Abstract
The study of isolated protein co7mplexes has greatly benefited from recent advances in mass spectrometry instrumentation and quantitative, isotope labeling techniques. The comprehensive characterization of protein complex components and quantification of their relative abundance relies heavily upon maximizing protein and peptide sequence information obtained from MS and tandem MS studies. Recent work has shown that using a metalloendopeptidase, Lys-N, for proteomic analysis of biological protein mixtures produces complementary protein sequence information compared to trypsin digestion alone. Here, we have investigated the suitability of Lys-N proteolysis for use with MALDI mass spectrometry to characterize the yeast Arp2 complex and E. coli PAP I protein interactions. Although Lys-N digestion resulted in an average decrease in protein sequence coverage of ~30% compared to trypsin digestion, CID analysis of singly-charged Lys-N peptides yielded a more extensive b-ions series compared to complementary tryptic peptides. Taking advantage of this improved fragmentation pattern, we utilized differential 15N/14N guanidination of Lys-N peptides and MALDI-MS/MS analysis to relatively quantify the changes in PAP I associations due to deletion of sprE, previously shown to regulate PAP I-dependent polyadenylation. Overall, this Lys-N/guanidination integrative approach is applicable for functional proteomic studies utilizing MALDI mass spectrometry analysis, as it provides an effective and economical mean for relative quantification of proteins in conjunction with increased sensitivity of detection and fragmentation efficiency.
Introduction
Mass spectrometry is a widely used technique for protein characterization in complex biological samples. Functional proteomic studies that employ mass spectrometry to investigate the dynamics of protein-protein interactions in response to changing environmental conditions can help reveal novel biological functions of cellular proteins. Recent work has shown that use of a hybrid MALDI LTQ-Orbitrap mass spectrometer is amenable to the study of protein complexes [1]. In this approach, protein complexes are isolated using cryogenic cell lysis and antibody-based affinity enrichment on magnetic beads [2]. The individual complex components are analyzed using a standard “bottom-up” approach, which involves SDS-PAGE and in-gel digestion with trypsin [2]. MALDI MS and MS/MS analysis of tryptic digests are performed in tandem to identify putative interacting proteins. Due to the generation of predominantly singly-charged ions from MALDI ionization, this approach benefits from decreased MS spectrum complexity when compared to spectra generated by electrospray (ESI). Yet, robust and informative MSn analyses of singly-charged ions remain a challenge. Collision induced dissociation (CID) of multiply-charged tryptic peptides, such as those produced by ESI, is aided by the presence of “mobile protons” that facilitate more uniform backbone cleavage compared to singly-charged tryptic peptides [3, 4]. Although CID spectra of singly-charged ions contain y- and b-ions, as well as ions from the neutral loss of water or ammonium, the C-terminal basic residue sequesters the lone proton, often generating spectra containing intense y-ions arising from preferential cleavages N-terminal to proline and C-terminal to acidic residues [5–7]. While these preferential cleavages are well-represented and can serve as diagnostic markers, the remaining ion series are often underrepresented, making these spectra less amendable to database search strategies or de novo sequencing.
Recent reports have described the use of the metalloendopeptidase Lysine N (Lys-N) for enzymatic digestion of proteins, which cleaves N-terminal to lysine, generating peptides containing lysine residues at the N-terminus [8]. Use of this enzyme in conjunction with MALDI-TOF mass spectrometry was shown to improve the sequence information obtained after CID fragmentation of singly-charged ions, resulting in a more prevalent series of N-terminal (b-) ions. In fact, in cases where a single basic residue was present in the peptide, a well-represented b-ion series was generated, making it suitable for de novo sequencing using MALDI-CID analysis [9].
In a typical MALDI MS experiment, arginine-containing peptides generated from trypsin digestion are more readily detected than those containing lysine residues due to increased gas phase basicity [10, 11]. Therefore, the conversion of lysine residues to homoarginine residues by guanidination using O-methylisourea was described as a useful tool to increase ionization efficiency and sensitivity of detection in MS [10, 12]. The use of guanidination was also reported to aid the fragmentation of doubly-charged ions in electron transfer dissociation (ETD) MS/MS analysis by increasing the intensity of the N-terminal c-ion series [13].
Guanidination by O-methylisourea has additional benefits, such as the ability to be synthesized from the relatively inexpensive urea and hence is readily adaptable to large-scale studies. Incorporation of heavy isotopes during the guanidination reaction permits relative quantification studies by differential peptide labeling with either [15N]- or [14N]-O-methylisourea [10]. Indeed, this approach has been successfully used to quantify relative changes in protein expression in hepatocytes during the de-differentiation and adaptation processes [12].
Here we have combined the use of Lys-N protein digestion with isotope-labeled guanidination for analysis of protein complexes isolated from yeast and bacterial cells by MALDI MS and MS/MS. Isolated complexes digested with trypsin or Lys-N were used for evaluating protein identification and sequence coverage, the behavior of singly-charged peptides following guanidination, and the utility of this approach for relative quantification studies. This complementary approach is applicable to studies utilizing MALDI mass spectrometry analysis for functional proteomics, as it provides an effective and economical mean to relatively quantify proteins and improve sensitivity of detection and fragmentation efficiency.
Materials and Methods
Bacterial and Yeast strains, media and growth conditions
The yeast strain expressing the chromosomal ARP2-GFP gene was purchased from Invitrogen. Yeast cells were grown to ~2 × 107 cells/mL, collected by centrifugation and frozen as pellets as described previously [2]. The bacterial strains expressing PAP I-GFP [14] were grown to stationary phase (optical density at 600 nm (OD600 2–4)), harvested and frozen as described [15]. Briefly, 2 L of stationary phase cells were centrifuged in four, 10 minute steps at 2000–2500 rcf to obtain a single pellet. To the weighed pellets, 100 µL of 20 mM HEPES, pH 7.5, 1.2% w/v polyvinyl-pyrrolidone (Sigma), 1/100 v/v protease inhibitor cocktail (Sigma) were added per gram.
Cryogenic cell lysis
Cryogenic cell lysis was carried out as described previously [2] using 25-mL jars, 20-mm stainless steel grinding balls (McMaster, NJ), and a Retsch MM 301 Mixer Mill (Retsch, Newtown, PA). Specific details of cryogenic lysis and optimized lysis buffer conditions for both the yeast and bacterial strains have been previously reported [2, 15]. Briefly, 3 g of cells of either the yeast or bacterial strains, were incubated with 15 mL lysis buffer. The lysis buffer for the Arp2 yeast strain was 20 mM HEPES, pH 7.4, 0.11 M KOAc, 2 mM MgCl2, 0.1% tween-20 (v/v), 1 µM ZnCl2, 1 µM CaCl2, 0.5% Triton X-100, 150 mM NaCl, 1:100 protease inhibitor cocktail (Sigma) and 1:200 phenylmethylsulphonyl fluoride (PSMF), while the lysis buffer for the PAP I bacterial cells was 20 mM HEPES, pH 7.4, 0.11 M KOAc, 2 mM MgCl2, 0.1% tween-20 (v/v), 1 µM ZnCl2, 1 µM CaCl2, 1% Triton X-100, 0.5% Deoxycholate, 150 mM NaCl, 1:100 protease inhibitor cocktail (Sigma) and 1:200 phenylmethylsulphonyl fluoride (PSMF). Following centrifugation at 4,000 rpm, 4°C for 10 min, the soluble fractions were used for the affinity purification experiments.
Immunoaffinity Purification
Single-step affinity purifications via the GFP tag were carried out as described previously [2]. Briefly, magnetic epoxy beads (Dynal Invitrogen), coated with purified polyclonal anti-GFP antibodies, were used to isolate protein complexes. Immunoaffinity purifications were carried out by incubation of the cell lysates with the conjugated beads for 1 hour for both bacterial cells and yeast cells. The magnetic beads were gathered by using a magnet (Magcraft). Proteins were eluted with 700 µL 0.5 N NH4OH, 0.5 mM EDTA solution, and dried by vacuum centrifugation (Speed Vac Plus SC110A with a gel pump GP110, Thermo Scientific). The dried pellets were resuspended with LDS-PAGE sample buffer (Invitrogen), alkylated with 100 mM iodoacetamide and separated on a 4–12% NuPAGE Novex bis-Tris gel (Invitrogen) according to the manufacturer’s instructions. Gels were immediately stained with GelCode Blue staining reagent (Thermo Scientific).
Protein Digestion
After staining, the gel lanes were cut into approximately 25 1-mm slices and prepared for analyses as described [1]. Proteins were digested with 12.5 ng/µL sequencing grade modified trypsin (Promega) or 12.5 ng/µL metalloendopeptidase Lys-N (Seikagaku Corp. (Tokyo, Japan)) in 50 mM ammonium bicarbonate (ABC) over night at 37°C. Lys-N digestion conditions were optimized for temperature and concentration of enzyme. BSA was digested with 6.25 ng/µL, 12.5 ng/µL and 18.75 ng/µL of Lys-N at both 25°C and 37°C. After mass spectrometric analysis, the digestions using 12.5 ng/µL and 18.75 ng/µL gave equivalent results, and the temperature was not a major factor. In view of the cost of the enzyme, we opted to use Lys-N at the concentration of 12.5 ng/µL for the following experiments. Peptides were extracted from the gel pieces on reverse phase resin (POROS 20 R2, Applied Biosystems). The samples were desalted and concentrated using ZipTips with C18 resin (Millipore), washed twice with 20 µL 0.1% (v/v) trifluoroacetic acid (TFA), and eluted with 4 µL of 20 mg/mL DHB (2,5-dihydroxybenzoic acid) in 50% (v/v) methanol, 20% (v/v) acetonitrile (ACN) and 0.1% (v/v) TFA, directly onto a MALDI target.
Guanidination
Guanidination was carried out using a modified version of the protocol described previously [12]. After peptides were in-gel digested with either trypsin or Lys-N, the reaction was stopped by adding TFA to a final concentration of 0.1%. Peptides were eluted from the gel pieces as described [15]. The guanidination reaction was carried out in 20 µL of 7N NH4OH and 1.5 µL of [N14]-O-methylisourea (prepared as described previously [12]). The samples were shaken overnight at room temperature, and the reaction was stopped by adding an equal volume of 1% (v/v) TFA. After drying, the samples were suspended in 25–50 µL of a solution of POROS beads in a 1:1 mixture of 5% formic acid, 0.2% TFA and 50 mM ABC. After incubation overnight, the samples were loaded onto zip tips as described above, or for quantification purposes they were mixed as described below.
Relative quantification following guanidination
For quantification purposes, the wild-type peptides were derivatized using [N15]-O-methylisourea following the same procedure described above. Digested peptides from equivalent bands from both the wild-type and ΔsprE PAP I-GFP cells were loaded onto the same zip tip and the procedure described above was carried out. Peptides from both the wild-type and ΔsprE cells could now be directly compared by the intensities from the peaks in the mass spectra. The heavy labeled peptides from wild-type cells would be shifted by +2 Thompson (Th) compared to the light peptides from the ΔsprE cells.
Mass Spectrometry
Mass spectrometric analyses were performed on a LTQ Orbitrap XL mass spectrometer equipped with a MALDI ion source (Thermo Electron, Bremen, Germany [16]). The MS, MS/MS analyses by collision-induced dissociation (CID), the parameters used for data acquisition, and the software used for spectra viewing and processing were as described previously [1]. Briefly, the instrument was operated in positive mode with a MALDI laser energy of 16 µJ. MS spectra were acquired for a mass range of m/z 700–4000 at resolution 60,000 (set at m/z 400). Additional parameters included: automated spectrum filter (ASF) off, 20 scans/step, automated gain control (AGC) off, 20–50 laser shots/scan. For Lys-N-digested samples, a prominent Lys-N autolytic peptide was used as an internal lock-mass standard to calibrate the MS spectra: KAPTTGTDSQAGTLVHESSHFTRN*GGT (*deamination), [M+H]+ at m/z 2758.303238. MALDI IT collision-induced dissociation (CID) was carried out using: precursor ion isolation in the linear ion trap, 4 mass units isolation width, normalized collision energy 35%, activation q 0.25, activation time 300 ms. For relative quantification studies, precursor ion selection was performed using an isolation window of ±6 mass units, centered at the midpoint m/z between the light and heavy monoisotopic peaks. The quantifications were performed utilizing the intensities of the monoisotopic precursor and fragment peaks. Database searching was limited to the use of the computer algorithm that allowed the specification of the Lys-N cleavage, PROWL (http://prowl.rockefeller.edu/prowl-cgi/) [17] and Mascot (www.matrixscience.com) [18]. The parameters used for database searching against the NCBI nonredundant protein database, version 06/10/16, were as described [1], with the following differences: the taxonomy was selected as either Saccharomyces cerevisiae (10452 sequences), or Escherichia coli (26383 sequences), Lys-N cleavage, and mass error limit of 5 ppm.
Results and Discussion
Protein sequence coverage following trypsin or Lys-N digestions
We aimed to incorporate the reported merits of proteolysis by Lys-N with guanidination and stable isotopic labeling for relative quantification of MALDI-generated singly-charged ions. Previous reports have described several different conditions (temperature, enzyme concentration and time) for in-solution and in-gel digestion using Lys-N [9, 13, 19, 20]. We tested several parameters (described in the Materials and Methods) and observed that a Lys-N concentration of 12.5 ng/µL in 50 mM ammonium bicarbonate solution at 37°C is suitable for digestion, in agreement with some of the previous reports [9]. We observed an extensive level of autolysis for Lys-N. Similar to the use of trypsin autolytic peaks [1], the Lys-N autolytic peptides could be utilized for internal calibration of MS spectra (see Materials and Methods), resulting in an average mass error and standard deviation of 0.40 ± 1.64 ppm.
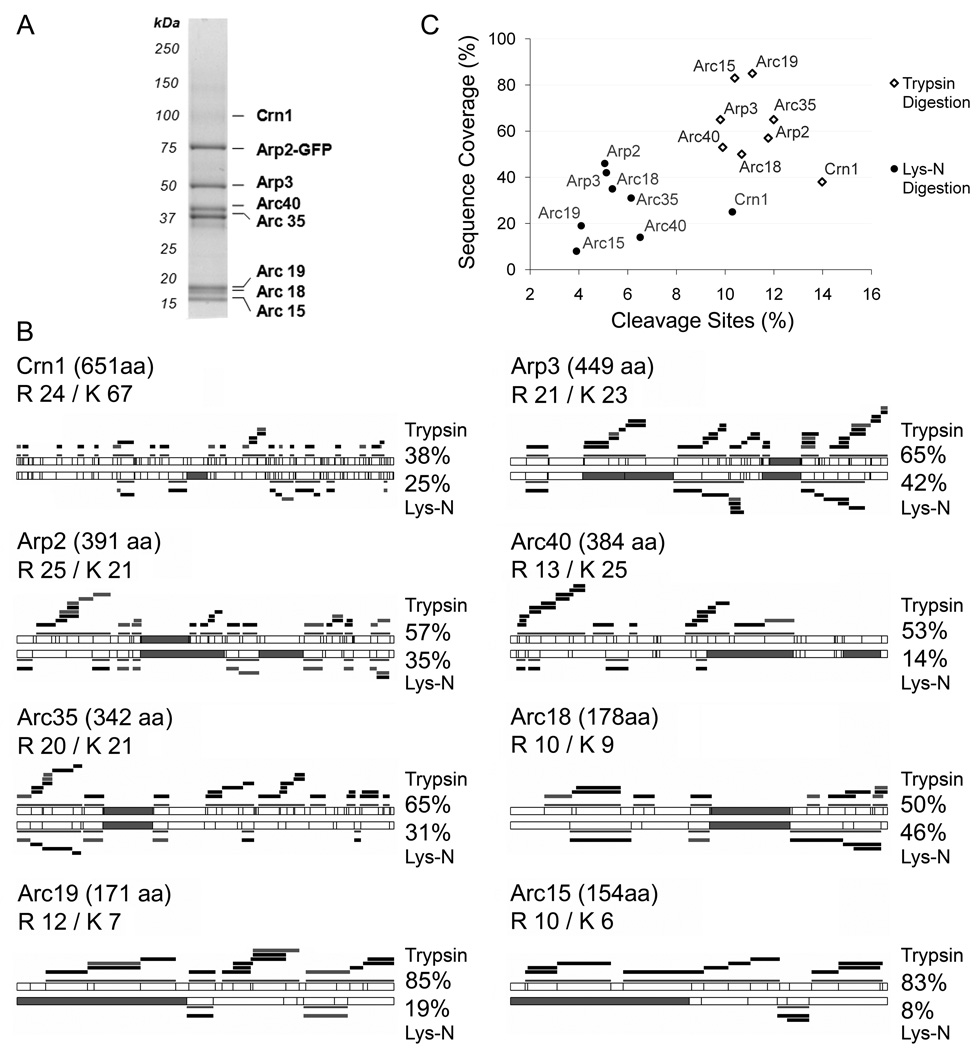
Previous studies have shown that trypsin and Lys-N digestions produced complementary results when analyzing unmodified or phosphorylated peptides from large data sets [19, 20]. The combination of peptides identified from Lys-N and trypsin digests of yeast extracts resulted in an increase in protein coverage of 1.7-fold over that obtained from trypsin digestion alone [19]. As these studies did not assess the variation in sequence coverage at the individual protein level or compare Lys-N and trypsin protein coverage directly, we asked whether the Lys-N digestion would be suitable for analysis of protein complexes by MALDI mass spectrometry. The protein coverage of complex components across a broad range of molecular weights can be critical for such studies. Towards this goal, we assessed the use of Lys-N for determining protein-protein interactions in both yeast and bacterial model organisms. First, we examined the well-characterized Arp2 complex in Saccharomyces cerevisiae cells (Fig. 1A). Arp2 is an essential protein, member of a highly conserved complex that initiates actin nucleation, and therefore crucial for numerous actin-controlled cellular processes [21–25]. One-step affinity purification of Arp2 genomically tagged with green fluorescent protein (GFP) was performed using magnetic beads conjugated to anti-GFP antibodies. As shown in Fig. 1A, this permitted the isolation of the previously reported interacting partners, Arp3, Arc35, Arc40, Arc19, Arc18, Arc15 and Crn1 [2, 26]. These associations were reproducible in triplicate experiments. The bands corresponding to these proteins were excised and in-gel digested with either trypsin or Lys-N. Protein sequence coverage was evaluated by MALDI MS and peptide mass fingerprinting database analysis. As Lys-N has fewer cleavage sites than trypsin, generating peptides of larger molecular weights, the sequence coverage is expected to depend on the lysine content, as well as the m/z of the resulting peptides. Overall, trypsin digestion yielded higher protein sequence coverage (62 ± 16%) compared to Lys-N (28 ± 13%) among the Arp2 complex members (Fig. 1B and C). We observed an inverse relationship between lysine content and percent reduction in protein sequence coverage. Yet, not all proteins followed this pattern, for example, both Arc18 and Arc19 contained relatively few lysines, 9 and 7, while the percent reduction in sequence coverage after Lys-N digestions was 4 % and 66 %, respectively. From these examples it is clear that the position of the lysine residues is a key determinant of individual sequence coverage. Since the upper limit of detection when using a MALDI LTQ Orbitrap is m/z 4000, Lys-N digestion can result in peptides that fall out the m/z range detected by the instrument (Fig. 1B, filled gray segments). Despite the reduced sequence coverage, Lys-N digestion resulted in the successful identification of all previously reported members of the Arp2 complex. Taken together with previous work comparing trypsin and Lys-N digestions [9, 13, 19, 20, 27], these results support the use of Lys-N as a proteolytic enzyme for protein identifications studies.
Figure 1. Comparison of protein sequence coverage following trypsin and Lys-N digestion.
(A) A representative SDS-PAGE gel from the immunopurification of Arp2-GFP from yeast. (B) The protein sequence coverages obtained following trypsin (top) and Lys-N (bottom) digestions are represented for the eight proteins from the isolated Arp2 complex. The total numbers of amino acids, R and K residues are indicated. The cleavage sites for trypsin and Lys-N are represented by vertical lines. Filled gray segments represent peptides with an m/z > 4000 that would not be detected by our MALDI LTQ Orbitrap instrument. Detected peptides are represented as black bars for unmodified peptides, and gray bars for peptides that are modified by oxidation of methionine. Projections of the observed peptides and corresponding sequence coverages are indicated. (C) Protein sequence coverage is plotted against the percentage of cleavage sites for the Arp2-isolated proteins following trypsin (open diamonds) and Lys-N (filled circles) digestions.
CID fragmentation of Lys-N-derived singly-charged peptides
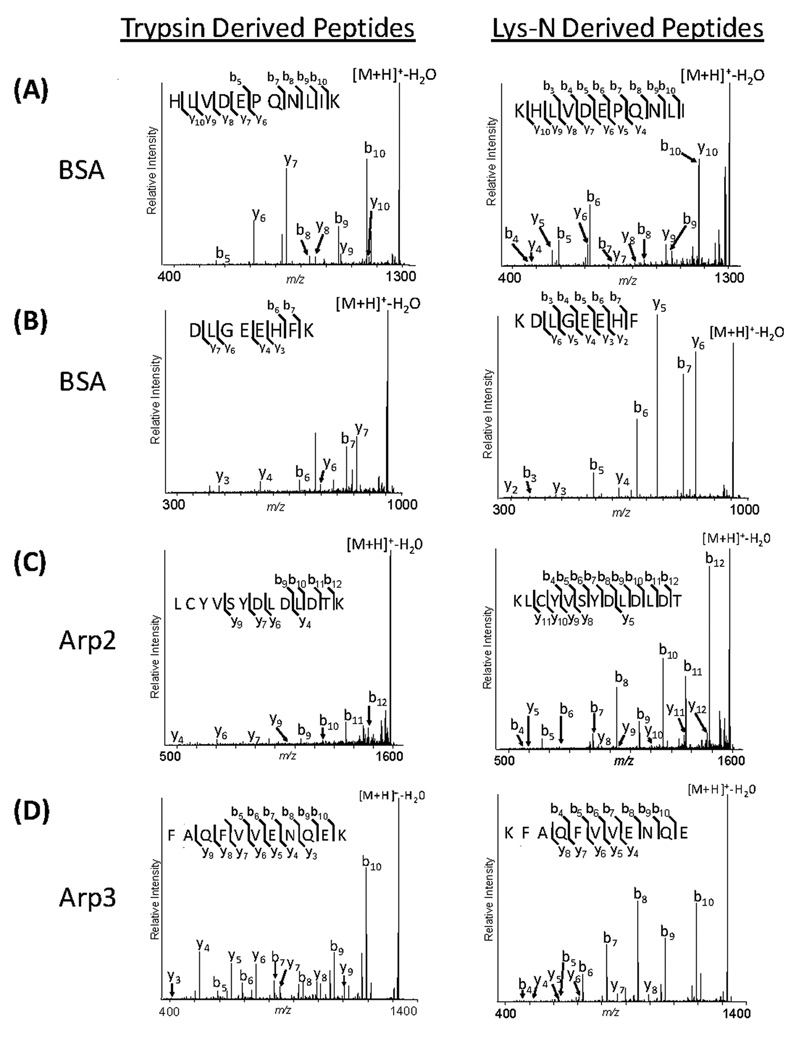
The fragmentation of peptides containing a single basic residue at the N-terminus, such as those generated from digestion with Lys-N, has been previously reported to produce simplified MS/MS spectra, containing prominent and readily distinguishable N-terminal ion series [9, 27]. To further assess this alternate proteolysis for studies of isolated protein complexes by MALDI mass spectrometry, we evaluated the CID fragmentation of unmodified, singly-charged ions corresponding to peptides generated by Lys-N or trypsin digestion of BSA as well as the Arp2 complex members (Fig. 1). We examined the fragmentation patterns of complementary trypsin or Lys-N peptides, i.e. peptides with the same sequence except for lysine residues at either the C-terminus or N-terminus, respectively. Representative MALDI IT CID spectra are illustrated in Fig. 2. The relative intensity and number of detected b-ions was higher for the Lys-N derived peptides than for tryptic peptides. CID fragmentation of peptides containing C-terminal lysine (trypsin) generated a mixture of y- and b-ions, with the most prominent product ions corresponding to preferential cleavages C-terminal to aspartate and glutamate residues and N-terminal to proline (e.g. y6 and y7 - Fig. 2A, y4 and y7 - Fig. 2B, y6 and b11 - Fig. 2C and y4 and b10 - Fig. 2D) [5, 6, 28, 29]. In comparison, peptides containing N-terminal lysine (Lys-N) gave rise to more complete b-ion series, but also retained preferential, charge-remote cleavages (e.g. b5 and b6 - Fig.2A) as was observed in CID spectra of tryptic peptides. Charge-remote fragmentation pathways are largely dependent on the degree of proton sequestration (proton mobility), with an increase in its occurrence as proton mobility decreases [30, 31]. Since the pairs of complementary tryptic and LysN peptides shown in figure 2 have similar ionizing sites (ε-amino side chain and N-terminal amino group) and similar proton mobilities (although influenced by the amino acid sequence) the presence of preferential cleavage in both cases was not unexpected. The presence of additional basicity due to histidine (Fig. 2A and B) provides an additional site for protonation and may result in increased y- or b-ion intensity depending on whether it is present at the C- or N-terminus, respectively, as previously established [30, 32]. This may explain the enhanced intensity of the y10 of KHLVDEPQNLI, Fig 2A when comparing the complementary Lys-N- and trypsin-derived peptides. The effect of the presence of both lysine and histidine residues can also be noticed on the fragmentation of the tryptic peptides HLVDEPQNLIK and DLGEEHFK (Fig. 2A and Fig. 2B). Both b-and y-ions are observed when the histidine and lysine are present at the opposite ends of the peptide (HLVDEPQNLIK), while mainly y-ions are observed when the basic residues are clustered at the C-terminus (DLGEEHFK). However, the effect of histidine on fragmentation may be minimized due to the limited proton mobility of singly-charged peptides. The reduced complexity of our sample (eight member protein complex generating reduced complexity spectra containing singly-charged ions) precluded us from further investigating this observation. Overall, the use of Lys-N proteolysis yielded more complete b-ion series by MALDI IT CID analysis compared to complementary tryptic peptides. The basis for these observations is most readily explained by Lys-N derived peptides achieving localization of the two most basic sites to the N-terminus (lysine side-chain and N-terminus amino group), which promotes charge transfer to b-ions following fragmentation and product ion dissociation. These results are consistent with those reported from large scale proteomic studies using a MALDI-TOF/TOF configuration [9] and those focusing on CID fragmentation of doubly-charged peptides [27]. A deeper mechanistic understanding of the processes involved in the fragmentation of singly-charged Lys-N derived peptides will require further investigation.
Figure 2. MALDI IT CID of trypsin- and Lys-N-derived peptides.
Equivalent peptides from BSA, Arp2, and Arp3 following trypsin (left column) or Lys-N (right column) digestion were selected for MALDI CID MS/MS analysis. The m/z values for the selected peptides were (A) 1304.70, (B) 973.450, (C) 1603.72 and (D) 1337.66.
Guanidination improves sensitivity of detection and fragmentation of singly-charged ions
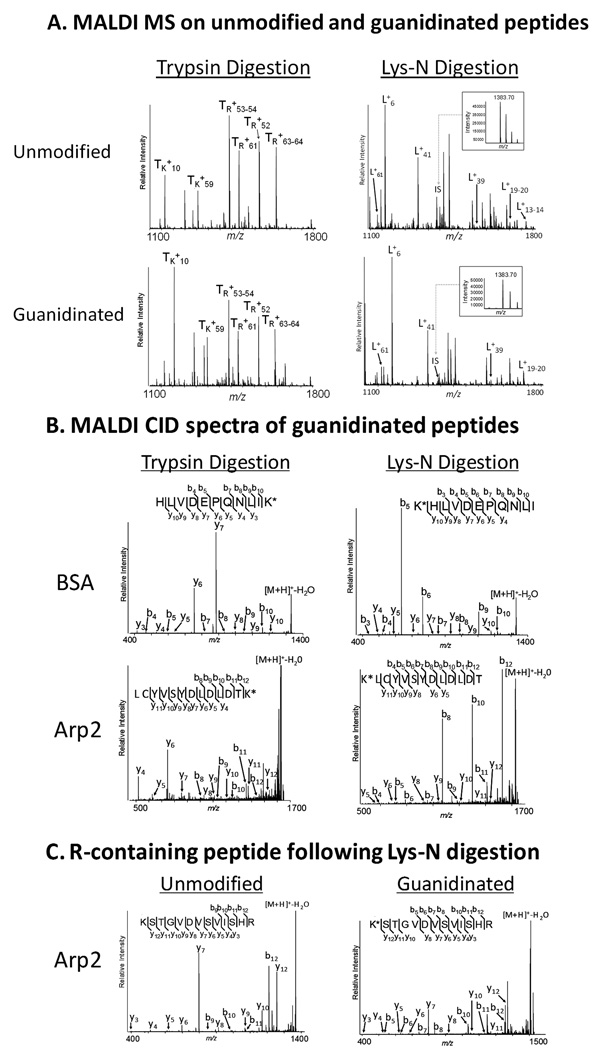
Guanidination converts lysine residues into the more basic homoarginine [10, 12], which was shown to increase the sensitivity of detection of lysine-containing peptides following digestion with trypsin. Recently, Lys-N digestion was coupled with guanidination to improve peptide fragmentation behavior for doubly-charged ions in ETD. We asked whether Lys-N digestion coupled with guanidination would also improve the sensitivity of detection for singly-charged ions, as well as the fragmentation along the peptide backbone in CID. BSA and Arp2-GFP were digested with either trypsin or Lys-N and subjected to guanidination. Representative MS spectra are shown from the digestion of BSA with trypsin and Lys-N (Fig. 3A). For the trypsin experiment, we assessed the differences in ionization efficiency of guanidinated versus unguanidinated peptides using R-containing tryptic peptides (denoted TR+n, Fig. 3A), which would not be modified by guanidination, as internal standards. This allowed the comparison of the relative intensities of unguanidinated and guanidinated K-containing peptides (denoted TK+n,, Fig. 3A). For the Lys-N experiment, untreated β-casein peptides were spiked at 50 fmol/µL in the guanidinated and umodified samples to serve as an internal standard (IS at m/z 1383.70, Fig. 3A inset). This was necessary as all the Lys-N digested peptides contain a N-terminal lysine residue that would be modified after guanidination. Our results indicated that the ionization efficiency and detection of lysine-containing tryptic peptides (TK+10 and TK+59) are improved following guanidination (compare with the 4 R-containing peptides, Fig. 3A), in agreement with previous studies [10, 13]. Similarly, guanidination of Lys-N peptides showed increased sensitivity of detection, as observed by comparison to the β-casein internal standard (compare all Lys-N peptides, denoted L+n, to the intensity of the internal standard, IS, before and after guanidination).
Figure 3. Guanidination improves sensitivity of detection and fragmentation of singly-charged ions.
(A) The MALDI MS spectra of trypsin or Lys-N digested BSA are illustrated for the m/z 1100–1800 range. TR+n indicates arginine-containing tryptic peptides, while the TK+n indicates lysine-containing peptides. All the peptides derived from Lys-N digestion (denoted L+n) contain lysine residues, and therefore are modified by guanidination. 50 fmol of tryptic β-casein peptides were spiked in the samples derived from the Lys-N digestion, serving as internal standards. The [M+H]+ peak at m/z 1383.70 from β-casein was included as a reference (labeled IS). (B) MALDI IT CID of T+59, L+41 BSA peptides (m/z 1346.71) and a selected Arp2 peptide (m/z 1645.72) are illustrated as representative MS/MS spectra. (C) MALDI IT CID of a Lys-N-derived Arp2 peptide (m/z 1383.75 unmodified and 1425.75 guanidinated) containing an N-terminus lysine and C-terminus arginine. K* represents homoarginine.
Guanidinated peptides were next subjected to MALDI IT CID. Fig. 3B illustrates the fragmentation of the guanidinated BSA and Arp2 peptides (see corresponding unmodified peptides in Fig. 2). The tryptic peptide T+59 and the corresponding Lys-N peptide, L+41, were selected for MS/MS analysis. In CID of tryptic peptides, guanidination decreased the overall intensity of the b-ion series, while increasing the coverage from the y-ion series (Fig. 3B, left panels). This observation is in agreement with ETD MS/MS analysis of doubly-charged ions, where the ion series containing the homoarginine is increased and the complementary ion series decreased [13]. The same trend can also be observed for the Lys-N digestion, where the N-terminal b-ions fragments containing the homoarginine have increased intensities, while the y-ions have reduced intensities (Fig. 3B, right panels). For example, the prominent y10 fragment of KHLVDEPQNLI (Fig 2A) has a reduced relative intensity following guanidination (Fig. 3B) consistent with the higher proton affinity of homoarginine compared to histidine. Also, guanidinated Lys-N peptides demonstrated more prominent preferential D/E cleavages (e.g. Fig. 3B, b5 and b6 in BSA peptide), compared to the corresponding unmodified Lys-N peptides (Fig. 2A and C). The increase in relative abundance of product ions from preferential cleavages following guanidination did not significantly impact the generation of relatively complete b-ion series as previously observed (Fig. 2), while maintaining relatively simplified MS/MS spectra. This may aid the confidence of protein identification by database searching strategies. These results are consistent with the mobile proton model, which predicts increased proton sequestration (due to increased gas phase basicity) of homoarginine compared to lysine, and therefore an augmentation in charge-remote fragmentation pathways. Based on the current data, a negative impact on percent of backbone cleavages due to guanidination of Lys-N peptides was not apparent. However, a quantitative approach using a diverse peptide population will be necessary to examine the relative contribution of increased ionization efficiency of homoarginine-containing peptides versus the effect of charge-remote pathways under CID fragmentation.
An interesting observation was obtained from the CID fragmentation of peptides containing both N-terminal lysine and C-terminal arginine (Fig. 3C). CID spectra of the unmodified peptide contained fragments clearly dominated by y-ions at the preferential cleavage sites, while b-ions were under represented. This was in contrast to the usual presence of prominent b-ions in CID of Lys-N-derived peptides. Following guanidination, some preferential cleavages were still detected as y-ions, but neither b- nor y-ions dominated the CID spectra. This suggested that the proton sequestration was balanced between the basic residues located at both the C- and N-termini of this peptide. Moreover, the presence of a histidine residue in the proximity of the arginine may also contribute to the C-terminus basicity and the observed y-ions. Consistent with the other results from guanidinated peptides (Fig. 3B), an increased number of b-ions was detected when compared to the unmodified peptide (Fig. 3C), which may reflect the increased basicity of homoarginine compared to lysine.
Integrating Lys-N digestion with guanidination for relative quantification
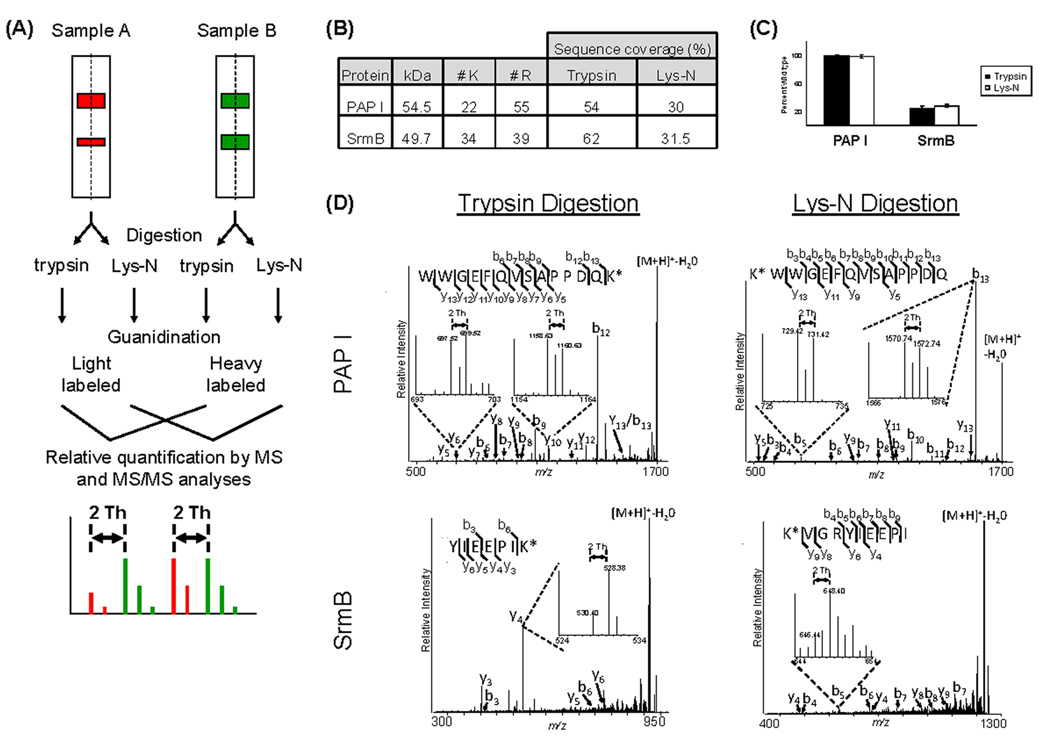
Derivatization of lysine residues via guanidination allows for differential isotope labeling, and therefore relative quantification studies, as reported for tryptic peptides [10, 12]. Lys-N is particularly well suited for incorporation of guanidination, as all the peptides generated by the digestion contain lysine residues. Also, the enhanced b ion series produced by Lys-N digestion may be advantageous for relative quantification performed at the MS/MS level. We illustrate its use for applications requiring relative quantification by focusing on a protein of interest—poly(A) polymerase I (PAP I) in E. coli. Polyadenylation of RNA in bacteria is believed to facilitate mRNA degradation [33], and this process is mainly carried out by PAP I [34–37]. Previous reports have shown that PAP I interacts with the SrmB RNA helicase [38], although the role of this association is not fully understood. We have previously reported that the response regulator SprE plays a role in the regulation of polyadenylation and mRNA stability [39]. We further characterized the PAP I interacting partners during exponential and stationary phases (Carabetta et al., manuscript in preparation). These studies indicated that SprE may play a key role in the maintenance of certain PAP I associations. To determine if Lys-N can be effectively used in conjunction with guanidination for quantifying the relative levels of the PAP I-SrmB association and its potential dependence on SprE, we performed affinity purifications of plasmid-borne PAP I-GFP in wild type and ΔsprE E. coli cells. The overall approach is illustrated in Fig. 4A, where sample A represents the SprE null cells and sample B the wild type cells. The samples were divided in two equal aliquots to compare the results obtained from trypsin and Lys-N digestions. The presence of SrmB in the PAP I isolates was detected by database searching using peptide mass fingerprinting and confirmed by MALDI IT MS/MS analyses in both trypsin and Lys-N experiments. Similar to our observations from analyzing the Arp2 complex (Fig. 1), the overall protein sequence coverage was also reduced for PAP I and SrmB when using Lys-N, likely related to the distribution and high number of lysine residues (Fig. 4B). Using the PAP I-GFP immunopurifications from wild-type and ΔsprE E. coli cells, we next compared the efficiency and reproducibility of the relative quantification using either trypsin or Lys-N. The peptides from the wild-type and ΔsprE samples were derivatized with [N15]- and [N14]-O-methylisourea, respectively, combined and at least four peptides were analyzed for relative quantification (Fig. 4A). The average relative intensities of y-and b-ion doublets were used for quantification of tryptic and Lys-N peptides, respectively, in wild type and ΔsprE E. coli cells (Fig. 4D). Consistent with the equal expression of PAP I-GFP measured in the two strains [39], both y-ion and b-ion doublets showed a 1:1 ratio. As expected, fragmentation of the Lys-N peptide yielded a greatly improved b-ion series (Fig. 4D). The relative quantification of PAP I-SrmB association at either the MS or MS/MS level demonstrated that in the absence of SprE, the interaction between SrmB and PAP-I decreased to 24.5% or 28% of the wild-type levels using trypsin or Lys-N, respectively (data not shown, and Fig. 4C). Similar results were obtained from the reverse derivatization experiment in which the wild type samples were light-labeled ([N14]-O-methylisourea) and the ΔsprE were heavy-labeled ([N15]-O-methylisourea). This comparison demonstrated that the pairs of fragment ions observed in the MS/MS spectra correspond to b ions and not to matrix or chemical noise (Supplementary Fig. S1). Additionally, obtaining equivalent results from this reciprocal experiment indicated that the isolation window of ±6 mass units did not bias precursor selection. Our results suggest that one possible role of SprE in the polyadenylation pathway is through the regulation of the association between PAP I and SrmB. We are currently investigating the biological significance of these findings. Overall, our results indicate that Lys-N digestion followed by guanidination can be effectively used as an alternative to trypsin in relative quantification studies.
Figure 4. Integrating Lys-N digestion and guanidination for relative quantification studies.
(A) Workflow of the relative quantification procedure. Proteins isolated with PAP I-GFP from E. coli cells were separated by 1D SDS PAGE, divided into equal aliquots and digested with trypsin or Lys-N. Peptides from the wild-type or ΔsprE cells were derivatized by guanidination with heavy (15N) or light (14N) labeled O-methylisourea, combined and analyzed using a MALDI LTQ Orbitrap. The intensities of the resulting doublets at MS or MS/MS levels of analysis were used for relative quantification. (B) Protein sequence coverage, number of lysine and arginine residues in PAP I and SrmB proteins. (C) Relative quantification of the association between PAP I-GFP and SrmB in ΔsprE cells. The ΔsprE peptides were normalized relative to the wild-type peptides. At least four peptides were used for each quantification. (D) Representative MALDI IT CID spectra of PAP I and SrmB peptides following trypsin and Lys-N digestions. Precursor ions were selected using an isolation window of ±6 mass units, which was centered at the midpoint m/z between the light and heavy monoisotopic peaks (m/z 1716.78 for illustrated PAP I peptide and m/z 933.47 and 1132.58 for the two SrmB peptides from trypsin and Lys-N digestions, respectively). The quantifications were performed utilizing the intensities of the monoisotopic fragment peaks averaging the resulting ratios from a minimum of 4 unique peptides. Inserts illustrate examples of y or b ion doublets used for relative quantification in trypsin or Lys-N studies, respectively.
Conclusions
We describe the use of Lys-N in conjunction with guanidination and isotope labeling for analyses of MALDI-generated singly-charged ions. This integrative approach provided increased sensitivity of detection at the MS level and improved fragmentation at the MS/MS level of analysis, as well as an economical mean for relative quantification. However, the observed reduction in protein sequence coverage of ~30% following Lys-N digestion, when compared to trypsin digestion, should be considered when implementing these approaches. Nevertheless, this methodology proved useful for the characterization and relative quantification of protein-protein interactions in yeast and bacteria, and we expect it to be applicable to various types of proteomic studies.
Supplementary Material
Acknowledgements
This work was supported by Award Number DP1DA026192 from the National Institute On Drug Abuse and Princeton University Start-up funding to IMC, and National Institute of General Medical Sciences GM65216 to TJS. The authors of the article have no competing financial interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute On Drug Abuse or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luo Y, Li T, Yu F, Kramer T, Cristea IM. Resolving the Composition of Protein Complexes Using a MALDI LTQ Orbitrap. J Am Soc Mass Spectrom. 2010;21(1):34–46. doi: 10.1016/j.jasms.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4(12):1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Burlet O, Orkiszewski RS, Ballard KD, Gaskell SJ. Charge promotion of low-energy fragmentations of peptide ions. Rapid Commun Mass Spectrom. 1992;6(11):658–662. doi: 10.1002/rcm.1290061106. [DOI] [PubMed] [Google Scholar]
- 4.Dongre AR, Somogyi A, Wysocki VH. Surface-induced dissociation: an effective tool to probe structure, energetics and fragmentation mechanisms of protonated peptides. J Mass Spectrom. 1996;31(4):339–350. doi: 10.1002/(SICI)1096-9888(199604)31:4<339::AID-JMS322>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Schilling B, Wang W, McMurray JS, Medzihradszky KF. Fragmentation and sequencing of cyclic peptides by matrix-assisted laser desorption/ionization post-source decay mass spectrometry. Rapid Commun Mass Spectrom. 1999;13(21):2174–2179. doi: 10.1002/(SICI)1097-0231(19991115)13:21<2174::AID-RCM771>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 6.Breci LA, Tabb DL, Yates JR, 3rd, Wysocki VH. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal Chem. 2003;75(9):1963–1971. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- 7.Tsaprailis G, Nair H, Somogyi A, Wysocki VH, Zhong W, Futrell JH, Summerfield SG, Gaskell SJ. Influence of Secondary Structure on the Fragmentation of Protonated Peptides. J Am Chem Soc. 1999;121(22):5142–5154. [Google Scholar]
- 8.Nonaka T, Hashimoto Y, Takio K. Kinetic characterization of lysine-specific metalloendopeptidases from Grifola frondosa and Pleurotus ostreatus fruiting bodies. J Biochem. 1998;124(1):157–162. doi: 10.1093/oxfordjournals.jbchem.a022074. [DOI] [PubMed] [Google Scholar]
- 9.Boersema PJ, Taouatas N, Altelaar AF, Gouw JW, Ross PL, Pappin DJ, Heck AJ, Mohammed S. Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol Cell Proteomics. 2009;8(4):650–660. doi: 10.1074/mcp.M800249-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brancia FL, Oliver SG, Gaskell SJ. Improved matrix-assisted laser desorption/ionization mass spectrometric analysis of tryptic hydrolysates of proteins following guanidination of lysine-containing peptides. Rapid Commun Mass Spectrom. 2000;14(21):2070–2073. doi: 10.1002/1097-0231(20001115)14:21<2070::AID-RCM133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Beardsley RL, Karty JA, Reilly JP. Enhancing the intensities of lysine-terminated tryptic peptide ions in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2000;14(23):2147–2153. doi: 10.1002/1097-0231(20001215)14:23<2147::AID-RCM145>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Warwood S, Mohammed S, Cristea IM, Evans C, Whetton AD, Gaskell SJ. Guanidination chemistry for qualitative and quantitative proteomics. Rapid Commun Mass Spectrom. 2006;20(21):3245–3256. doi: 10.1002/rcm.2691. [DOI] [PubMed] [Google Scholar]
- 13.Hennrich ML, Boersema PJ, van den Toorn H, Mischerikow N, Heck AJ, Mohammed S. Effect of chemical modifications on peptide fragmentation behavior upon electron transfer induced dissociation. Anal Chem. 2009;81(18):7814–7822. doi: 10.1021/ac901108g. [DOI] [PubMed] [Google Scholar]
- 14.Carabetta V, Mohanty BK, Kushner SR, Silhavy TJ. The response regulator SprE (RssB) modulates polyadenylation and mRNA stability in Escherichia coli. J Bacteriol. 2009 doi: 10.1128/JB.00870-09. doi:10.1128/JB.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carabetta VJ, Silhavy TJ, Cristea IM. The response regulator SprE (RssB) is required for maintaining PAP I-degradosome association during stationary phase. J Bacteriol. 2009 doi: 10.1128/JB.00300-10. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strupat K, Kovtoun V, Bui H, Viner R, Stafford G, Horning S. MALDI Produced Ions Inspected with a Linear Ion Trap-Orbitrap Hybrid Mass Analyzer. J Am Soc Mass Spectrom. 2009;20(8):1451–1463. [Google Scholar]
- 17.Beavis R, Fenyo D. Finding protein sequences using PROWL Chapter 13. Curr Protoc Bioinformatics. 2004;Unit 13 2 doi: 10.1002/0471250953.bi1302s7. [DOI] [PubMed] [Google Scholar]
- 18.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Hohmann L, Sherwood C, Eastham A, Peterson A, Eng JK, Eddes JS, Shteynberg D, Martin DB. Proteomic analyses using Grifola frondosa metalloendoprotease Lys-N. J Proteome Res. 2009;8(3):1415–1422. doi: 10.1021/pr800774h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 21.Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8(7):1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11(1):117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 23.Schwob E, Martin RP. New yeast actin-like gene required late in the cell cycle. Nature. 1992;355(6356):179–182. doi: 10.1038/355179a0. [DOI] [PubMed] [Google Scholar]
- 24.Boldogh IR, Yang HC, Nowakowski WD, Karmon SL, Hays LG, Yates JR, 3rd, Pon LA. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc Natl Acad Sci U S A. 2001;98(6):3162–3167. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goode BL, Rodal AA. Modular complexes that regulate actin assembly in budding yeast. Curr Opin Microbiol. 2001;4(6):703–712. doi: 10.1016/s1369-5274(01)00272-7. [DOI] [PubMed] [Google Scholar]
- 26.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 27.Taouatas N, Drugan MM, Heck AJ, Mohammed S. Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat Methods. 2008;5(5):405–407. doi: 10.1038/nmeth.1204. [DOI] [PubMed] [Google Scholar]
- 28.Yu W, Vath JE, Huberty MC, Martin SA. Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-Xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1993;65(21):3015–3023. doi: 10.1021/ac00069a014. [DOI] [PubMed] [Google Scholar]
- 29.Gu C, Tsaprailis G, Breci L, Wysocki VH. Selective gas-phase cleavage at the peptide bond C-terminal to aspartic acid in fixed-charge derivatives of Asp-containing peptides. Anal Chem. 2000;72(23):5804–5813. doi: 10.1021/ac000555c. [DOI] [PubMed] [Google Scholar]
- 30.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. J Mass Spectrom. 2000;35(12):1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O'Hair RA, Speed TP, Simpson RJ. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem. 2003;75(22):6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 32.Tabb DL, Huang Y, Wysocki VH, Yates JR., 3rd Influence of basic residue content on fragment ion peak intensities in low-energy collision-induced dissociation spectra of peptides. Anal Chem. 2004;76(5):1243–1248. doi: 10.1021/ac0351163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum E, Carpousis AJ, Higgins CF. Polyadenylation promotes degradation of 3'-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274(7):4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 34.Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184(17):4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. discussion 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34(5):1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 36.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34(19):5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92(6):1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raynal LC, Carpousis AJ. Poly(A) polymerase I of Escherichia coli:characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol Microbiol. 1999;32(4):765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 39.Carabetta VJ, Mohanty BK, Kushner SR, Silhavy TJ. The response regulator SprE (RssB) modulates polyadenylation and mRNA stability in Escherichia coli. J Bacteriol. 2009;191(22):6812–6821. doi: 10.1128/JB.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.