes methylation during progression of the disease, leading to dysregulation of AR targets, including the IGF1R gene, at advanced metastatic stages.
Keywords: insulin-like growth factor-I receptor (IGF1R), androgen receptor, DNA methylation, prostate cancer, epigenetic regulation
Introduction
Prostate cancer is a major health issue in the Western world. Approximately 80–90% of primary prostate tumors are strictly dependent on androgen action for tumor growth and development. The androgen receptor (AR), a member of the nuclear steroid receptor superfamily, is a key component of the androgen transduction cascade in responsive tissues [1]. Alterations in AR structure and expression are, in part, responsible for the progression of the tumors from an organ-confined, androgen-sensitive disease to a more aggressive, hormone-refractory, androgen-independent disease [2, 3]. In addition, the progression of prostate cancer to advanced, metastatic stages is associated with dysregulation of AR-regulated target genes.
The insulin-like growth factors (IGFs) are a family of growth factors, binding proteins, and receptors that play a key role in regulating growth, resistance to apoptosis, and differentiation. The biological actions of the IGFs are mediated by their activation of the IGF1R, a transmembrane heterotetramer linked to the ras-raf-MAPK and PI3K-PKB/Akt signal transduction cascades [4–6]. The involvement of the IGF axis in tumorigenesis in general, and in prostate cancer in particular, has been the subject of extensive research [7–9]. Furthermore, the contribution of IGF1 action to prostate cancer development is supported by epidemiological studies showing a positive correlation between serum IGF1 values and prostate cancer risk [10, 11]. While most studies suggest an important role for IGF1 action in prostate cancer initiation, clinical and experimental evidence indicates that the progression of prostate cancer from an androgen-sensitive disease to a metastatic one is associated with a significant decrease in local IGF1R mRNA and protein levels [12, 13]. Other studies, however, showed sustained up regulation of the IGF1R in metastases [14] and correlation between up-regulation of IGF axis components and tumor grade [15].
Our comprehension of the joint regulation of the androgen and IGF1 signaling pathways in prostate cancer is, however, limited. Recent studies have identified the IGF1R gene as a downstream target for AR action in the prostate (Schayek et al, manuscript submitted). Specifically, activated wild type (wt), but not mutant, AR was shown to stimulate IGF1R expression. Furthermore, results of chromatin immunoprecipitation assays revealed that the mechanism of action of AR involves binding to the proximal IGF1R promoter. In addition, Pandini et al [16] have shown that androgens selectively upregulate the IGF1R in AR positive cells through the activation of a non-genomic AR signaling pathway. On the other hand, a number of studies have established that IGF1 may affect AR signaling. Specifically, activation of the MAPK pathway by IGF1 was shown to sensitize the AR transcriptional complex to subphysiologic levels of androgens in LnCaP cells [17]. Analyses of the complex interactions between the IGF1R and AR pathways identified a number of transcription factors and signaling molecules involved in the control of this bi-directional hormonal interplay [18]. The involvement of epigenetic mechanisms in the regulation of the AR-IGF1R interactions in the prostate has not yet been investigated.
DNA methylation is a major epigenetic alteration affecting gene expression. Methylation involves the addition of methyl groups, catalyzed by DNA methyltransferase, to the 5-carbon of deoxycytosines in the palindromic dinucleotide CpG. Methylation of CpG islands leads to inactivation of gene transcription [19, 20] and plays a critical role during development. CpG islands are mostly unmethylated in normal tissues and hypermethylated in various cancers [19, 21, 22]. Promoter CpG island hypermethylation of tumor suppressor genes is a common hallmark of all human cancers and affects most cellular pathways. AR promoter hypermethylation and gene inactivation have been detected in about 8–28% of prostate tumors [23, 24]. AR hypermethylation has been usually associated with advanced stages of the disease. However, little information exists regarding the impact of AR methylation on downstream targets expression.
Given the important roles of androgens, AR, and the IGF1 system in prostate cancer initiation and progression [25], we examined in the present study the hypothesis that methylation of the AR promoter constitutes a key event in prostate cancer progression, with important pathological consequences as a result of dysregulation of AR target genes. In addition, our study was aimed at elucidating the mechanism/s, including potential epigenetic changes, responsible for IGF1R silencing at advanced prostate cancer stages. Results obtained indicate that progression of prostate cancer from a benign, non-tumorigenic stage to an aggressive, metastatic one in a cellular model of prostate cancer is associated with specific AR promoter methylation. On the other hand, IGF1R gene silencing in tumorigenic and metastatic prostate cancer cells is not correlated with DNA hypermethylation of CpG dinucleotides in the proximal IGF1R promoter. Taken together, our data is consistent with a model in which IGF1R silencing, with ensuing impairment of IGF1 signaling, constitutes an important pathological outcome of AR promoter methylation.
Materials and methods
Cell cultures
Generation of the P69-derived series of prostatic carcinoma cell lines has been previously described [26, 27]. Briefly, the P69 cell line was obtained by immortalization of prostate epithelial cells isolated from the prostate gland of a 63-yr old man with SV40 T antigen. P69 cells are responsive to IGF1 and are rarely tumorigenic. Cell lines M2205, M2182, and M12 were derived by injection of P69 cells into athymic nude mice and serial reimplantation of tumor nodules into nude mice. Cell lines M2205 and M2182 are tumorigenic but rarely to non-metastatic. M12 cells are highly metastatic and exhibit a reduced IGF1 responsiveness. Cells were cultured in serum-free conditions in RPMI-1640 medium. Cell lines were provided by Dr. Joy L. Ware (Medical College of Virginia). Human prostate cancer cell lines PC3, DU145, and C4-2 were obtained from the American Type Culture Collection.
5-Aza-2'-deoxycytidine analyses
To evaluate the methylation status of the IGF1R and AR genes, cells were cultured at low density for 24 hr, after which treatment with the demethylating agent 5-Aza-2'-deoxycytidine (5-Aza; 1 µg/ml; Sigma-Aldrich) was initiated. Cells were treated with 5-Aza for 3 days, with daily medium changes. Cells were then harvested and total protein was prepared for Western blots. All experiments were conducted in triplicate dishes and repeated at least three times.
Western immunoblots
5-Aza-treated cells were harvested with phosphate buffered saline containing 5 mM EDTA, and lysed in the presence of protease inhibitors. Samples (80 µg protein) were subjected to 10% SDS-PAGE, followed by transfer of the proteins to nitrocellulose membranes. Membranes were blocked with 5% milk and then incubated with anti-IGF1R β-subunit (C20; Santa Cruz Biotechnology), anti-human AR (sc-7305), and anti-tubulin. Membranes were washed and incubated with a horseradish peroxidase-conjugated secondary antibody. Proteins were detected using the SuperSignal West Pico® Chemiluminescent Substrate (Pierce).
DNA extraction and modification by sodium bisulfite
DNA was extracted from prostate cancer cell lines using a QIAamp DNA Mini kit. A total of 2 µg of genomic DNA was treated with sodium bisulfite as described [28]. Briefly, following DNA denaturation in 0.3 M NaOH at 50° C for 20 min, sodium bisulfite and hydroquinone were added at final concentrations of 2.5 M and 125 mM, respectively. The bisulfite modification reaction was performed at 55° C for 16 hr, after which the DNA was purified with a Wizard DNA purification system (Promega), ethanol-precipitated, dried, and resuspended in 100 µl of distilled water. Deamination of Cs and conversion into Ts following sodium bisulfite treatment indicates that the C nucleotide was unmethylated. Methylated Cs remain unaltered following sodium bisulfite treatment.
Sodium bisulfite-PCR methylation analysis
For the analysis of the methylation status of the 5'-regulatory regions of the IGF1R and AR genes, sodium bisulfite-PCR was performed using a primer set (U) that anneals to unmethylated DNA and another primer set (M) that anneals to methylated DNA. Unmodified DNA was amplified with a wt primer set, which serves as a positive control for PCR. PCR reactions contained 20 ng of modified DNA, 1.5 mM MgCl2, 200 µM dNTPs, 0.3 µM primers F and R, and 0.5 U of AmpliTaq Gold DNA polymerase. Amplifications were performed using the following conditions: 94° C for 10 min, followed by 35 cycles (94° C for 30 sec, 43° C−65° C for 1 min, 72° C for 30 sec) and then 72° C for 5 min. The details of primers, PCR conditions, and PCR product size for the AR and IGF1R promoters are listed in Table 1 and Table 2, respectively. Sodium bisulfite-PCR products were directly sequenced using the BigDye terminator method (Perkin-Elmer). In addition, the methylation status of the following genes was assessed: estrogen receptor (ER)-α, progesterone receptor (PR)-A and -B, breast cancer gene-1 (BRCA1), and Kruppel-like factor-6 (KLF6).
Table 1.
Primer sets and PCR conditions for AR promoter methylation analysis
| Primer | Sequence | Annealing(°C) | Product size |
|---|---|---|---|
| AR-WT F | CGCCCCCTCCGAGATCCCG | 65 | 213 bp |
| AR-WT R | CGGGCGGCGGCTTCGAAGCCG | ||
| AR-UF | TGTTTTTTTTGAGATTTTG | 43 | 213 bp |
| AR-UR | CAAACAACAACTTCAAAACCA | ||
| AR-MF | CGTTTTTTTCGAGATTTCG | 50 | 213 bp |
| AR-MR | CGAACGACGACTTCGAAACCG |
Table 2.
Primer sets and PCR conditions for IGF1R promoter methylation analysis
| A. Set 1 | |||
|---|---|---|---|
| Primer | Sequence | Annealing(°C) | Product size |
| IGF1R -1WT-F | TATTTTGCAACAGCTGCAAGAAACAATGAA | 65 | 178 bp |
| IGF1R -1WT-R | GGCAGGGGTGGGTAGCCAGGGAAAG | ||
| IGF1R -1MF | TATTTTGTAATAGTTGTAAGAAATAATGAA | 50 | 178 bp |
| IGF1R -1MR | AACAAAAATAAATAACCAAAAAAAA | ||
| B. Set 2 | |||
|---|---|---|---|
| Primer | Sequence | Annealing(°C) | Product size |
| IGF1R-2WT-F | TCTTGGGGAACCGGGCTCCGGTTTTTTG | 65 | 201 bp |
| IGF1R-2WT-R | GGTAAACAAGAGCCCCAGCCT | ||
| IGF1R-2U-F | TTTTGGGGAATTGGGTTTTGGTTTTTTG | 65 | 201 bp |
| IGF1R-2U-R | GGTAAATAAGAGTTTTAGTTT | ||
| IGF1R-2M-F | TTTTGGGGAATCGGGTTTCGGTTTTTTG | 65 | 201 bp |
| IGF1R-2M-R | GGTAAATAAGAGTTTTAGTTT | ||
Treatment with flutamide and dihydrotestosterone (DHT)
To evaluate the relationship between AR methylation and IGF1R mRNA levels, M12 cells were treated with 1 µg/ml of 5-Aza for 72 h. Cells were then treated with DHT (10−9 M) and/or with the AR inhibitor flutamide (10−5 M) for 24 h. At the end of the incubation period the cells were harvested and total RNA was prepared for Quantitative Real-time PCR. All experiments were conducted in triplicate dishes and repeated at least three times.
Quantitative Real-time PCR
Quantitative Real-time PCR was done using Power SYBR green PCR master (Applied Biosystems). An ABI Prism 7000 Sequence Detection System was used. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were measured to normalize the IGF1R mRNA values. The primers sequences used for IGF1R mRNA were: sense, GAAGTGGAACCCTCCCTCTC; antisense, CTTCTCGGCTTCAGTTTTGG. The primers sequences used for GAPDH mRNA were: sense, GAAGGTGAAGGTCGGAGTC; antisense, GAAGATGGTGATGGGATTTC. Amplification was carried out after an incubation of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles at 95°C for 15 s, 1 min at 55°C, and 30 s at 72°C. The number of PCR cycles to reach the fluorescence threshold was the cycle threshold (Ct). Each cDNA sample was tested in triplicate and mean Ct values are reported. Furthermore, for each reaction, a "no template" sample was included as a negative control. Fold-differences were calculated using the method.
Results
Analysis of AR gene methylation in prostate cancer cells
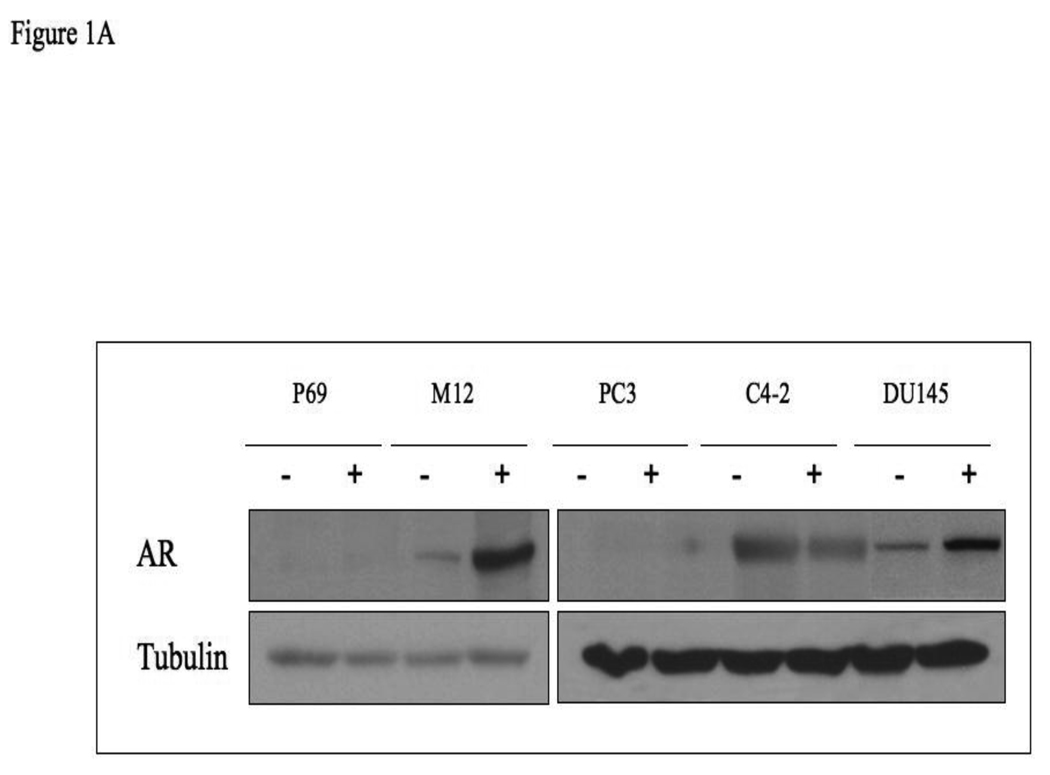
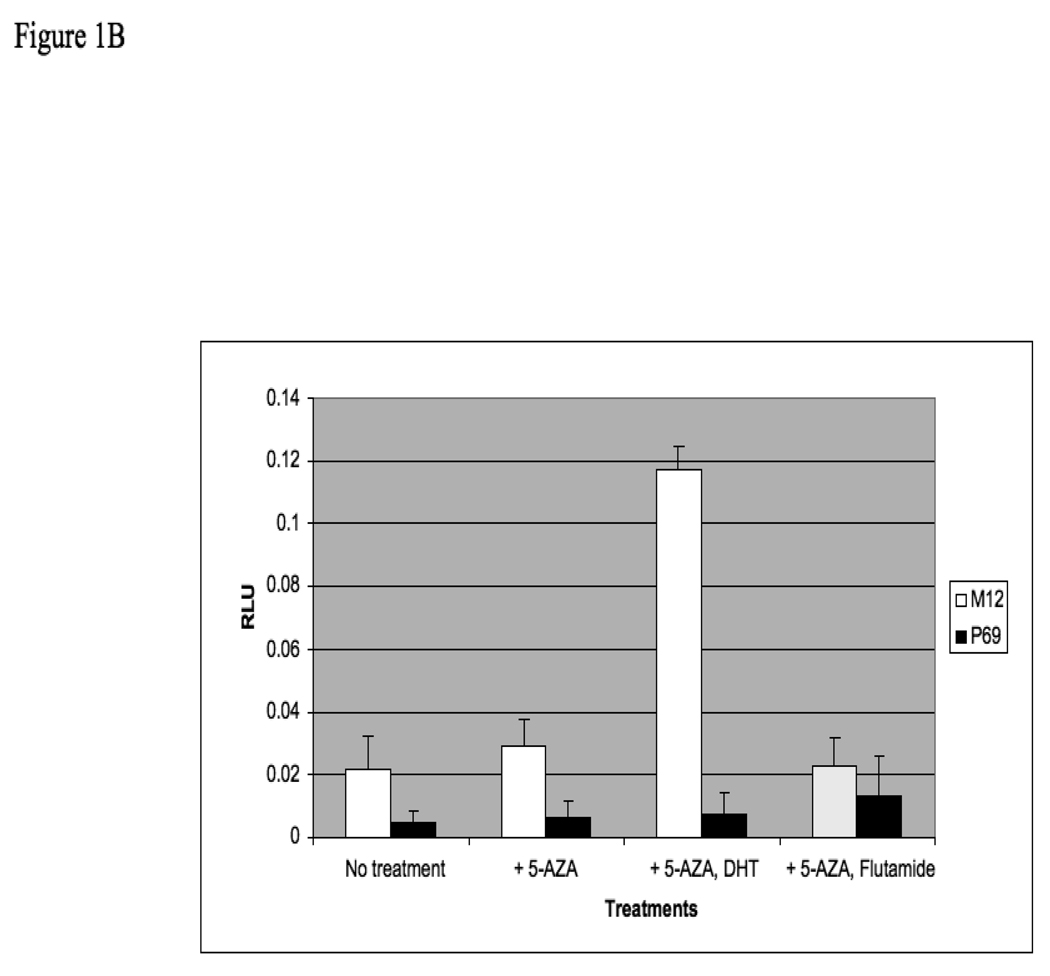
The AR gene has been shown to undergo methylation in ∼8–28% of prostate cancer cases. To evaluate in a systematic manner the potential epigenetic regulation of the AR promoter during the progression of prostate cancer from benign to metastatic stages, the P69-derived series of syngeneic prostate cancer cell lines was employed. This series includes the P69 (immortalized, non-tumorigenic), M2205 and M2182 (tumorigenic but rarely to non-metastatic), and M12 (metastatic) cell lines. In addition, AR promoter methylation was examined in the PC3, C4-2, and DU145 prostate cancer cell lines. In initial experiments, P69, M12, PC3, C4-2 and DU145 cells were treated with the demethylating agent 5-Aza for 72 hr, after which the cells were lysed and AR levels were measured by Western blots. As shown in Fig. 1A, 5-Aza treatment resulted in a marked increase in AR levels in the M12 and DU145 cell lines. To establish whether this increase was associated with an elevation in AR activity, P69 and M12 cells were treated with 5-Aza (or vehicle) for 72 h. Cells were then transfected with an AR luciferase reporter vector including a tandem probasin promoter sequence (AAR3). After 24 h, cells were treated with 10−9 M DHT (or vehicle) in the absence or presence of flutamide, an AR inhibitor. Luciferase assays revealed that 5-Aza plus DHT led to a significant increase in AAR3 –luc activity (p<0.001) compared to 5-Aza-treated cells without DHT (Fig. 1B). This activity was blocked by flutamide. No responses were seen in P69 cells and there was also no response in M12 cells when DHT was added if not treated with AZA ( data not shown).
Figure 1. AR expression analysis in prostate cancer cells after 5-Aza treatment.
(A) P69, M12, PC3, C4-2 and DU145 cells were treated with 5-Aza (1 µg/ml) for 72 hr (+), or left untreated (−), after which cells were lysed and AR levels were measured by Western blots. After stripping, the blot was reprobed with a tubulin antibody for control purposes. (B) P69 and M12 cells were treated with 5-Aza or vehicle for 72 h. Cells were then transiently transfected with an AR luciferase reporter construct (AAR3) including a tandem probasin promoter sequence using lipofectin. After 24 h, DHT (10−9 M) was added in the absence or presence of flutamide. After 5 h, cells were harvested and luciferase assays were performed. RLU, relative luciferase units. Results are controlled for transfection efficiency with β-galactosidase cotransfection. The experiment was performed in triplicates and the error bars represent ± 2 SD.
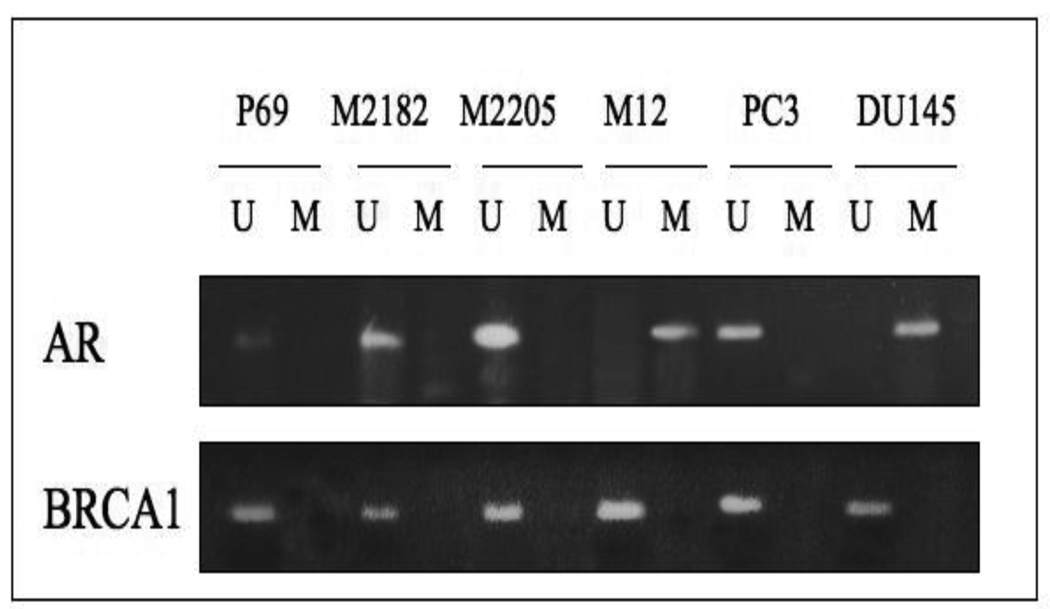
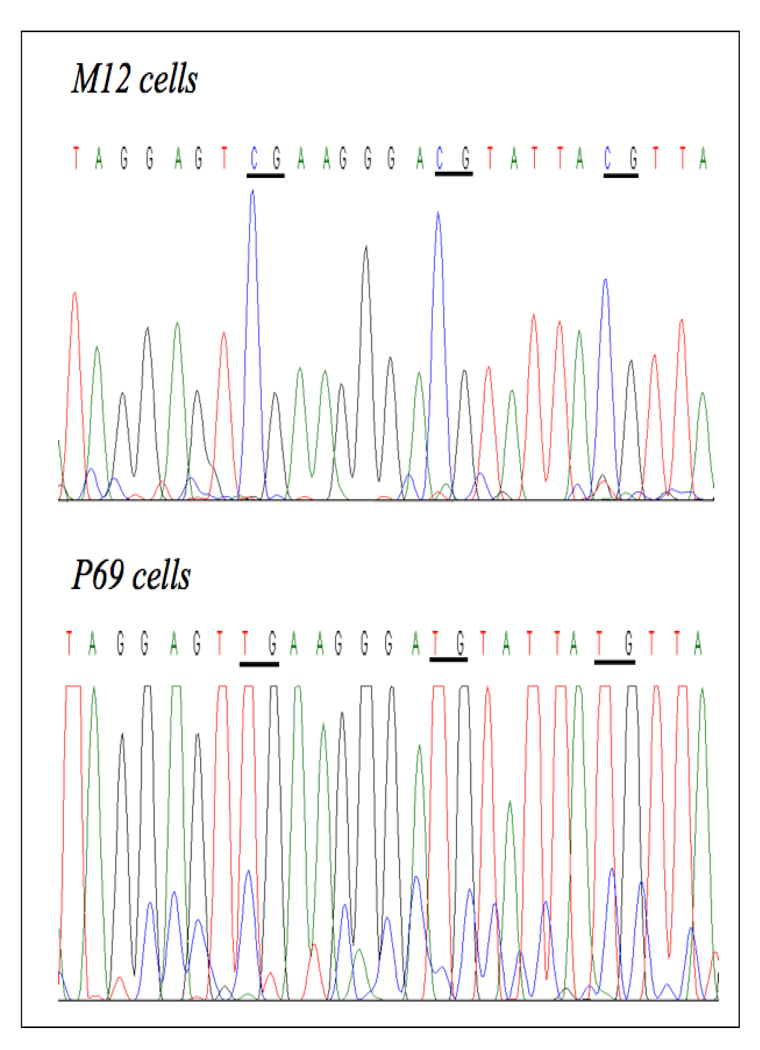
Next, we determined the methylation status of the AR promoter using methylation-specific PCR (MSP) and direct DNA sequencing. A 213-bp fragment located approximately 400 bp upstream of the transcription start site of the AR gene (NCBI Accession number M23263) was selected [23]. This promoter fragment includes 21 CpG loci. The details of primers and PCR conditions are listed in Table 1. As shown in Fig. 2, unmethylated specific primers were able to amplify PCR products in DNA obtained from the P69, M2182, M2205, and PC3 cell lines, but not in DNA from the M12 and DU145 cell lines. Conversely, methylated specific primers were able to generate PCR products in DNA from M12 and DU145, but not from the other, cells. Direct DNA sequencing of sodium bisulfite-treated DNA confirmed that the AR gene is methylated in the metastatic M12 and DU145 cell lines but unmethylated in the other cell lines (Fig. 3).
Figure 2. Assessment of AR promoter methylation in prostate cancer cell lines using methylation-specific PCR.
PCR was performed using unmethylated (U) and methylated (M) AR specific primers. PCR products in lanes U, but not in lanes M, indicate the presence of an unmethylated allele in DNA from the P69, M2128, M2205, PC3 and C4-2 cell lines whereas PCR products in lanes M, but not in lanes U, indicate the presence of a methylated allele in DNA from the M12 and DU145 cell lines. For control purposes we evaluated the methylation status of the BRCA1 gene. BRCA1 was unmethylated in all cell lines examined.
Figure 3. Assessment of AR promoter methylation using the sodium bisulfite method.
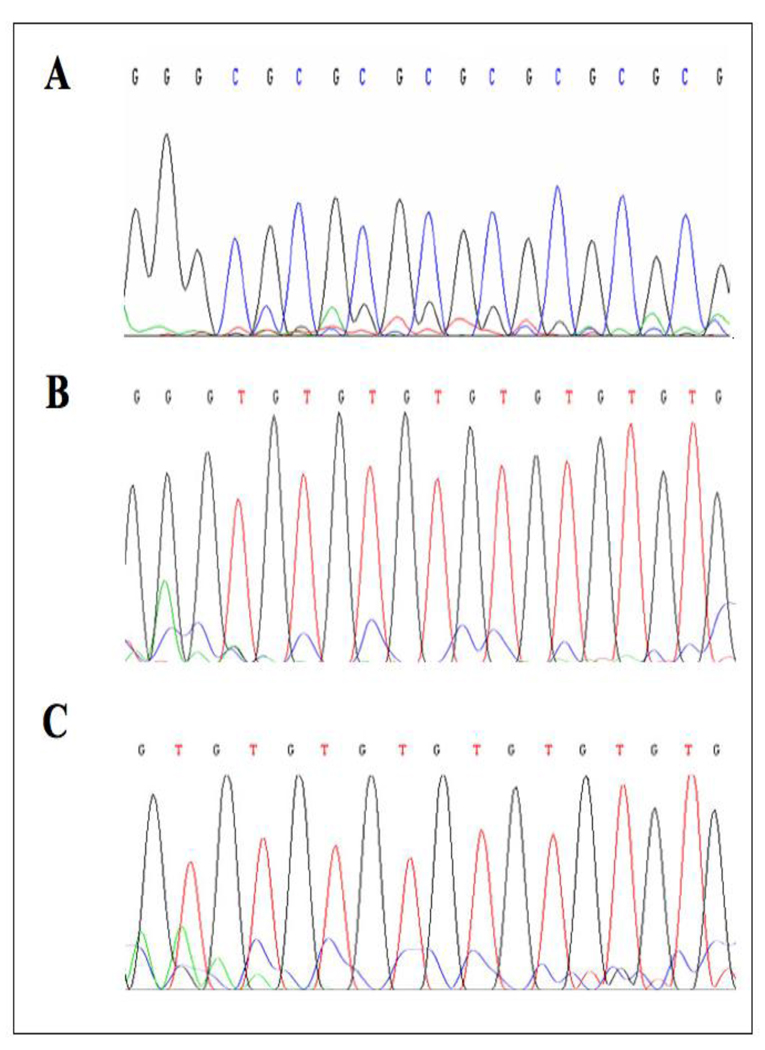
Genomic DNA was obtained from M12 and P69 cells, followed by sodium bisulfite treatment and direct DNA sequencing. Examples of DNA sequencing chromatograms are shown here. CpG sites are underlined. All cytosines (C) are deaminated and converted to thymines (T) in the P69 cell line, but 5-methylcytosines (shown as C) remained unaltered in the M12 cell line, meaning that the AR promoter is methylated in M12, but unmethylated in P69, cells.
Search for IGF1R methylation in prostate cancer progression
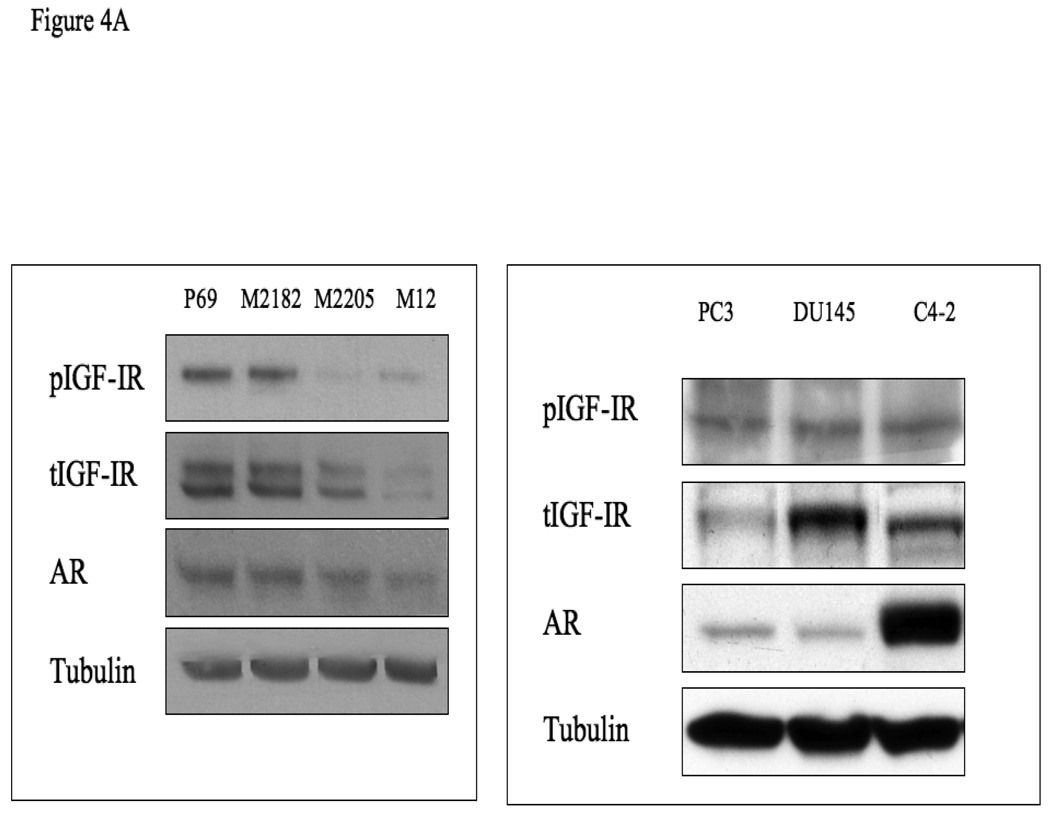
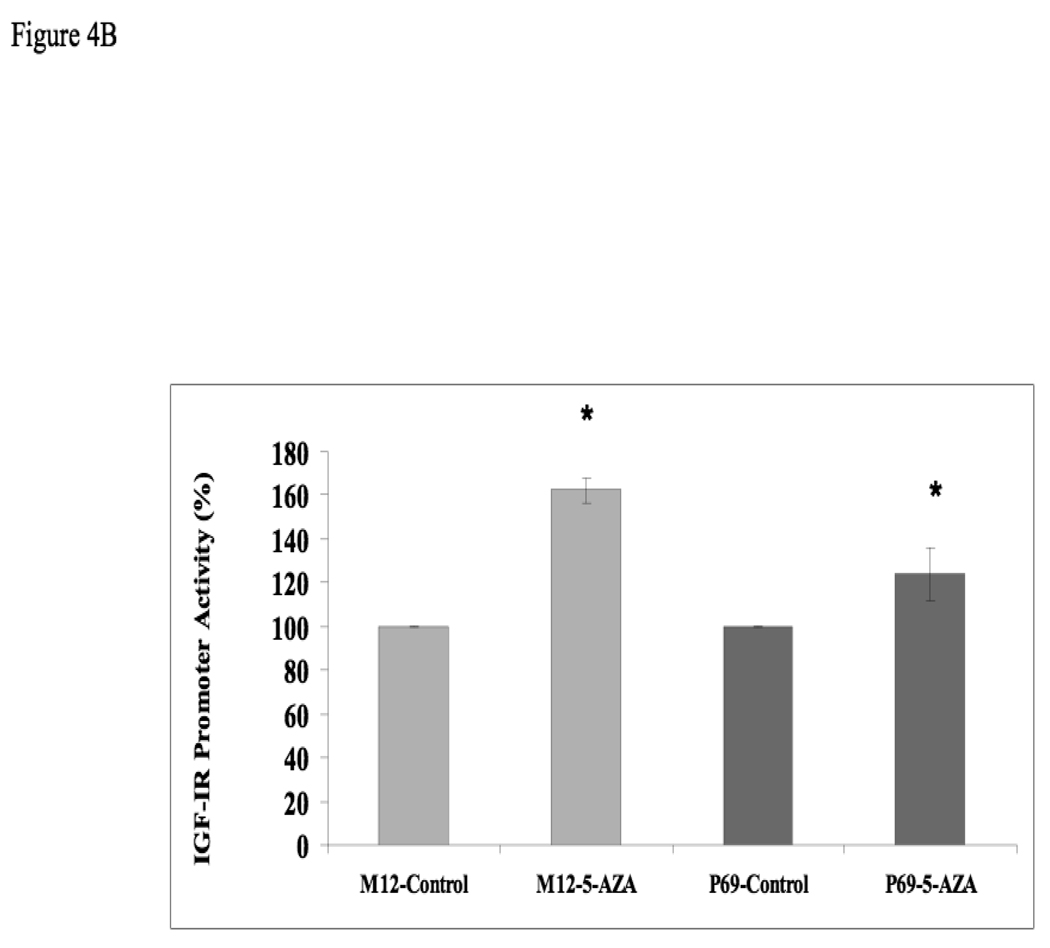
To evaluate the regulation of IGF1R expression during prostate cancer progression, total IGF1R levels were measured in the P69-derived cell lines. Results of Western blots showed that progression towards metastatic stages was correlated with a drastic reduction in total IGF1R levels (Fig. 4). This decline was correlated with a concomitant reduction in basal phospho-IGF1R values, reflecting a decrease in IGF1R activation. Specifically, P69 cells express high levels of total- and phospho-IGF1R, M2205 and M2182 cells express intermediate levels, and M12 cells express very low levels of both total and phosphorylated (activated) receptor. AR levels were very low in all four cell lines, though a consistent decrease in AR levels was seen in M12 cells. Similar results were seen using immunoprecipitation assays (data not shown). For comparative purposes, IGF1R expression and activation in additional prostate cancer cell lines (PC3, DU145, and C4-2) is shown in the right panel of Fig. 4A. Some of these results replicate previously reported findings [29]. To evaluate the potential effect of 5-Aza on IGF1R promoter activity, P69 and M12 cells were treated with 5-Aza (or left untreated) and, after 24 h, cells were transiently transfected with a proximal IGF1R promoter-luciferase reporter construct [p(−476/+640)LUC], along with a β-galactosidase plasmid. Cells were harvested 48 h after transfection for luciferase and β-galactosidase assays. Results of promoter assays revealed a ∼162% increase in IGF1R promoter activity in M12 cells, in comparison to a ∼124% increase in P69 cells (Fig. 4B).
Figure 4. Expression of AR and IGF1R in prostate cancer cell lines.
(A) Cells were lysed in the presence of protease inhibitors, as indicated under Materials and methods. Equal amounts of protein (80 µg) were separated by 10% SDS-PAGE, transferred onto nitrocellulose filters, and blotted with anti-IGF1R, anti-phospho (p)-IGF1R, anti-AR, and anti-tubulin. (B) Effect of 5-Aza treatment on IGF1R promoter activity. P69 and M12 cells were treated with 5-Aza (or left untreated) and, after 24 h, cells were transiently transfected with a proximal IGF1R promoter-luciferase reporter construct [p(−476/+640)LUC], along with a β-galactosidase plasmid. Cells were harvested 48 h after transfection for luciferase and β-galactosidase assays. * p<0.02 versus respective control.
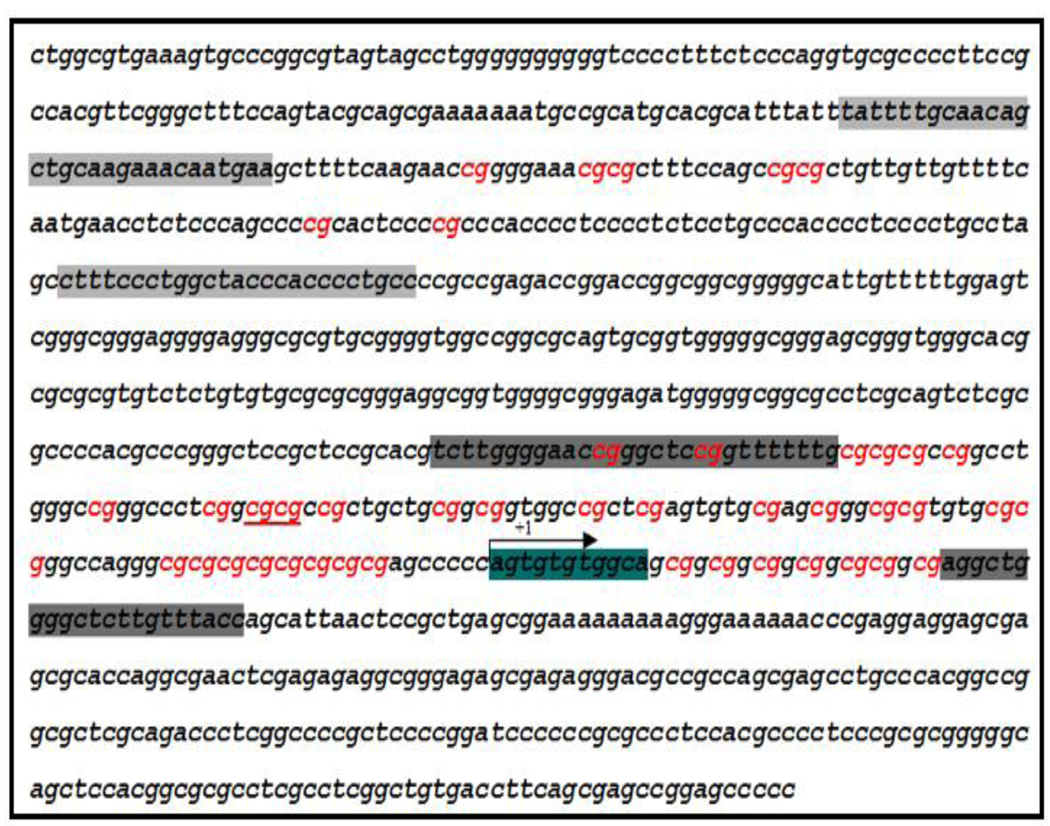
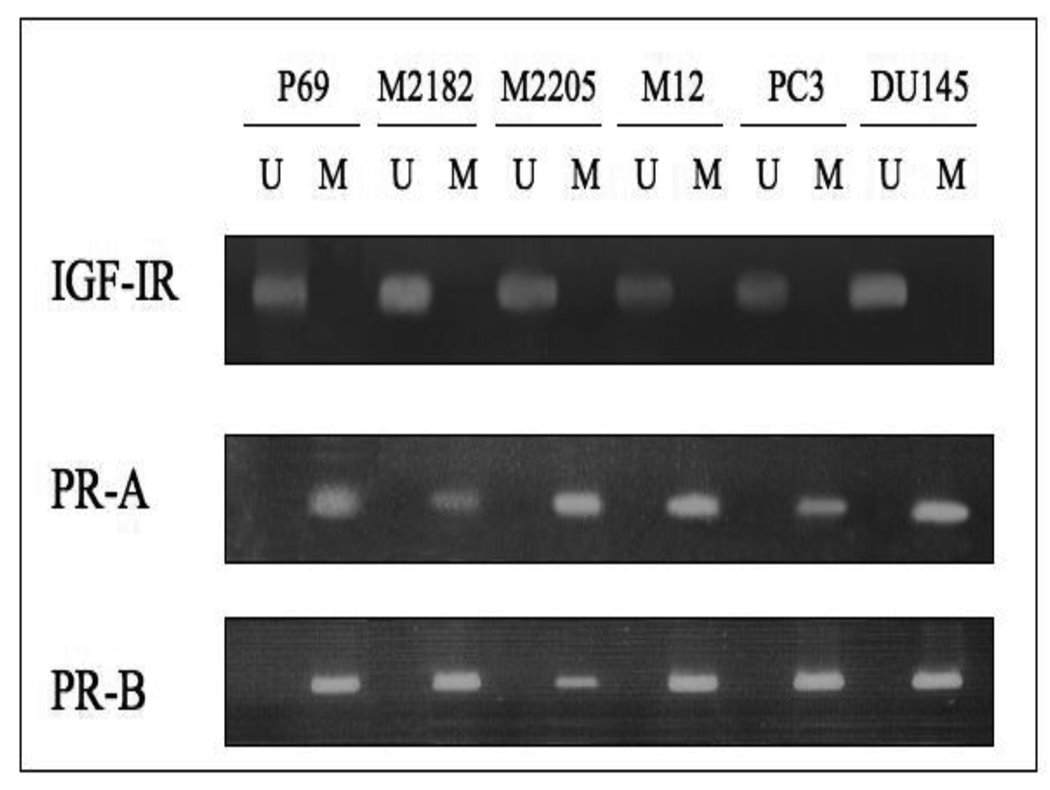
The IGF1R promoter region is extremely GC-rich (approximately 70–75% GC content in the human, rat, and mouse genes). Bioinformatic analysis of the human promoter region revealed the presence of multiple CpG dinucleotides (Fig. 5). To establish whether the decrease in IGF1R levels in prostate cancer metastatic cells was associated with DNA methylation-induced IGF1R gene silencing, we evaluated the methylation status of the IGF1R gene in all four P69-derived cell lines and in the PC3, C4-2 and DU145 prostate cancer cells using MSP and direct DNA sequencing. For this purpose, the IGF1R promoter sequence (NCBI Accession number NM_000875) was searched, and two CpG island-containing fragments were selected for further analysis. Fragment 1 is a 173-bp fragment located approximately 400 bp upstream of the transcription start site, which includes 7 CpG loci. Fragment 2 is a 201-bp fragment which begins at position −150 in the 5’ flanking region and overlaps the transcription start site. This fragment includes 36 CpG loci (Fig. 5). The details of primers, PCR conditions, and PCR product sizes for Fragments 1 and 2 are listed in Tables 2A and 2B, respectively. MSP and direct DNA sequencing was performed with DNA obtained from all seven cell lines (P69, M2128, M2205, M12, PC3, C4-2, and DU145). Results of MSP showed DNA amplification when using unmethylated specific primers whereas no amplification was seen when using methylated specific primers (Fig. 6). Furthermore, PCR sequencing analysis of both fragments showed that all cytosines (C) were converted to thymines (T) in all 43 CpG loci. An example of direct DNA sequencing of Fragment 2 is shown in Fig. 7. All eight Cs in this particular fragment were deaminated and converted to Ts in both cell lines. Hence, this data indicates that the IGF1R promoter is unmethylated in all of the prostate cancer cell lines examined.
Figure 5. Schematic diagram of the IGF1R proximal promoter showing the location of primer set 1 and primer set 2.
Set 1 (light gray) is a 173-bp fragment located ∼400 bp upstream of the transcription start site (marked as +1). Set 2 (dark gray) is a 201-bp fragment which begins at position −150 in the 5’ flanking region and overlaps the transcription start site (arrow). Region 1 contains 7 CpG loci (marked in red) and region 2 contains 36 CpG loci.
Figure 6. Assessment of IGF1R promoter methylation in prostate cancer cell lines using methylation-specific PCR.
PCR was performed using unmethylated (U) and methylated (M) IGF1R specific primers (set 2). PCR products in lanes U, but not in lanes M, indicate the presence of an unmethylated allele in all prostate cancer cell lines tested. As positive controls we are showing the PCR products of the progesterone receptors A and B (PR-A, PR-B) promoters, showing methylation in all of the cell lines.
Figure 7. Sodium bisulfite-DNA sequencing analysis of the IGF1R promoter.
Genomic DNA was extracted from P69 and M12 cells and modified with sodium bisulfite as described in Materials and methods. PCR was performed using a set of primers encompassing Region 2 of the proximal IGF1R promoter. Sodium bisulfite-PCR products were sequenced using the BigDye terminator method. (A) Unmodified DNA obtained from P69 cells. (B) Sodium bisulfite-modified DNA from P69 cells. (C) Sodium bisulfite-modified DNA from M12 cells.
Is AR gene methylation associated with loss of function of AR in M12 cells?
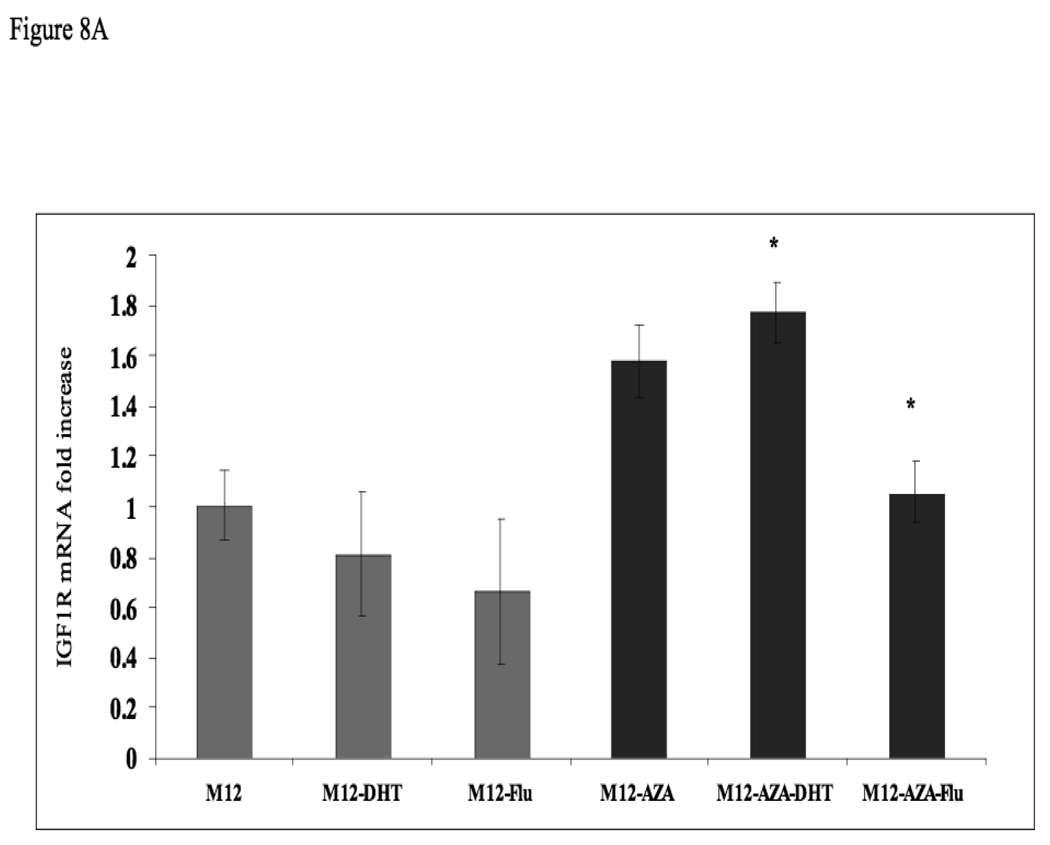
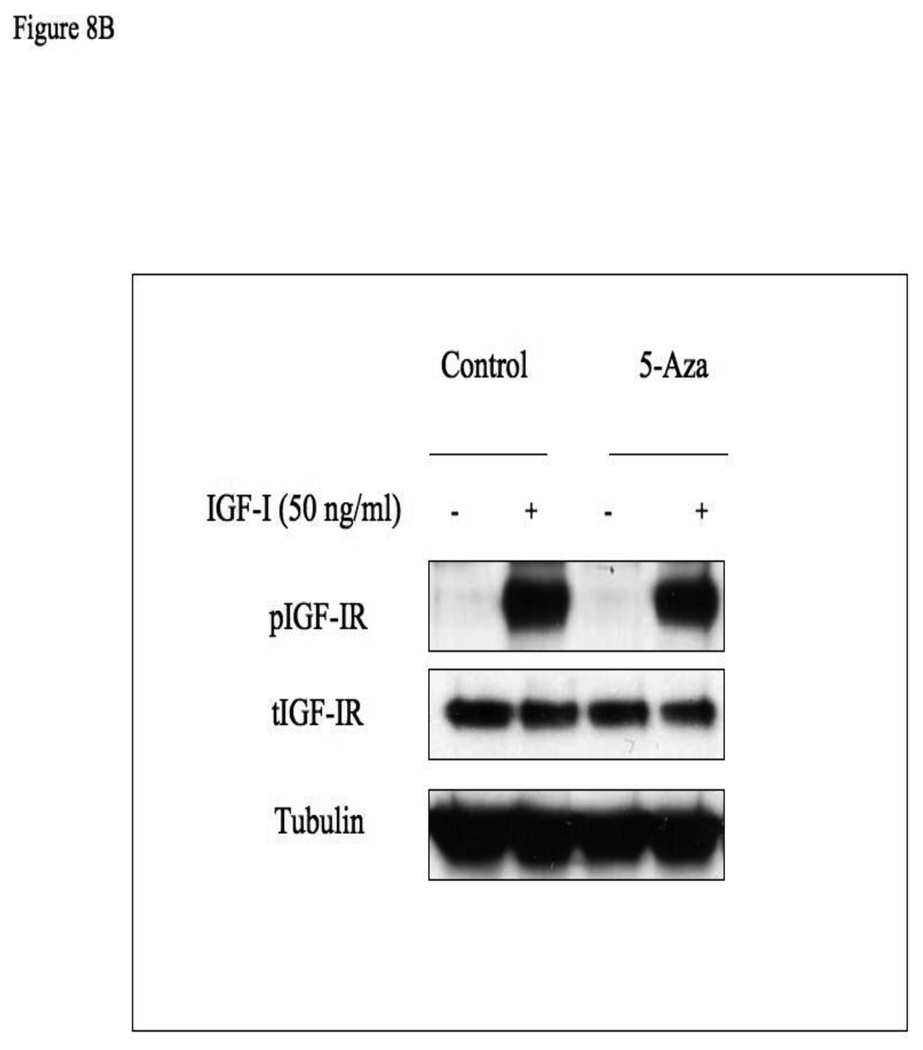
Given that IGF1R levels and IGF1 responsiveness are reduced in M12 cells (expressing a methylated AR promoter) in comparison to P69 cells (expressing an unmethylated AR gene), we postulated that AR methylation may lead to AR silencing, with ensuing downregulation of the IGF1R gene, a bona fide AR target. In order to evaluate whether AR promoter methylation alters AR function, M12 cells were treated with 5-Aza for 72 hr. Cells were further incubated with or without DHT (10−9 M), in the absence or presence of the AR inhibitor flutamide for 24 h. The rationale for this experiment resides in the fact that DHT binds to AR, leading to transcriptional activation of the IGF1R gene (Schayek et al, manuscript submitted). The AR inhibitor was expected to abolish IGF1R gene transactivation. Following incubation, cells were harvested and IGF1R mRNA levels were measured by qPCR. No significant change in IGF1R mRNA level as a result of DHT or flutamide addition was seen in untreated M12 cells (Fig. 8A). In contrast, 5-Aza treatment, which activated AR function, caused a significant (∼50%) increase in basal IGF1R mRNA level, while DHT addition leads to a further increase (∼70%) in mRNA level. However, the addition of flutamide, which inhibits AR function, decreased IGF1R mRNA values to the basal levels measured prior to the 5-Aza treatment. Finally, we evaluated the effect of 5-Aza on the IGF1-stimulated IGF1R phosphorylation. For this purpose, M12 cells were treated with 5-Aza (or left untreated, control) for 48 h, after which cells were treated with IGF1 for an additional 24 h. As shown in Fig. 8B, IGF1 treatment led to a marked induction of IGF1R phosphorylation in both 5-Aza treated and control cells.
Figure 8. Effect of AR methylation on IGF1R mRNA levels and IGF1-induced IGF1R activation.
(A) M12 cells were treated with 5-Aza (1 µg/ml) for 72 hr (black bars), or left untreated (gray bars), after which cells were treated with or without DHT (10−9 M) and/or flutamide (10−5 M) for 24 h. Twenty-four hours after the treatment, cells were harvested, and IGF1R mRNA levels were measured by qPCR. *, p <0.02 versus M12-AZA cells. (B) Effect of 5-Aza on IGF1-induced IGF1R phosphorylation. M12 cells were treated with 5-Aza (or left untreated, control) for 48 h, after which cells were treated with IGF1 for an additional 24 h. Total and phospho-IGF1R levels were measured by Western immunoblots.
Discussion
DNA methylation is a major epigenetic alteration affecting gene expression. Methylation involves the addition of methyl groups to the 5-carbon of deoxycytosines in the palindromic dinucleotide CpG. Methylation of CpG islands leads to inactivation of gene transcription [19, 20] and plays a critical role during development by establishing haploid gene dosage and, in the long-term, repression of selected genes. CpG islands are mostly unmethylated in normal tissues and hypermethylated in various human cancers [19, 21, 22]. Promoter CpG island hypermethylation of tumor suppressor genes is a common hallmark of all human cancers and affects most cellular pathways. In addition to classical antioncogenes, methylation involves genes in DNA repair pathways, microRNAs, and genes involved in premature aging. In the specific case of prostate cancer, DNA methylation of the ERα and β promoters appears to play a role in the inactivation of the ER gene [30, 31]. Aberrant CpG methylation in prostate cancer was also described for GSTP1 in ∼90% of the cases, for RASSF1A in ∼63%, and for RARβ2 in ∼79% [32, 33]. Other examples of genes frequently silenced in prostate cancer are APC, MGMT, and MDR1 [34, 35]. AR promoter methylation and gene inactivation have been detected in about 8–28% of prostate tumor samples [23, 24]. Kinoshita el al. reported that 4 of 15 tumors obtained from men who had died from hormone-independent prostate cancer demonstrated a significant loss of AR expression and two (50%) of these AR-negative tumors contained AR methylation.
The present study identifies AR promoter methylation as a defined critical event in the progression of prostate cancer epithelial cells from an early, benign stage to an advanced, metastatic stage. Specifically, AR promoter methylation was detected in the metastatic M12 and DU145 cell lines, whereas tumorigenic but non-metastatic (M2205 and M2218) as well as benign (P69) cells, display an unmethylated AR promoter. PC3 cells, usually regarded as metastatic, do not fit this interpretation as they were shown to exhibit an unmethylated AR. In addition, we showed that 5-Aza treatment of M12 cells, which caused demethylation of the AR promoter, leads to a significant increase in IGF1R mRNA levels, whereas addition of the AR inhibitor flutamide decreased the mRNA levels to the basal values measured prior to the 5-Aza treatment. IGF1R gene silencing is probably one of the critical consequences of the AR methylation-induced dysregulation of AR targets. The fact that changes in IGF1R mRNA levels are not seen at the protein level could probably be the result of differential expression of various splice variants which have been shown to differ in their degradation rates. Alternatively, the fact that IGF1R protein is constitutively present at high levels in cancer cells may obscure the visualization of further increments in protein amounts.
In the context of the IGF system, DNA methylation plays an important role in IGF2 gene regulation. The IGF2 gene constitutes one of the classical examples of imprinted genes. Loss-of-imprinting (LOI) leads to biallelic expression of the IGF2 gene, thus providing a proliferative advantage to transformed cells by elevating the levels of available IGF2 ligand. IGF2 LOI is an important mechanism in the etiology of various overgrowth syndromes (e.g., Beckwith-Wiedemann) and neoplasia (e.g., Wilms' tumors) [36, 37]. Likewise, the IGF2/mannose-6-phosphate receptor gene is also methylated, being its expression dependent on an intronic CpG island [38]. The IGF2 and IGF1R genes include GC-rich, TATA-less promoters. Furthermore, a number of transcription factors, including Sp1, p53, Wilms’ tumor-1 (WT1), and others, were shown to be involved in regulation of gene expression of both genes. Despite this overlap in transcriptional mechanisms, and in spite of the overall similarity in IGF2 and IGF1R promoters architectures, our results showed that the IGF1R promoter is unmethylated at all stage of the disease.
The interplay between the androgen and IGF1 systems is of major importance in prostate cancer. However, the mechanisms by which IGF1R signaling interacts with AR action, and vice versa, are still a matter of debate [29]. The hypothesis that growth factors can substitute for signaling from the AR and be the driving force in androgen-independent prostate cancer was postulated more than a decade ago [39]. However, the finding that AR is consistently increased in androgen-independent prostate cancer led to the question as to what was stimulating AR signaling if the patient had been castrated and testosterone was no longer present [40]. A number of reports have identified several cytokines that are able to activate AR in the absence of androgens. Considerable data suggest that in the absence of androgens, signaling through the IGF1R can enhance AR transcriptional activity [41, 42]. In addition, IGF1R activation leads to AR phosphorylation and increased nuclear translocation. Inhibition of IGF signaling, on the other hand, may lead to cytoplasmic AR retention, with ensuing changes in expression and/or activation of androgen-regulated genes. Interestingly, androgens were shown to control IGF1R expression via genomic and non-genomic pathways [16]. The present study identifies epigenetic mechanisms as a novel level of regulation in the control of the AR-IGF1R loop in prostate cancer progression.
In addition, we evaluated the methylation status of the BRCA1 and KLF6 promoters in prostate cancer. The rationale for these analyses was the fact that these transcription factors were previously identified as upstream regulators of the IGF1R gene in prostate cells [18, 43, 44]. In addition, a number of studies have shown that BRCA1 and KLF6 are involved in prostate cancer biology [18, 45, 46]. Our analyses, however, provide no evidence of methylation of the BRCA1 gene (Fig. 2) and KLF6 gene (data not shown) in any prostate cancer cell line examined. Likewise, the ERα promoter was unmethylated in all of the cell lines assayed (not shown). On the other hand, the PR-A and PR-B promoters were methylated in all prostate cancer cell lines, regardless of their tumorogenicity and metastatic capacity (Fig. 6). Hence, the methylation status of the PR genes cannot explain the change in IGF1R levels during disease progression.
In summary, our results demonstrate that progression of prostate cancer from a benign stage (P69 cells) to a metastatic one (M12 cells) is associated with hypermethylation of the AR gene. On the other hand, the low IGF1R levels seen at metastatic prostate cancer stages are most probably not caused by direct methylation of the IGF1R promoter. Given that the IGF1R gene has been identified as a downstream target for AR action, our data is consistent with a model in which AR gene methylation during progression of the disease leads to dysregulation of AR targets, including the IGF1R gene, at metastatic stages.
Acknowledgements
This work was performed in partial fulfillment of the requirements for a Ph.D. degree by Hagit Schayek in the Sackler Faculty of Medicine, Tel Aviv University. The authors wish to thank Dr. Joy L. Ware for providing cell lines and Ms. Tal Ohayon for help with manuscript preparation. This research was supported by Grant 2003341 of the United States-Israel Binational Science Foundation (to H.W. and S.R.P.) and CA97186-06 and Veterans Affairs Research Service (to S.R.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gelmann EP. Molecular biology of the androgen receptor. J. Clin. Oncol. 2002;20:2001–2015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen-refractory prostate cancer. J. Natl. Cancer Inst. 2001;93:1687–1697. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- 3.Shi XB, Ma AH, Xia L, Kung HJ, de Vere White RW. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer Res. 2002;62:1496–1502. [PubMed] [Google Scholar]
- 4.LeRoith D, Werner H, Beitner-Johnson D, Roberts CT., Jr Molecular and cellular aspects of the insulin-like growth factor I receptor. Endocrine Rev. 1995;16:143–163. doi: 10.1210/edrv-16-2-143. [DOI] [PubMed] [Google Scholar]
- 5.Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B. The D receptor in cell growth, transformation and apoptosis. Biochim. Biophys. Acta. 1997;1332:F105–F126. doi: 10.1016/s0304-419x(97)00007-3. [DOI] [PubMed] [Google Scholar]
- 6.Werner H, LeRoith D. New concepts in regulation and function of the insulin-like growth factors: implications for understanding normal growth and neoplasia. Cell. Mol. Life Sci. 2000;57:932–942. doi: 10.1007/PL00000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocrine Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 8.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nature Rev. Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 9.Baserga R. The insulin-like growth factor-I receptor as a target for cancer therapy. Expert Opin. Ther. Targets. 2005;9:753–768. doi: 10.1517/14728222.9.4.753. [DOI] [PubMed] [Google Scholar]
- 10.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, Hennekens CH, Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 11.Renehan AG, Zwahlen M, Minder C, O'Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 12.Tennant MK, Thrasher JB, Twomey PA, Drivdahl RH, Birnbaum RS, Plymate SR. Protein and mRNA for the type 1 insulin-like growth factor (IGF) receptor is decreased and IGF-II mRNA is increased in human prostate carcinoma compared to benign prostate epithelium. J. Clin. Endocrinol. Metab. 1996;81:3774–3782. doi: 10.1210/jcem.81.10.8855837. [DOI] [PubMed] [Google Scholar]
- 13.Chott A, Sun Z, Morganstern D, Pan J, Li T, Susani M, Mosberger I, Upton MP, Bubley GJ, Balk SP. Tyrosine kinases expressed in vivo by human prostate cancer bone marrow metastases and loss of type 1 insulin-like growth factor receptor. Am. J. Pathol. 1999;155:1271–1279. doi: 10.1016/S0002-9440(10)65229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellawell GO, Turner GD, Davies DR, Poulsom R, Brewster SF, Macaulay VM. Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 2002;62:2942–2950. [PubMed] [Google Scholar]
- 15.Liao Y, Abel U, Grobholz R, Hermani A, Trojan L, Angel P, Mayer D. Up-regulation of insulin-like growth factor axis components in human primary prostate cancer correlates with tumor grade. Human Pathol. 2005;36:1186–1196. doi: 10.1016/j.humpath.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Pandini G, Mineo R, Frasca F, Roberts CT, Jr, Marcelli M, Vigneri R, Belfiore A. Androgens up-regulate the insulin-like growth factor-I receptor in prostate cancer cells. Cancer Res. 2005;65:1849–1857. doi: 10.1158/0008-5472.CAN-04-1837. [DOI] [PubMed] [Google Scholar]
- 17.Bakin RE, Gioeli D, Sikes RA, Bissonette EA, Weber MJ. Constitutive activation of the Ras/mitogen-activated protein kinase signaling pathway promotes androgen hypersensitivity in LNCaP prostate cancer cells. Cancer Res. 2003;63:1981–1989. [PubMed] [Google Scholar]
- 18.Schayek H, Haugk K, Sun S, True LD, Plymate SR, Werner H. Tumor suppressor BRCA1 is expressed in prostate cancer and control IGF1-R gene transcription in an androgen receptor-dependent manner. Clin. Cancer Res. 2009;15:1558–1565. doi: 10.1158/1078-0432.CCR-08-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics join genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 20.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim. Biophys. Acta. 2004;1704:87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 22.Li LC, Zhao H, Shiina H, Kane CJ, Dahiya R. PGDB: a curated and integrated database of genes related to the prostate. Nucleic Acids Res. 2003;31:291–293. doi: 10.1093/nar/gkg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita H, Shi Y, Sandefur C, Meisner LF, Chang C, Choon A, Reznikoff CR, Bova GS, Friedl A, Jarrard DF. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60:3623–3630. [PubMed] [Google Scholar]
- 24.Sasaki M, Tanaka Y, Perinchery G, Dharia A, Kotcherguina I, Fujimoto S, Dahiya R. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J. Natl. Cancer Inst. 2002;94:384–390. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 25.Wu JD, Haugk K, Woodke L, Nelson P, Coleman I, Plymate SR. Interaction of IGF signaling and the androgen receptor in prostate cancer progression. J. Cell. Biochem. 2006;99:392–401. doi: 10.1002/jcb.20929. [DOI] [PubMed] [Google Scholar]
- 26.Bae VL, Jackson-Cook CK, Brothman AR, Maygarden SJ, Ware JL. Tumorigenicity of SV40 T antigen immortalized human prostate epithelial cells: association with decreased epidermal growth factor receptor (EGFR) expression. Int. J. Cancer. 1994;58:721–729. doi: 10.1002/ijc.2910580517. [DOI] [PubMed] [Google Scholar]
- 27.Bae VL, Jackson-Cook CK, Maygarden SJ, Plymate SR, Chen J, Ware JL. Metastatic sublines of an SV40 large T antigen immortalized human prostate epithelial cell line. Prostate. 1998;34:275–282. doi: 10.1002/(sici)1097-0045(19980301)34:4<275::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plymate SR, Tennant MK, Culp SH, Woodke L, Marcelli M, Colman I, Nelson PS, Carroll JM, Roberts CT, Jr, Ware JL. Androgen receptor (AR) expression in AR-negative prostate cancer cells results in differential effects of DHT and IGF-I on proliferation and AR activity between localized and metastatic tumors. Prostate. 2004;61:276–290. doi: 10.1002/pros.20099. [DOI] [PubMed] [Google Scholar]
- 30.Li LC, Chui R, Nakajima K, Oh BR, Au HC, Dahiya R. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer Res. 2000;60:702–706. [PubMed] [Google Scholar]
- 31.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60:3175–3182. [PubMed] [Google Scholar]
- 32.Singal R, van Wert J, Bashambu M. Cytosine methylation represses glutathione S-transferase P1 (GSTP1) gene expression in human prostate cancer cells. Cancer Res. 2001;61:4820–4826. [PubMed] [Google Scholar]
- 33.Kuzmin I, Gillespie JW, Protopovov A, Geil L, Dreijerink K, Yang Y, Vocke CD, Duh F-M, Zabarovsky E, Minna JD, Rhim JS, Emmert-Buck MR, Linehan WM, Lerman MI. The RASSF1A tumor suppressor gene is inactivated in prostate tumor and suppresses growth of prostate carcinoma cells. Cancer Res. 2002;62:3498–3502. [PubMed] [Google Scholar]
- 34.Fiori AR, Steinhoff C, Muller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br. J. Cancer. 2004;91:985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang GH, Lee SW, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J. Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 36.Vu TH, Hoffman A. Alterations in the promoter-specific imprinting of the IGF-2 gene in Wilms' tumor. J. Biol. Chem. 1996;271:9014–9023. doi: 10.1074/jbc.271.15.9014. [DOI] [PubMed] [Google Scholar]
- 37.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumor. Nature Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 38.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Inprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 39.Culig Z, Hobisch A, Cronauer M, Radmayr C, Hittmair A, Zhang J, Thurnher M, Bartsch G, Klocker H. Regulation of prostatic growth and function by peptide growth factors. Prostate. 1996;28:392–405. doi: 10.1002/(SICI)1097-0045(199606)28:6<392::AID-PROS9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 40.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen receptor signaling axis. J. Clin. Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 41.Lin HK, Yeh S, Kang HY, Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. USA. 2001;98:7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orio F, Terouanne B, Georget V, Lumbroso S, Avances C, Siatka C, Sultan C. Potential action of IGF-I and EGF on androgen receptor nuclear transfer and transactivation in normal and cancer human prostate cell lines. Mol. Cell. Endocrinol. 2002;198:105–114. doi: 10.1016/s0303-7207(02)00374-x. [DOI] [PubMed] [Google Scholar]
- 43.Rubinstein M, Idelman G, Plymate SR, Narla G, Friedman SL, Werner H. Transcriptional activation of the IGF-I receptor gene by the Kruppel-like factor-6 (KLF6) tumor suppressor protein: potential interactions between KLF6 and p53. Endocrinology. 2004;145:3769–3777. doi: 10.1210/en.2004-0173. [DOI] [PubMed] [Google Scholar]
- 44.Abramovitch S, Glaser T, Ouchi T, Werner H. BRCA1-Sp1 interactions in transcriptional regulation of the IGF-IR gene. FEBS Lett. 2003;541:149–154. doi: 10.1016/s0014-5793(03)00315-6. [DOI] [PubMed] [Google Scholar]
- 45.Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, Hod E, Leake D, Friedman SL, Hall SJ, Chinnaiyan AM, Gerald WL, Rubin MA, Martignetti JA. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J. Clin. Invest. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosen EM, Fan S, Goldberg ID. BRCA1 and prostate cancer. Cancer Invest. 2001;19:396–412. doi: 10.1081/cnv-100103134. [DOI] [PubMed] [Google Scholar]