Abstract
Epidemiological studies of DNA methylation (DNAm) profiles may hold substantial promise for identifying mechanisms through which genetic and environmental factors jointly contribute to disease risk. Different cell types are likely to have different DNAm patterns. We investigate the DNAm differences between two types of biospecimens available in many genetic epidemiology studies. We compared DNAm patterns in two different DNA samples from each of 34 participants in the Genetic Epidemiology Network of Arteriopathy study (20 Caucasians and 14 African-Americans). One was extracted from peripheral blood cells (PBC) and the other from transformed B-lymphocytes (TBL). The genome-wide DNAm profiles were compared at over 27,000 genome-wide methylation sites. We found that 26 out of the 34 participants had correlation coefficients higher than 0.9 between methylation profiles of PBC and TBL. Although a high correlation was observed in the DNAm profile between PBC and TBL, we also observed variation across samples from different DNA resources and donors. Using principal component analysis of the DNAm profiles, the two sources of the DNA samples could be accurately predicted. We also identified 3,723 autosomal DNAm sites that had significantly different methylation statuses in PBC compared to TBL (Bonferroni corrected p value <0.05). Both PBC and TBL provide a rich resource for understanding the DNAm profiles in humans participating in epidemiologic studies. While the majority of DNAm findings in PBC and TBL may be consistent, caution must be used when interpreting results because of the possibility of cell type-specific methylation modification.
Introduction
Both environmental and genetic effects contribute to the development and progression of common human diseases. Understanding the interplay between environmental and genetic factors is critical for unraveling the complex etiology and pathology of many of these diseases. Several studies have shown that environmental modulation of genetic effects may act at the level of gene expression regulation through epigenetic mechanisms (Davis and Ross 2007; Herceg 2007). The modern definition of epigenetics is the study of heritable changes in gene expression levels that are unrelated to changes in the underlying DNA sequence (Bird 2007; Richards 2006). DNA methylation (DNAm), the covalent addition of a methyl (CH3-) group to the nucleotide cytosine number 5, is the best understood and most easily measured mechanism of epigenetic modification. DNAm sites located at CpG islands in the promoter region of a gene are known to play a key role in regulating gene expression level (Suzuki and Bird 2008). DNA methylation is mediated by DNA methyltransferases (DNMT) and in eukaryotes, at least three functional DNMTs have been identified (DNMT1, DNMT3A, and DNMT3B). DNMT1 is the primary enzyme involved in maintenance of methylation patterns during replication, while DNMT3A and DNMT3B have more de novo methylation activity and are active during embryogenesis.
During fetal development, tissue-specific DNAm profiles have been observed and are thought to be a central mechanism underlying cellular differentiation that manifests through differential gene expression (Ehrlich 2003; Meissner et al. 2008; Namihira et al. 2008). It is also well established that alterations in DNAm occur in cancer, including hypomethylation of oncogenes and hypermethylation of tumor suppressor genes (Christensen et al. 2009; Irizarry et al. 2009). Ladd-Acosta et al. (2007) also reported that DNAm profile of CpG sites can classify different human brain tissues and that these tissue-specific DNA methylation patterns affect gene transcription. There is also evidence for an age-related loss of normal epigenetic patterns, which may contribute to diseases with later onset (Bjornsson et al. 2008).
Epidemiological studies of DNAm profile may hold substantial promise for identifying mechanisms through which genetic and environmental factors jointly contribute to disease risk (Foley et al. 2009). However, the tissue-specificity of DNAm profiles raises key questions about what cell populations are appropriate for epidemiological studies of disease association. In most genetic epidemiology studies, DNA samples are extracted from fresh or frozen peripheral blood cells (PBC) obtained directly from a blood draw. In more established epidemiological cohorts, it is also not uncommon to have transformed B-lymphocytes (TBL) samples on individuals that have been developed to supply the epidemiological study with an ‘unlimited’ supply of DNA for future genetic studies. Because DNAm patterns are known to be tissue-specific and may be affected by the transformation process in TBL, it is important to understand the reproducibility and variability between the DNAm profiles from the two sources of DNA likely to be used in large epigenetic studies of common human diseases. To address these questions, we obtained two DNA samples from each of 34 participants in the Genetic Epidemiology Network of Arteriopathy (GENOA) study (20 Caucasians and 14 African-Americans), one extracted from PBC and the other extracted from TBL, to compare the genome-wide DNAm profiles between the two sources of DNA. In addition, we randomly selected four of these participants to have replicate measures of their DNAm profiles assayed on one of the two DNA sources.
Methods
Sample
The GENOA study is a community-based study of hypertensive sibships that aims to identify genes influencing blood pressure (Daniels et al. 2004; FBPP Investigators 2002). GENOA data were collected in two phases. Phase I (1996–1999) and Phase II (2000–2004) data consist of demographic information, medical history, clinical characteristics, lifestyle factors, and blood samples for genotyping and biomarker assays. The GENOA study was approved by the Institutional Review Boards of all participating institutions. Each participant gave written informed consent. For this study, participants’ stored samples were selected based on race (Caucasians and African-Americans), gender (male and female), and leukoariosis score. All participants were hypertensive at the time of collection. In addition, four individuals (one from each race–sex category) were randomly selected to assess the reproducibility of the Illumina chip's assessment of methylation status.
Sample preparation
The DNA from each selected participant's transformed B-lymphocytes and peripheral blood cell pellets were extracted by utilizing an automated platform (AutoGen FlexStar Qiagen Chemistries), including quantification by UV absorbance and quality control by 260/280 OD ratio. Samples were stored in Tris–EDTA buffer at concentration of 250 ng/ul. The genomic DNA samples were bisulfite converted, and then subjected to methylation profiling of 27 K CpG loci at the Mayo Clinic Genotyping Resource (GSR) facility. The EZ DNA Methylation Gold Kit (Zymo Research, Orange, CA, USA) was used for bisulfite conversion of all DNA samples (1 μg of genomic DNA per sample), according to the manufacturer's recommendations. An aliquot of the converted DNA (corresponding to 250 ng starting gDNA) was then used to assay up to 27,578 methylation sites simultaneously on an array. The DNA extracted for the four individuals chosen for the replication experiment was the same across chips (replicate 1 vs. replicate 2) in order to assess chip-to-chip variation rather than variation in sample preparation.
Genome-wide methylation assay
The Illumina Infinium HumanMethylation27 BeadChip (Illumina, San Diego, CA, USA) was used to measure 27,578 methylation sites. Bisulfite-converted DNA samples were whole-genome amplified, enzymatically fragmented, and purified. Samples then were hybridized in batches of 12 to the BeadChip, which contains locus-specific DNA oligomers. The allele specific extension reaction, washing, and staining were carried out in a TECAN Te-Flow Chamber. The arrays were fluorescently stained, scanned, and assessed for fluorescence intensities at the methylated and unmethylated bead sites using Illumina iScan system. The intensity data were then loaded into the GenomeStudio Methylation Module for analysis.
Data processing and methylation quantitation
Each methylation site is represented by the fluorescent signals from the methlyated and unmethylated alleles on the bead chip. The raw fluorescence data from the scanner were processed in BeadStudio software to yield β values, which are continuous variables ranging from 0 to 1. The beta value (β) is used to estimate the methylation level of the locus using the ratio of intensities between methylated and unmethylated alleles. For the Infinium Methylation Assay, β is calculated as:
where SignalA and SignalB are produced by two different bead types and reported in the same color.
Illumina chose the calculation above to provide an estimate of the intensity of the methylated signal (SignalB) as a percentage of the overall signal. As such, β values range from 0 to 1 and can be analyzed statistically using a Beta distribution. A constant bias of 100 was added to regularize β when both the methylated and unmethylated signals are small (Illumina 2009).
Statistical methods
We calculated the Pearson product–moment correlation coefficient between the β values (i.e., the DNAm profiles) of each cell type (PBC vs. TBL) for each individual in order to estimate the covariation in methylation states of each of the ~27,000 methylation sites measured. The correlation between the DNAm signals for each of the four replicates (replicate 1 vs. replicate 2) was also estimated using the Pearson product–moment correlation.
In order to identify the DNAm sites which differentiate the two DNA sources, PBC versus TBL, we first calculated the log2 ratio of the two β-values from a single participant at each DNAm site. Then we tested H0: βPBC = βTBL which is equivalent to log2(βPBC/βTBL) = 0 for each measured DNAm site across individuals using t test. A p value threshold of 1.8 × 10–6 (equivalent to 0.05 after correcting for multiple testing using Bonferroni correction) was used to identify log2β-ratios that significantly deviated from 0 (i.e., levels of methylation derived from the fluorescence measurement are different between the two DNA sources).
We used principal component analysis (PCA) to study the underlying variability of the full set of DNAm sites measured by the HumanMethylation27 BeadChip (27,578 sites) including X and Y chromosomes, as well as the autosomal DNAm sites only (26,486 sites). The raw beta-values were directly used for computing the principal components (PCs) without centering or rescaling the data. We first calculated the PCs of all DNAm sites and antosomal DNAm sites separately using the pooled sample of 34 pairs of DNAm measurements. We presented the patterns of the top five PCs among all 68 samples using a parallel coordinates plot implemented in R package “lattice”. We tested the association of the top five PCs of 26,486 autosomal DNAm sites with age, sex, race, and body mass index (BMI) using a linear mixed model for the continuous traits and a generalized estimating equations (GEE) model for the categorical traits to adjust for repeated measurements. We then estimated the PCs of the autosomal DNAm sites using the 34 DNA samples from each of the two cell types, PBC and TBL (separately) and tested the association of the top five PCs from each cell type with age, sex, race, and BMI using linear regression for the continuous traits and logistic regression for the categorical traits.
Based on chromosomal location (NCBI 36.1), we identified the genes that were significantly different between PBC and TBL in their DNAm status. We then used gProfile (Reimand et al. 2007) to estimate the most over-represented Gene Ontology terms associated with these genes.
All statistical analyses were performed with R statistical environment version 2.9.0 from R Project (http://www.r-project.org/). The authors had full access to the data and take responsibility for its integrity. All authors have read and agree to the manuscript as written.
Results
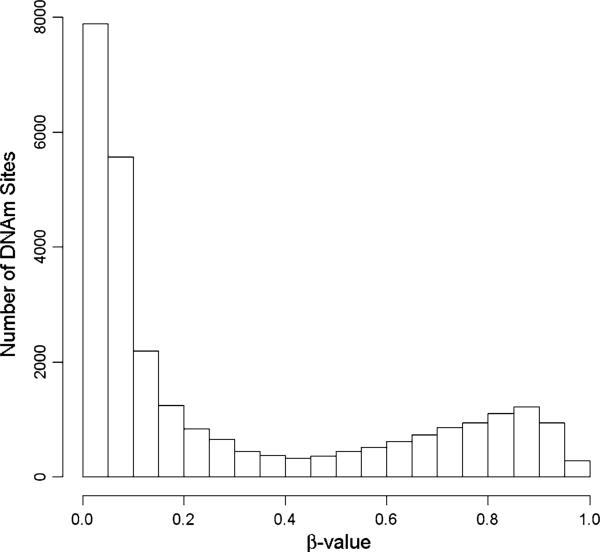
In Fig. 1, we present the histogram of β values—a continuous measurement of the methylation status of each CpG site—from a single Illumina HumanMethylation27 BeadChip randomly chosen from the 72 samples. The majority of the measured DNAm sites were unmethylated with the β value close to 0. The bimodal distribution also indicates a cluster of methylated DNAm sites with a β value close to 1 (Fig. 1). This bimodal distribution is consistent across all 72 chips measured in this study (data not shown).
Fig. 1.
Histogram of β-value distribution from a single sample
For each of the 34 pairs of DNAm profiles from the two cell types of the same individual, we calculated the correlation coefficient using all 27,578 measured DNAm sites. Table 1 is a summary of the correlation results by race and sex. The average correlation coefficients of male Caucasians, female Caucasians, male African-Americans and female African-Americans are 0.916, 0.934, 0.910, and 0.888, respectively (Table 1). There were 26 pairs of DNAm profiles (76.5%) with correlation coefficient larger than 0.9. The correlation coefficients of the four pairs of DNA replicates (one pair each in the four race–cell type combinations) ranged from 0.985 to 0.996.
Table 1.
Summary of correlation of DNAm status between peripheral blood cells and transformed B-lymphocytes
| Caucasians |
African-Americans |
|||
|---|---|---|---|---|
| Male (N = 10) | Female (N = 10) | Male (N = 7) | Female (N = 7) | |
| Mean of correlation | 0.916 | 0.934 | 0.910 | 0.888 |
| SD of correlation | 0.026 | 0.031 | 0.043 | 0.081 |
| Median of correlation | 0.919 | 0.948 | 0.921 | 0.920 |
| Max of correlation | 0.949 | 0.960 | 0.953 | 0.956 |
| Min of correlation | 0.865 | 0.861 | 0.823 | 0.721 |
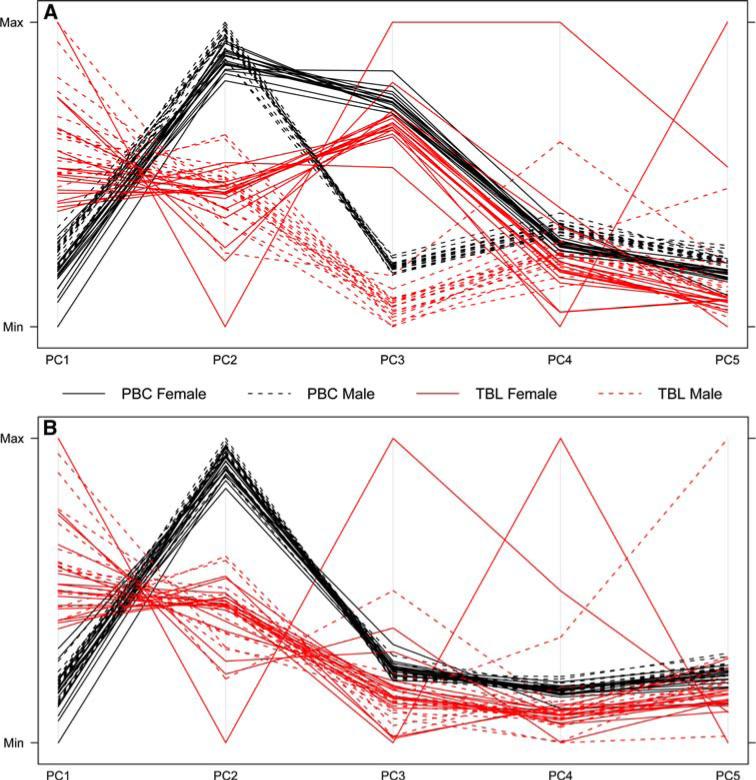
By comparing the top PCs from the 27,578 DNAm sites (including X and Y chromosomes) and 26,486 DNAm sites (autosomes only), we found that the top PCs divided the samples into distinct clusters (Fig. 2). The first two PCs of the DNAm profiles, PC1 and PC2, discretely separated the DNA samples into two clusters, one from PBC and the other from TBL, using all DNAm sites (Fig. 2a) or only the autosomal DNAm sites (Fig. 2b). When the DNAm sites located on chromosome X and Y were included in the full genomic set of DNAm sites, the samples from the males and females were separated by PC3 (Fig. 2a); however, the effect of sex on the DNAm profile was not observed when only the autosomal DNAm sites (Fig. 2b) were included. The first PC of the 26,486 autosomal DNAm sites explained 96.4% of the total variability, and the top five PCs explained 98.3% of the total variability altogether.
Fig. 2.
The clustering of the DNA samples by the top PCs using a all DNAm sites including chromosome X and Y, and b DNAm sites on autosomes (chromosomes 1–22)
In the regression analysis using the PCs of the autosomal DNAm sites, we did not identify any significant associations of the top five PCs with age or BMI (Table 2) in either the pooled samples (PBC + TBL) or the cell type specific samples. We did not observe any significant associations of top five PCs with sex or race except the association of PC2 with race in PBC (Table 2). The associations of PC2 and PC5 with sex in PBC were also significant at an alpha level of 0.05 (Table 2); however, they were not statistically significant after correcting for multiple testing.
Table 2.
Summary of the association tests of top five PCs from 26,486 autosomal DNAm sites with age, BMI, sex, and race
|
p value |
|||||||
|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | |||
| Age | 65.2 ± 2.7 | PBC + TBL | 0.272 | 0.064 | 0.417 | 0.380 | 0.818 |
| PBC | 0.810 | 0.317 | 0.678 | 0.974 | 0.538 | ||
| TBL | 0.454 | 0.706 | 0.840 | 0.614 | 0.837 | ||
| BMI | 32.7 ± 5.4 | PBC + TBL | 0.906 | 0.316 | 0.722 | 0.436 | 0.815 |
| PBC | 0.296 | 0.892 | 0.902 | 0.944 | 0.560 | ||
| TBL | 0.963 | 0.696 | 0.856 | 0.128 | 0.627 | ||
| Sex | 50% | PBC + TBL | 0.263 | 0.143 | 0.278 | 0.300 | 0.062 |
| PBC | 0.476 | 0.010* | 0.873 | 0.843 | 0.028* | ||
| TBL | 0.265 | 0.457 | 0.272 | 0.380 | 0.967 | ||
| Race | 41% | PBC + TBL | 0.909 | 0.115 | 0.121 | 0.427 | 0.211 |
| PBC | 0.780 | 0.006** | 0.559 | 0.560 | 0.172 | ||
| TBL | 0.997 | 0.360 | 0.378 | 0.693 | 0.154 | ||
p value less than 0.05
p value less than 0.01
Using the log2β-ratio test described in the “Methods” section, we identified 3,723 (14.1%) autosomal DNAm sites which had significantly different methylation status in PBC and TBL (Supplementary Table 1). Among these cell-specific DNAm sites, 81.7% exhibited a lower level of methylation in TBL than in PBC. There are ~3,000 genes associated with the 3,723 cell-specific DNAm sites. By searching the Gene Ontology (GO) database through g:Profiler (Reimand et al. 2007), which uses cumulative hypergeometric p values to identify the most enriched terms corresponding to the input set of genes, we found the most over-represented GO terms (p < 10–20) among these genes are GO:0005576 (extracellular region), p value of 1.78 × 10–54; GO:0005886 (plasma membrane), p value of 1.97 × 10–30; GO:0009605 (response to external stimulus), p value of 7.44 × 10–29; GO:0044459 (plasma membrane part), p value of 1.31 × 10–28; GO:0006952 (defense response), p value of 4.30 × 10–27; GO:0050896 (response to stimulus), p value of 1.12 × 10–24; GO:0002376 (immune system process), p value of 3.68 × 10–24; GO:0005615 (extracellular space), p value of 9.57 × 10–24; GO:0005887 (integral to plasma membrane), p value of 1.81 × 10–23; GO:0031226 (intrinsic to plasma membrane), p value of 3.80 × 10–23; GO:0009611 (response to wounding), p value of 8.43 × 10–23; and GO:0006955 (immune response), p value of 1.03 × 10–21.
Discussion
Environmental agents may modify gene expression independently of the primary DNA sequence through epigenetic modifications, which are mitotically heritable chemical/structural changes that regulate gene activity in the absence of underlying changes to DNA sequence. These modifications are the likely mediators of gene–environment interaction, and aberrant epigenetic modification can have downstream genetic consequences. The primary DNA sequence is generally fixed at conception, but epigenetic marks are dynamic and modifiable, probably throughout the entire lifespan.
Blood draws are the primary sources of genetic information for epidemiologic studies because they are inexpensive and non-invasive. DNA from blood draws is obtained directly from leukocytes present in the blood (PBC) or after transformation into immortalized cell lines of B-lymphocytes (TBL). The cellular composition of peripheral blood leukocytes has been shown to exhibit significant inter-individual variation, but is composed primarily of neutrophils (mean = 57% in a sample of elderly men), lymphocytes (mean = 30%), and monocytes (mean = 9%) (Moverare-Skrtic et al. 2009). DNAm profiles have been shown to differ among these cell types (Moverare-Skrtic et al. 2009) which may account for the detection of sites that are not fully methylated or fully unmethylated in PBC (sites with β-value significantly different from 0 or 1). Cell types such as neutrophils and B-lymphoctyes undergo relatively few mitotic divisions and have been shown to exhibit less methylation at CpG rich regions (tag sites) that begin unmethylated at infancy than T-lymphocytes which replicate frequently throughout life (Chu et al. 2008). The same phenomenon of increased methylation at tag sites has been observed in tissues that undergo frequent mitotic division (for example, epithelial tissues) compared to tissues that undergo infrequent mitotic division (for example, brain and heart tissues) (Chu et al. 2007). Due to the differences in DNAm profiles among the different cell types in PBC, adjustment for individual cell-type composition is necessary when analyzing DNAm profiles from PBC in epidemiologic studies (Moverare-Skrtic et al. 2009).
TBL is a convenient source of DNA for epidemiologic studies because they are immortalized cell lines that provide an essentially endless source of genetic material. Although the primary DNA sequence of these cells remains unchanged during the immortalization process and through subsequent cellular division, the epigenetic changes to immortalized cells are not fully understood. Changes in DNAm at key cell cycle regulating genes are known to be a necessary process in the immortalization of cancerous cells (Fridman and Tainsky 2008). In addition, there is emerging evidence that changes in the methylation of promoter regions of genes that mediate the cell cycle are an important regulator of cell division (Metivier et al. 2008).
Recent study by Brennan et al. (2009) compared the DNAm profile of the promoter regions of 320 genes in peripheral blood leukocytes versus TBL from six Caucasians. They identified ~8% of the promoter regions that revealed different DNAm profile in the DNA samples from two cell types. In our DNAm study of 14,475 human genes using a larger sample size (34 pairs vs. 6 pairs), a larger proportion (14.1%) of the DNAm sites revealed statistically different methylation status in DNA samples from PBC and TBL. Our DNAm investigation provides a more comprehensive coverage of DNAm on human genome and a finer map of DNAm sites with different methylation status in the two resources of DNA.
PBC and TBL are both excellent resources for DNAm profiles associated with chronic diseases that involve inflammation and immunity, as leukocytes play a key role in the inflammatory process and immune response. Epigenetic profiles have been shown to influence the expression of genes involved in inflammation and immunity, and epigenetic dysregulation has been proposed as a mechanism leading to an increase in inflammatory/immune diseases with age (Dong et al. 2002; Wilson 2008). Lower levels of DNAm at the promoter regions of several inflammatory cytokines have been shown to be associated with decreased gene expression in leukocytes, including interferon γ (IFN-γ) and interleukin 3 (IL-3) in mouse CD8+ T-cells (Fitzpatrick et al. 1998) interleukin 2 (IL-2) in mouse CD4+ T-cells (Bruniquel and Schwartz 2003), and tumor necrosis factor (TNF) in human monocytes (Kochanek et al. 1991; Wilson 2008). Changes in methylation patterns at promoter regions in T-cells in response to inflammatory cytokines have also been shown to immediately follow T-cell activation (Bruniquel and Schwartz 2003; Northrop et al. 2006; Pearce and Shen 2006), and these changes are stably transmitted to activated T-cell progeny (Fitzpatrick et al. 1998).
It is notable that the GO categories that were over-represented among the significant DNAm differences detected between PBC and TBL are those involved in the extracellular region, plasma membrane, response to stimulus/wounding/defense, and immune system process/response. These types of differences are expected between PBC and TBL because they are different types of leukocyte cell populations, and thus differentially express genes related to immune response. DNA methylation is known to be involved in regulating the differentiation of immune cells into specific cell types such as neutrophils, lymphocytes, and monocytes and maintaining the differentiated state of the cells (Fitzpatrick and Wilson 2003). Some key differences between differentiated types of immune cells are the proteins expressed on the plasma membrane and in the extracellular matrix, specifically proteins responsible for cell migration and binding such as integrins and selectins, as well as their response to environmental stimulus, such as the release of cytokines (Cotran et al. 1999).
The method of GEE (Liang and Zeger 1986) has been developed to analyze repeated measures. Use of empirical covariance estimators in a GEE analysis provides consistent estimation of regression parameters and standard errors of these estimates even when the correlation model is mis-specified. However, use of the empirical estimator tends to underestimate the true variance in small samples and results in inflated type I error rates (Kauermann and Carroll 2001; O'Brien and Fitzmaurice 2004). Lu et al. (2007) reported that no bias adjustment is required for binary outcomes when the size of the cluster is larger than 40. Although GEE does tend to have liberal estimates (i.e., smaller standard errors and lower p values) for sample sizes less than 40, the size of the cluster in this study is 34 (only slightly smaller than 40). Importantly, we did not observe any statistically significant associations between the top PCs and the binary outcomes (i.e., sex and race). Therefore, the potential bias caused by the small sample size is unlikely to change the statistical significance of the PC association with sex and race.
Results from this study indicate that the overall DNAm profiles of PBC and TBL are similar. The correlation between the overall methylation between the two cell types was generally high (most samples exhibited a correlation greater than 0.9), and no significant difference in DNAm status was observed at 86.5% of the assayed sites. However, the 3,723 autosomal sites that differed between the two cell types indicate that there are important differences in the DNAm profiles that cannot be ignored in the data analysis phase. This study provides a resource for beginning to understand the way in which methylation patterns may be affected during the transformation process and may be a starting point for epidemiologic researchers that wish to extrapolate findings from one cell type to another or investigate the source of cell type-specific differences. Overall, these results indicate that both PBC and TBL provide a rich resource for assaying the DNAm profiles of leukocytes in epidemiologic studies. However, our findings of differential methylation of 3,000 genes suggest that comparison of DNAm results across tissues and cell types requires serious consideration and careful interpretation.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health grant NS041558 and HL100185.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-010-0810-y) contains supplementary material, which is available to authorized users.
Conflict of interest statement The authors declare that they have no competing interests.
Contributor Information
Yan V. Sun, Department of Epidemiology, School of Public Health, University of Michigan, 1415 Washington Heights #4629, Ann Arbor, MI 48109, USA
Stephen T. Turner, Division of Nephrology and Hypertension, Mayo Clinic, Rochester, MN, USA
Jennifer A. Smith, Department of Epidemiology, School of Public Health, University of Michigan, 1415 Washington Heights #4629, Ann Arbor, MI 48109, USA
Pamela I. Hammond, Division of Nephrology and Hypertension, Mayo Clinic, Rochester, MN, USA
Alicia Lazarus, Department of Epidemiology, School of Public Health, University of Michigan, 1415 Washington Heights #4629, Ann Arbor, MI 48109, USA.
Jodie L. Van De Rostyne, Division of Nephrology and Hypertension, Mayo Clinic, Rochester, MN, USA
Julie M. Cunningham, Division of Experimental Pathology, Mayo Clinic, Rochester, MN, USA
Sharon L. R. Kardia, Department of Epidemiology, School of Public Health, University of Michigan, 1415 Washington Heights #4629, Ann Arbor, MI 48109, USA
References
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan EP, Ehrich M, Brazil DP, Crean JK, Murphy M, Sadlier DM, Martin F, Godson C, McKnight AJ, van den Boom D, Maxwell AP, Savage DA. Comparative analysis of DNA methylation profiles in peripheral blood leukocytes versus lymphoblastoid cell lines. Epigenetics. 2009;4:159–164. doi: 10.4161/epi.4.3.8793. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, Karagas MR, Wrensch MR, Yeh RF, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Bueno R, Sugarbaker DJ, Kelsey KT. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res. 2009;69:227–234. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, Kanel GC, Tavare S, Shibata D. Lack of increases in methylation at three CpG-rich genomic loci in non-mitotic adult tissues during aging. BMC Med Genet. 2007;8:50. doi: 10.1186/1471-2350-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M, Siegmund KD, Hao QL, Crooks GM, Tavare S, Shibata D. Inferring relative numbers of human leucocyte genome replications. Br J Haematol. 2008;141:862–871. doi: 10.1111/j.1365-2141.2008.07142.x. [DOI] [PubMed] [Google Scholar]
- Cotran RS, Kumar V, Collins T, Robbins SL. Pathologic basis of disease. WB Saunders; Philadelphia: 1999. [Google Scholar]
- Daniels PR, Kardia SL, Hanis CL, Brown CA, Hutchinson R, Boerwinkle E, Turner ST. Genetic Epidemiology Network of Arteriopathy study (2004) Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) Study. Am J Med. 116:676–681. doi: 10.1016/j.amjmed.2003.12.032. [DOI] [PubMed] [Google Scholar]
- Davis CD, Ross SA. Dietary components impact histone modifications and cancer risk. Nutr Rev. 2007;65:88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Dong C, Yoon W, Goldschmidt-Clermont PJ. DNA methylation and atherosclerosis. J Nutr. 2002;132:2406S–2409S. doi: 10.1093/jn/132.8.2406S. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- FBPP Investigators Multi-center genetic study of hypertension: the family blood pressure program (FBPP). Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol. 2003;109:37–45. doi: 10.1016/s1521-6616(03)00205-5. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DR, Shirley KM, McDonald LE, Bielefeldt-Ohmann H, Kay GF, Kelso A. Distinct methylation of the interferon gamma (IFN-Gamma) and Interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-Gamma promoter demethylation and mRNA expression are heritable in CD44(High)CD8+ T cells. J Exp Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, Saffery R. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27:5975–5987. doi: 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- Illumina Illumina GenomeStudio methylation module v1.0 user guide, Part # 11319130. 2009.
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauermann G, Carroll RJ. A note on the efficiency of sandwich covariance matrix estimation. J Am Stat Assoc. 2001;96:1387–1396. [Google Scholar]
- Kochanek S, Radbruch A, Tesch H, Renz D, Doerfler W. DNA methylation profiles in the human genes for tumor necrosis factors alpha and beta in subpopulations of leukocytes and in leukemias. Proc Natl Acad Sci USA. 1991;88:5759–5763. doi: 10.1073/pnas.88.13.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lu B, Preisser JS, Qaqish BF, Suchindran C, Bangdiwala SI, Wolfson M. A comparison of two bias-corrected covariance estimators for generalized estimating equations. Biometrics. 2007;63:935–941. doi: 10.1111/j.1541-0420.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Moverare-Skrtic S, Mellstrom D, Vandenput L, Ehrich M, Ohlsson C. Peripheral blood leukocyte distribution and body mass index are associated with the methylation pattern of the androgen receptor promoter. Endocrine. 2009;35:204–210. doi: 10.1007/s12020-009-9153-7. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363:2099–2109. doi: 10.1098/rstb.2008.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-Gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177:1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- O'Brien LM, Fitzmaurice GM. Analysis of longitudinal multiple-source binary data using generalized estimating equations. J R Stat Soc Ser C Appl Stat. 2004;53:177–193. [Google Scholar]
- Pearce EL, Shen H. Making sense of inflammation, epigenetics, and memory CD8+ T-cell differentiation in the context of infection. Immunol Rev. 2006;211:197–202. doi: 10.1111/j.0105-2896.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.