Abstract
A hybrid linear ion-trap Fourier-transform ion cyclotron resonance mass spectrometer was used for top-down characterization of the abundant human salivary Cystatins, including S, S1, S2, SA, SN, C, and D, using collisionally activated dissociation (CAD) after chromatographic purification of the native, disulfide intact proteins from saliva. Post-translational modifications and protein sequence polymorphisms arising from single nucleotide polymorphisms (SNPs) were assigned from precursor and product ion masses at a tolerance of 10 ppm allowing confident identification of individual intact mass tags. Cystatins S, S1, S2, SA and SN were cleaved of a N-terminal 20 amino-acid signal peptide, and Cystatin C a 26-residue peptide, to yield a generally conserved N-terminus. In contrast, Cystatin D isoforms with 24 and 28 amino-acid residue N-terminal truncations were found such that their N-termini were not conserved. Cystatin S1 was phosphorylated at Ser3, while S2 was phosphorylated at Ser1 and Ser3 of the mature protein, in agreement with previous work. Both Cystatin D isoforms carried the polymorphism C46R (SNP: rs1799841). The 14328 Da isoform of Cystatin SN previously assigned with polymorphism P31L due to a SNP (rs2070856) was found only in whole saliva. Parotid secretions contained no detectable Cystatins while whole saliva largely mirrored the contents of submandibular/sublingual (SMSL) secretions. Top-down high-resolution mass spectrometry is a powerful tool for the identification and characterization of potential protein biomarkers in saliva.
Introduction
High-resolution mass spectrometry (MS) of intact proteins led to the inception of the ‘top-down’ approach ten years ago [1]. Intact proteins are introduced into the gas phase by electrospray ionization (ESI) for high-resolution mass measurements of intact protein precursor ions prior to direct dissociation for product ion mass measurements. The top-down precursor and product ion datasets are then reconciled with the primary structure of the protein, including all modifications that affect mass. The advent of top-down MS was proceeded by cornerstone developments in the field of mass spectrometry including the application of high-resolution Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) to larger biomolecules [2], the ability to ionize intact proteins and dissociate them in the mass spectrometer [3], and the ability to identify proteins from such experiments using sequence tags [4], as well as the availability of genomic sequence data. However, limited availability of high-resolution instrumentation and relatively low throughput issues compared with bottom-up shotgun approaches have limited the contribution that top-down MS has made to the field of proteomics. The biomarker field has embraced intact protein screening for several years now though progress in this arena has been hampered by the use of low-resolution instrumentation and the difficulties of subsequent unequivocal identification of promising candidates. High-resolution tandem mass spectrometry offers the ability to overcome these issues since intact mass tags from candidate biomarker can be directly analyzed by the top-down strategy. We show that simple top-down experiments can reliably link a protein identification to a low-resolution (or high-resolution) intact mass tag.
Saliva is produced in and secreted from three major pairs of salivary glands (submandibular, sublingual and parotid), and is composed of water and other compounds including electrolytes, mucus, antibacterial molecules and a variety of proteins [5]. Saliva has a variety of protective functions including lubrication, antimicrobial activity, mucosal integrity, lavage/cleansing, buffering and remineralization, as well as other oral functions such as digestion, taste and speech [6]. There is considerable interest in saliva because of its easy accessibility compared to nearly all other body fluids and the possibility that it might yield useful biomarkers or biosignatures of health and disease. The Cystatins are a family of cysteine protease inhibitors found in human saliva with highest concentrations found in submandibular secretions and little present in parotid [7, 8]. Included within this family are Cystatins S, S1, S2, SA, SN, C, and D. These proteins typically have between 120 - 121 amino acids after signal sequence cleavage, molecular weights in the 13 - 14kDa range and two conserved disulfide bonds [9, 10]. The Cystatins are known to inhibit cysteine proteases of both host and microbial origin, thereby preventing harmful proteolysis of the tissues of the oral cavity that could otherwise facilitate microbial infection [11]. A growing body of literature reports Cystatins as putative biomarkers of human disease and expression levels of various members of the family found not only in saliva but also in blood, cerebrospinal fluid and urine, have been correlated with a range of conditions. Cystatin C is most widely discussed, with links to many different medical conditions including cancer [12], diabetes and kidney disease [13], heart disease [14], neurotrauma [15] and neurodegeneration [16]. Cystatin SN has shown some promise as a urinary biomarker for colorectal cancer [17] and Cystatin SA as a saliva marker for oral cancer [18]. Interestingly, studies where the intact proteins were measured have suggested that it was an abnormally truncated form of the protein that was the marker; loss of an extra three amino acids from the N-terminus of Cystatin SA-1 (SN) [18] or loss of an extra eight amino acids from the N-terminus of Cystatin S [19].
Bearing in mind these observations, we report a high-resolution mass spectrometry study of the native forms of human salivary Cystatins. Collisionally activated dissociation (CAD) was used to identify and fully describe selected intact mass tags corresponding to the major detectable forms of Cystatins isolated from human saliva. Interpretation of the data required consideration of post-translational modifications including N-terminal signal peptide removal, disulfide formation and phosphorylation, as well as protein sequence polymorphisms arising from single nucleotide polymorphisms (SNPs). The ability to directly identify potential biomarkers should empower intact protein biomarker screening.
Experimental
Chemicals
All chemicals were purchased from Fisher Scientific.
Sample collection
Adult saliva donors of various ethnic and racial backgrounds, ranging in age from 22 to 30 years, were recruited from the general population, and samples were collected at the UCLA Medical Center with full donor consent using procedures in accord with the Medical Institution Review Board and the Office of Protection for Research Subjects, as previously described [20]. Whole saliva (WS) was collected in an unstimulated fashion, while parotid (P), submandibular (SM), and sublingual (SL) secretions were collected after application of an aqueous citric acid solution (2 %), as previously described [20]. Collected samples were centrifuged (10,000 × g, 15 minutes, 4°C), the supernatant was then treated with protease/phosphatase inhibitors (aprotinin, 1 μL/mL saliva, 10 mg/mL; sodium orthovanadate, 3 μL/mL saliva, 400 mM; phenylmethyl sulfonyl fluoride, 10 μL/mL saliva, 10 mg/mL) added promptly while the sample was on ice. Samples were then aliquoted and stored at −80°C prior to MS analyses.
Reverse-phase chromatography with online electrospray-ionization mass spectrometry and fraction collection (LC-MS+). Pooled samples (1 mL) were dried by centrifugal evaporation, re-dissolved in 400 mL 6 M guanidine-HCl and centrifuged (10,000 × g, 5 min, room temperature). Aliquots (4 × 100mL) of the resulting supernatants were then injected onto a reverse-phase HPLC column (PLRP/S 5μm, 300 Å, 2.1 mm × 150 mm, Varian Inc.) equilibrated in water/acetonitrile/TFA (95/5/0.1, vol/ vol) and eluted (100 μL/minute, 40°C) with an increasing concentration of acetonitrile (min/% acetonitrile; 0/5, 5/5, 10/20, 70/50, 90/90). The eluent was passed through a UV detector (280nm) prior to a flow splitter with fused silica capillaries to transfer liquid to the low-resolution ESI source (50cm) and the fraction collector (25 cm). Fractions, collected into microcentrifuge tubes at 1 min intervals, were stored at −80 °C prior to off-line high-resolution nanospray analysis.
2.4 Low resolution electrospray ionization mass spectrometry
LC–MS+ experiments were performed using a triple quadrupole instrument (API III+, Applied Biosystems) tuned and calibrated using a PEG mixture as described previously [21]. Spectra were recorded by scanning from m/z 600 - 2300 with the orifice voltage ramped with mass (60 - 120) using a 0.3 Da step size and a scan speed of 6 sec. Data were processed using MacSpec 3.3, or BioMultiview 1.3.1 software (Applied Biosystems).
2.5 High-resolution electrospray ionization mass spectrometry
Top-down mass spectrometry was performed on a hybrid linear ion-trap 7 T FTICR mass spectrometer (LTQ-FT Ultra, Thermo Fisher Corporation, San Jose, USA) fitted with an off-line nanospray source. HPLC fractions were individually loaded into 2 μm i.d. externally coated nanospray emitters (Proxeon, Cambridge, MA, USA) and desorbed using a spray voltage of 1.8 kV (versus the inlet of the mass spectrometer). These conditions produced a flow rate of 20 – 50 nL/min. Ion transmission into the linear trap and further to the ICR cell was automatically optimized for maximum ion signal. The ion count targets for the full scan and MS2 experiments were 2 × 106. The m/z resolving power of the instrument was set at 100,000 (defined by m/Δm 50% at m/z 400). Individual charge states of the multiply protonated molecular ions were selected for isolation and collisional activation in the linear ion trap followed by the detection of the resulting fragments in the ICR cell. Helium is used as collision gas in the LTQ mass spectrometer, which was operated in the standard mass range of m/z 300 – 2000. Precursor ions were isolated with widths of m/z 4 - 8 in order to maximize homogeneity of the ion while maintaining maximal signal strength. Precursors were activated using collision energy settings between 12 and 15 at the default activation q-value of 0.25 [22].
2.6 Data analysis with Prosight PC
All top-down FT-ICR spectra were obtained by averaging between 50 and 200 transient signals. Precursor masses were calculated using Xtract Version 3.0.1.1 (Thermo Scientific, Bremen, Gremany) with S/N threshold of 2, minimum intensity of 2, minimum fit of 30 and a remainder threshold of 3. Product ion spectra were processed using Prosight PC (version 2; Thermo Scientific, Bremen, Germany) to produce monoisotopic mass lists (S/N = 2; minimum RL value 0.9). Where identity was not known sequence tags were compiled for sequence tag searching to generate candidates for further manual fitting. The absolute mass search mode was used for refinement of primary structure to maximize agreement of precursor and product ions matched. Mass tolerance was set at 10 ppm and the deltamass feature was deactivated. All protein sequence databases were taken from SwissProt entries.
Results and Discussion
Chromatographic Separation of Proteins from Whole (WS), Submandibular/Sub-lingual (SMSL), and Parotid (PR) saliva
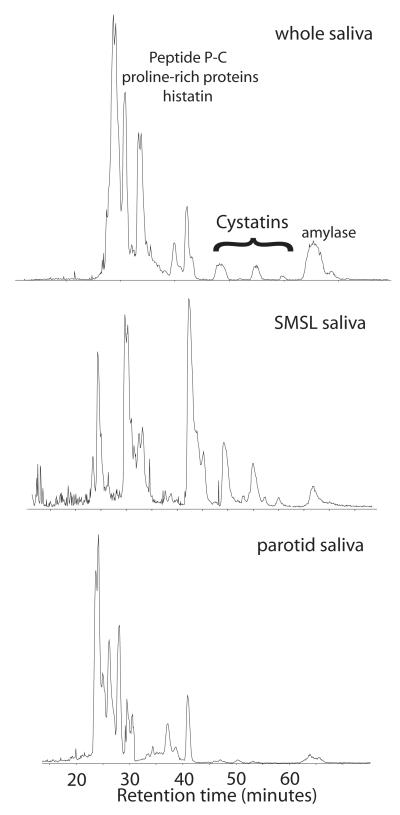
Pooled human saliva samples were dried and dissolved in 6 M guanidine prior to immediate reversed-phase chromatography with online ESI mass spectrometry and concomitant fraction collection LC-MS+ [23]. The results of the low-resolution mass analysis were used to select fractions and protein-specific ions for further top-down high-resolution MS analysis. The three saliva samples collected from the 3-primary glandular sources produced moderately complex, partially super-imposable total ion chromatograms (Fig. 1). While the profiles were similar, it was readily apparent that the Cystatin family of proteins, that eluted in the range 45 - 60 minutes, were poorly represented in PR secretions compared to SMSL and WS, in agreement with previous studies [24, 25]. Top-down analyses were then performed on stored fractions from the original LC-MS+ experiment in order to characterize this family of proteins as thoroughly as possible, especially with respect to post-translational modifications and variants due to single-nucleotide polymorphisms (SNPs) that were previously detected in a preliminary high-resolution analysis of salivary proteins [26]. In Table 1, the average mass determined in the original low-resolution LC-MS+ is included to allow facile comparison with other mass spectral data on Cystatins in the literature. Average mass is used for traditional ‘intact mass tags’ [27] whereas monoisotopic mass is used for high-resolution intact mass tags.
Figure 1. Reversed-phase chromatography of human salivary proteins.
Whole saliva, as well as parotid and sub-mandibular/sub-lingual ductal secretions were dried and dissolved in 6M guanidine prior to separation by HPLC and online ESI mass spectrometry with fraction collection (LC-MS+; see methods). Typical total ion chromatograms are shown highlighting the region where the Cystatin family elutes. Intact Cystatins were found in only whole saliva and SMSL secretions.
Table 1.
Human salivary Cystatins
| Cystatin | Swiss- Prot |
# amino acids |
Modification | Measured average mass (Da) |
Calculated monoisotopic mass after modification (Da) |
Experimental monoisotopic mass (FTMS) (Da) |
Delta (Da) |
Delta RMSa | (ppm)(ppm) |
|---|---|---|---|---|---|---|---|---|---|
| S | P01036 | 121 | 1-20 removed, 2 disulfide bonds |
14186 | 14175.8005 | 14175.8569 | 0.0564 | 3.98 | 3.69 |
| S1 | P01036 | 121 | 1-20 removed, 2 disulfide bonds, phosphorylation |
14266 | 14255.7668 | 14255.8567 | 0.0899 | 6.30 | 4.26 |
| S2 | P01036 | 121 | 1-20 removed, 2 disulfide bonds, 2 phosphorylation |
14346 | 14335.7335 | 14335.8110 | 0.0775 | 5.40 | 4.75 |
| SA | P09228 | 121 | 1-20 removed, 2 disulfide bonds |
14347 | 14337.0014 | 14336.9856 | 0.0158 | 1.10 | 6.55 |
| SN | P01037 | 121 | 1-20 removed, 2 disulfide bonds |
14313 | 14303.2228 | 14303.1553 | 0.0675 | 4.72 | 5.00 |
| SN (SNP) |
P01037 | 121 | 1-20 removed, 2 disulfide bonds, rs2070856 (P31L) |
14328 | 14319.1187 | 14319.171b | 0.0523 | 3.65 b | 7.07 b |
| C | P01034 | 120 | 1-26 removed, 2 disulfide bonds |
13345 | 13334.5969 | 13334.5829 | 0.0140 | 1.05 | 3.90 |
| D (SNP) | P28325 | 114 | 1-28 removed, 2 disulfide bonds, rs1799841 (C46R) |
13165 | 13154.4776 | 13154.4675 | 0.0101 | 0.77 | 2.47 |
| D (SNP) | P28325 | 118 | 1-24 removed, 2 disulfide bonds, rs1799841 (C46R) |
NDc | 13596.7064 | 13596.7015 | 0.0049 | 0.36 | 0.35 |
Root mean square of error on product ion assignments (within 10 ppm tolerance).
Values taken from reference 26.
Detected by FT-ICR-MS only
Protein Identification
Attempts to identify Cystatins in ProsightPC’s ‘absolute mass’ mode typically fail when full genomic translations are used in the database. This is because N-terminal residues need to be removed before b-ions are matched and two disulfides oxidized before y-ions are matched. Fortunately, the N-terminal half of the Cystatins frequently yields good b- and y-ion series resulting in sequence tags that can be used to identify them using this functionality within ProsightPC. Custom annotated databases are then created with corrected N-termini, post-translational modifications and protein sequence polymorphisms.
Cystatin S, S1, and S2 The Cystatins have an N-terminal signal peptide that is cleaved during maturation of the protein. In the case of Cystatin S (P01036), the first twenty amino acids are cleaved to give a 121 amino acid protein with a calculated monoisotopic mass of 14179.8005 Da (Table 1). The experimentally determined monoisotopic mass of 14175.8569 Da for the HPLC peak eluting at 48 min differs from this calculated mass by −3.9431 Da consistent with a mass loss associated with the formation of a pair of disulfide bonds (−4.0313 Da). Oxidation of the four Cys residues in Cystatin S to form two disulfide bonds between Cys94-Cys104 and Cys118-Cys138 in conservation with all the salivary Cystatins brings coincidence of measured and calculated masses to better than 4 ppm (Δ = 0.0564 Da, 3.98 ppm). Analysis of the CAD dataset for the disulfide-oxidized N-truncated protein using ProSight PC 2.0 (tolerance of 10 ppm; deltamass mode off) yielded 10 matching b- ions. After manual modification of the four Cys residues to account for the formation of two disulfide bonds (−1.0078 each residue) the software analysis yielded 11 b- and 27 y- product ions, agreement of calculated and measured mass within 10 ppm and a P-Score of 1.71E-57 (Figure 2, Table 1).
Figure 2. Top-down mass spectrometry of Cystatins S, S1, and S2.
Appropriate fractions were analyzed by static nanospray on a 7 T LTQFT Ultra. CAD experiments on isolated precursor ions yielded complex tandem mass spectra that were interpreted using Prosight PC software (see methods). The precursor ion isolation used for each experiment is shown on the left (m/z) while the b- and y- product ion assignments observed at 10 ppm tolerance (deltamass feature deactivated) are shown superimposed on the sequence to the right. A twenty amino-acid signal peptide has been removed from the precursor sequence and each Cys residue has been oxidized (−1.0078 Da) to reflect the intact pair of disulfide bonds of the native protein. For Cystatin S 41% of product ions were matched to b- and y- ions giving a Pscore of P-Score of 1.71E-57. For Cystatin S1 a single phosphorylation (+79.9663 Da) was added at position 3 of the mature protein with 31% of product ions matched and a Pscore of 4.75E-42. For Cystatin S2 a second phosphorylation was added at position 1 of the mature protein with 26% of product ions matched and a Pscore of 8.06E-32.
Cystatin S is known to be monophosphorylated at Ser23 (position 3 of the mature form) to produce a post-translationally modified form, Cystatin S1 [28-30]. The experimentally determined monoisotopic mass of 14255.8567 Da for the HPLC peak eluting at 48 minutes is within 10 ppm of the calculated mass of the mature Cystatin S1 protein, including two disulfide bonds and a single phosphorylation (14255.7668 Da; Δ = 0.0899 Da, 6.30 ppm). An absolute mass search with ProSightPC 2.0 yielded only one artifactual matching y-fragment ion until oxidation of the four cysteines (2 disulfide bonds; −4.03130036 Da) and phosphorylation of Ser23 (+79.9663 Da) were introduced yielding 14 b- and 28 y- product ions matched, and a P-Score of 4.75E-42 (Figure 2, Table 1). The detection of the y119 product ion provides definitive evidence that the singly phosphorylated form is modified at Ser23, although this may not be exclusive; some modification at Ser21 or Ser22 cannot be ruled out based on the observed coverage.
Cystatin S is also known to be diphosphorylated at Ser21 and at Ser23 [28] to produce another post-translationally modified form, Cystatin S2. The experimentally determined monoisotopic mass of 14335.8110 Da for the HPLC peak eluting at 48 min is in good agreement with the calculated mass for the mature protein, including two disulfide bonds and two phosphorylations (14335.7335 Da; Δ = 0.0775 Da, 5.40 ppm). The CAD product ion peaklist could only be matched to the primary structure of Cystatin S after manual modification of the four cysteine residues (2 disulfides; −4.0313 Da) and phosphorylation of Ser21 and Ser23 (+159.9326 Da), protein sequence coverage increased significantly with 5 b- and 19 y-fragment ions matched to the CAD data set and a P-Score of 8.06E-32 (Fig. 2, Table 1). Some modification of Ser22 cannot be ruled out based on the observed coverage
All three experiments on the Cystatin S family yielded b- and y- fragments from the region between Cys104 and Cys118 supporting disulfide crosslinking of Cys94 to Cys104 and Cys118 to Cys138 rather than some other arrangement. Based upon this observation no further analysis of disulfide bonding was deemed necessary. Annotation of human salivary Cystatin disulfide crosslinking has generally been achieved by similarity to the original work on egg-white Cystatin and human Cystatin C [31]. The minor adduct seen on each ion isolation has a delta mass of 12 Da and is as yet unidentified.
Identification and Characterization of Cystatin SA, SN, and C
Cystatins SA and SN have a signal peptide consisting of the first twenty amino acids while that of Cystatin C consists of the first twenty-six amino acids. The mature forms of Cystatin SA and SN contain 121 amino acids while Cystatin C has 120 amino acids (Table 1). Cystatins SA, SN, and C all contain two disulfide bonds in homology with the family (see Figure 5). Cystatins SA and SN have these bonds between Cys94-Cys104 and Cys118-Cys138, while Cystatin C has these bonds between Cys99-Cys109 and Cys123-Cys143. The mature form of Cystatin SA in the peak eluting at 48 min has an experimentally determined monoisotopic mass of 14336.9856 Da, in agreement with the calculated monoisotopic mass of 14337.0014 Da, including the two disulfide bonds (Δ = 0.0158, 1.10 ppm). A top-down CAD experiment confirmed the identity of Cystatin SA with 6 b- and 27 y- product ions as well as the precursor matched within 10 ppm giving a P-Score of 2.81E-46 (Figure 3; Table 1). The mature form of Cystatin SN in the peak eluting at 42 min has an experimentally determined monoisotopic mass of 14303.1553 Da, in agreement with the calculated monoisotopic mass of 14303.2228 Da, including two disulfide bonds (Δ = 0.0675, 4.72 ppm). A top-down CAD experiment confirmed the identity of Cystatin SN with 8 b- and 13 y- product ions as well as the precursor matched within 10 ppm giving a P-Score of 4.30E-29 (Figure 3; Table 1). The mature form of Cystatin C in the peak eluting at 45 minutes has an experimentally determined monoisotopic mass of 13334.5829 Da, in agreement with the calculated monoisotopic mass of 13334.5969 Da, including two disulfide bonds (Δ = 0.0140 Da, 1.05 ppm). A top-down CAD experiment confirmed the identity of Cystatin C with 9 b- and 18 y- product ions as well as the precursor matched within 10 ppm giving a P-Score of 1.25E-34 (Figure 3; Table 1).
Figure 5. Sequence alignment of the abundant human salivary Cystatins.
ClustalW was used for the alignment [36].
Figure 3. Top-down mass spectrometry of Cystatins SA, SN and C.
Appropriate fractions were analyzed as in Figure 2. For Cystatin SA and SN a twenty amino-acid signal peptide was removed while for Cystatin C a twenty-six amino-acid peptide was removed, conserving the N-terminus of the mature protein. Each Cys residue has been oxidized (−1.0078 Da) to reflect the intact pair of disulfide bonds of the native protein. For Cystatin SA 34% of product ions were matched giving a Pscore of 2.8E-46. For Cystatin SN 30% of product ions were matched giving a Pscore of 4.3E-29. For Cystatin C 22% of product ions were matched giving a Pscore of 1.25E-34.
Truncation of Cystatin D with a Protein Sequence Polymorphism
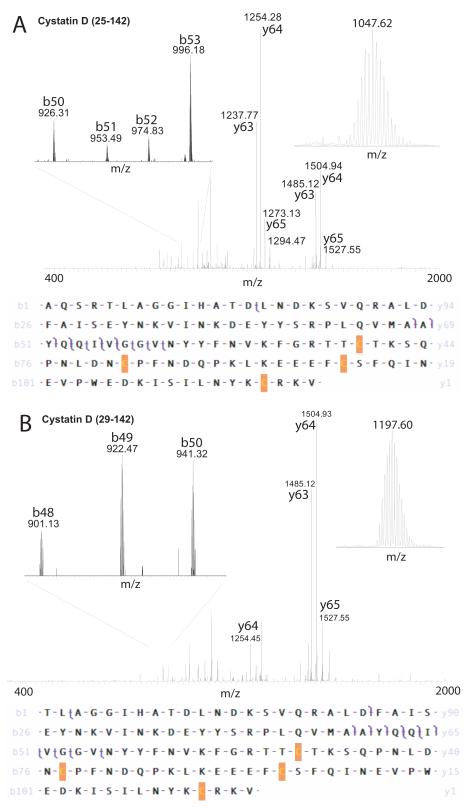
As was the case for the other Cystatins, Cystatin D has a signal peptide reported to consist of the first twenty amino acids that is cleaved to form the mature 122 amino acid protein, and like the other Cystatins, Cystatin D contains two disulfide bonds, between Cys95-Cys105 and Cys119-Cys139 by homology to the family. In our experiments, two previously undescribed forms of Cystatin D were identified both with a single nucleotide polymorphism (SNP; rs1799841) that resulted in the protein sequence polymorphism C46R, and, unlike any of the other Cystatins, both were differentially truncated at their N-terminii resulting in two distinct mature proteins, with residues 25-142 and 29-142 (Table 1). Top-down CAD experiments confirmed the primary structure of the two novel isoforms of Cystatin D (Figure 4). The larger form had an experimental monoisotopic mass of 13596.7015 Da in good agreement with the calculated monoisotopic mass of 13596.7064 Da (Δ = 0.0049 Da, 0.36 ppm) with the CAD experiment yielding 6 b- and 7 y- fragment ions matched for a P-Score of 3.44E-18 (Figure 4A). The smaller form had an experimental monoisotopic mass of 13154.4675 Da in good agreement with the calculated monoisotopic mass of 13154.4776 Da (Δ = 0.0101 Da, 0.77 ppm) with the CAD experiment yielding 7 b- and 6 y- fragment ions matched for a P-Score of 2.00E-22 (Figure 4B). No evidence was found for Cystatin D (with C46R) cleaved at the reported signal peptide site (21-142) with calculated monoisotopic mass of 13845.7368 Da. Common y-ions that dominate the product ion spectrum can be seen in both experiments while unique b-ions distinguishing the two isoforms are present, some of which are shown expanded in Figure 4.
Figure 4. Top-down mass spectrometry of two isoforms of Cystatin D.
A. A protein of average mass 13605 Da that was not detected by LC-MS+ was analyzed by top-down CAD of the 13+ precursor ion (inset top right) giving the product ion spectrum shown. The precursor and product ion masses were accounted for by removing 24 amino-acids from the N-terminus of the human Cystatin D sequence, oxidizing the 4 Cys residues to reflect two disulfide bonds and making the C46R polymorphism arising from a known SNP (Table 1). 31% of product ions were matched giving a Pscore of 3.44E-18.
B. A protein of average mass 13165 Da that was detected by LC-MS+ was analyzed by top-down CAD of the 11+ precursor ion (inset top right) giving the product ion spectrum shown. The precursor and product ion masses were accounted for by removing 28 amino-acids from the N-terminus of the human Cystatin D sequence, oxidizing the 4 Cys residues to reflect two disulfide bonds and making the C46R polymorphism arising from a known SNP (Table 1). 51% of product ions were matched giving a Pscore of 2.0E-22. A small section of the product ion spectrum was expanded to show unique b-ions (top left in A and B).
Origin of Cystatins
The measurements described were obtained on samples from either whole saliva or a mixture of submandibular/sublingual (SMSL) ductal secretions. The chromatogram shown for parotid saliva (Figure 1) shows only small inflections in the region where the Cystatins are known to elute. Examination of the low resolution ESI-MS data collected during LC-MS+ showed no detectable intact mass tags for any of the Cystatin family (Table 2). While there are measurable differences in abundance between proteins in SM versus SL salivary secretions [32], the Cystatin profiles looked similar when analyzed by LC-MS+ (data not shown). It is noted that the static nanospray FT-ICR-MS experiments are more sensitive than LC-MS+ and that the larger form of Cystatin D with calculated average mass of 13605 Da was only detected by FT-ICR-MS. An alignment of the human Cystatin family is shown in Figure 5.
Table 2.
Relative abundance of human salivary Cystatins
| Cystatin | Swiss-Prot accession# | Average Mass (Da) |
Whole salivaa | SM/SLa | Parotida | Elution time (min.) |
|---|---|---|---|---|---|---|
| S | P01036 | 14186 | + | ++ | − | 55 |
| S1 | P01036 | 14266 | +++ | ++++ | − | 55 |
| S2 | P01036 | 14346 | ++ | + | − | 55 |
| SA | P09228 | 14347 | +++ | + | − | 60 |
| SN | P01037 | 14313 | ++++ | +++ | − | 48 |
| SN+SNP |
P01037 rs2070856 |
14328 | + | − | − | 48 |
| C | P01034 | 13345 | ++ | ++ | − | 52 |
| D (SNP) 29-142 |
P28325 rs1799841 |
13165 | − | + | − | 50 |
| D (SNP) 25-142 |
P28325 rs1799841 |
(13605)b | − | (+)b | − | 50 |
+, ++,+++,++++, relative abundance; −, not detected.
detected by FT-ICR-MS only
The experimental measurements described define high-resolution intact mass tags (IMTs) for eight human salivary Cystatins, supplementing our previous top-down study that revealed the P31L polymorphism of Cystatin SN [26]. Extensive mass spectrometry studies of human saliva proteins have established a database of knowledge with respect to the primary structure and post-translational modifications of the intact salivary proteome [25]. Most of this previous work was performed using low-resolution electrospray-ionization instruments that generally provide a convenient means to follow different abundant salivary proteins. High-resolution top-down MS provides a significant advantage however, in cases where lower resolution instruments might yield ambiguous results. For example, the top-down MS analysis of the P31L isoform of Cystatin SN allowed us to distinguish this isoform from a previously described oxidized form of this protein [9, 25]. Both precursor and product ion assignments were significantly better using a delta of CH4 (calculated mass = 16.0313 Da) rather than O (15.9949 Da) and product ion assignments from CAD and ECD experiments localized the modification at position 11 of the mature protein, illustrating the power of the top-down, high resolution MS approach. It is likely that other researchers will more frequently use high-resolution approaches as MS instrumentation becomes widely available and, for example, it is noted that a recent review reports the presence of acrylonitrile adducts (+53 Da) on the P-B peptide based upon analysis of human saliva using an orbitrap analyzer [33]. Once IMTs are defined they can be used to monitor changes in abundance in different samples. For example, Messana and coworkers used IMTs to track changes in saliva proteins across subjects of different ages, and to compare normal subjects to autism patients [34, 35].
Previously, human Cystatin D was reported to have an intact mass of 13848 Da, supporting a 20 amino-acid signal peptide and conservation of the N-terminus with other Cystatins [9], while a subsequent study from the same group failed to detect it in a range of samples including whole saliva as well as ductal secretions from parotid and SMSL [25]. In the present study, two truncated isoforms of Cystatin D were characterized in molecular detail, with removal of either 24 or 28 amino-acids from the N-terminus, as well as the known C46R polymorphism. Since protease inhibitors were used and other Cystatins had N-termini in agreement with the literature it is concluded that Cystatin D is unusually sensitive to N-terminal proteolysis resulting in the truncated forms. A previous study reported detection of proteins in the mass range 12582 – 13904 that were hypothetically matched to N-terminally trimmed Cystatins including D [9]. The masses reported here are in agreement with those previously reported for Cystatins S, S1, S2, SA, SN and C [9]. Recently, Toyoshima’s group reported that a truncated form of Cystatin SA with three extra amino acids removed from the N-terminus showed significant differential expression in patients with oral squamous cell carcinoma [18].
Bottom-up strategies have been used to extensively map the human salivary proteome [20] and detected all of the Cystatins described here as well as Cystatin B that has yet to be characterized in its intact state. Cystatin B is a smaller sized protein and lacks the paired disulfide motif of the Cystatins described here. While none of the Cystatins were detected by top-down MS analysis of parotid saliva in this study, it is noted that all members were detected in the bottom-up study and also in the intact protein analysis of parotid saliva [25].
Conclusions
The top-down MS analyses described herein were adequate to confidently assign an intact mass tag (IMT) to a gene and to fully confirm the primary structure in the context of a genomic translation. Novel or labile post-translational modifications in regions of the protein with poor bond cleavage could require more detailed experiments including reduction of the disulfides and additional dissociation modes such as ECD, as was used in the description of the P31L isoform of Cystatin SN [26]. The hybrid linear ion-trap FT-ICR mass spectrometer used for the top-down MS experiments achieved mass accuracy on precursor and product ions that was typically better than 5 ppm. This and similar high-resolution instruments such as the Fourier-transform orbitrap represent powerful tools for top-down proteomics and biomarker discovery. Moreover, we envision that there will be a substantial future for monitoring intact saliva proteins for biomarkers and biosignatures of human disease.
Supplementary Material
Acknowledgements
We congratulate Neil Kelleher for his achievements in top-down mass spectrometry and his award of the 2009 Biemann Medal. Financial support from NIH-NIDCR (U01 DE016275-01; T32 DE07296-13) is gratefully acknowledged. The LTQ-FT was purchased with NIH-NCRR support (S10 RR023045).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelleher NL, L H, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J. Am. Chem. Soc. 1999;121:806–807. [Google Scholar]
- 2.Henry KD, Williams ER, Wang BH, McLafferty FW, Shabanowitz J, Hunt DF. Fourier-transform mass spectrometry of large molecules by electrospray ionization. Proc. Natl. Acad. Sci. U S A. 1989;86:9075–9078. doi: 10.1073/pnas.86.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loo JA, Edmonds CG, Smith RD. Primary sequence information from intact proteins by electrospray ionization tandem mass spectrometry. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 4.Mortz E, O’Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M. Sequence tag identification of intact proteins by matching tanden mass spectral data against sequence data bases. Proc. Natl. Acad. Sci. U S A. 1996;93:8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar WM. Saliva: its secretion, composition and functions. Br Dent J. 1992;172:305–312. doi: 10.1038/sj.bdj.4807861. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman E, Lamster IB. The diagnostic applications of saliva--a review. Crit. Rev. Oral Biol. Med. 2002;13:197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 7.Rathman WM, Van Zeyl MJ, Van den Keybus PA, Bank RA, Veerman EC, Amerongen A. V. Nieuw. Isolation and characterization of three non-mucinous human salivary proteins with affinity for hydroxyapatite. J. Biol. Buccale. 1989;17:199–208. [PubMed] [Google Scholar]
- 8.Aguirre A, Testa-Weintraub LA, Banderas JA, Dunford R, Levine MJ. Levels of salivary cystatins in periodontally healthy and diseased older adults. Arch. Oral. Biol. 1992;37:355–361. doi: 10.1016/0003-9969(92)90018-4. [DOI] [PubMed] [Google Scholar]
- 9.Lupi A, Messana I, Denotti G, Schinina ME, Gambarini G, Fadda MB, Vitali A, Cabras T, Piras V, Patamia M, Cordaro M, Giardina B, Castagnola M. Identification of the human salivary cystatin complex by the coupling of high-performance liquid chromatography and ion-trap mass spectrometry. Proteomics. 2003;3:461–467. doi: 10.1002/pmic.200390060. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Fernandez M, Liang YH, Abrahamson M, Su XD. Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile. J. Biol. Chem. 2005;280:18221–18228. doi: 10.1074/jbc.M411914200. [DOI] [PubMed] [Google Scholar]
- 11.Shah A, B B. Cystatins in Health and Diseases. Journal of Peptide Research and Therapeutics. 2009;15:43–48. doi: 10.1007/s10989-008-9160-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terpos E, Katodritou E, Tsiftsakis E, Kastritis E, Christoulas D, Pouli A, Michalis E, Verrou E, Anargyrou K, Tsionos K, Dimopoulos MA, Zervas K. Cystatin-C is an independent prognostic factor for survival in multiple myeloma and is reduced by bortezomib administration. Haematologica. 2009;94:372–379. doi: 10.3324/haematol.2008.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chudleigh RA, Ollerton RL, Dunseath G, Peter R, Harvey JN, Luzio S, Owens DR. Use of cystatin C-based estimations of glomerular filtration rate in patients with type 2 diabetes. Diabetologia. 2009;52:1274–1278. doi: 10.1007/s00125-009-1379-7. [DOI] [PubMed] [Google Scholar]
- 14.Naruse H, Ishii J, Kawai T, Hattori K, Ishikawa M, Okumura M, Kan S, Nakano T, Matsui S, Nomura M, Hishida H, Ozaki Y. Cystatin C in acute heart failure without advanced renal impairment. Am. J. Med. 2009;122:566–573. doi: 10.1016/j.amjmed.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 15.Hanrieder J, Wetterhall M, Enblad P, Hillered L, Bergquist J. Temporally resolved differential proteomic analysis of human ventricular CSF for monitoring traumatic brain injury biomarker candidates. J. Neurosci. Methods. 2009;177:469–478. doi: 10.1016/j.jneumeth.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 16.Tsuji-Akimoto S, Yabe I, Niino M, Kikuchi S, Sasaki H. Cystatin C in cerebrospinal fluid as a biomarker of ALS. Neurosci. Lett. 2009;452:52–55. doi: 10.1016/j.neulet.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 17.Yoneda K, Iida H, Endo H, Hosono K, Akiyama T, Takahashi H, Inamori M, Abe Y, Yoneda M, Fujita K, Kato S, Nozaki Y, Ichikawa Y, Uozaki H, Fukayama M, Shimamura T, Kodama T, Aburatani H, Miyazawa C, Ishii K, Hosomi N, Sagara M, Takahashi M, Ike H, Saito H, Kusakabe A, Nakajima A. Identification of Cystatin SN as a novel tumor marker for colorectal cancer. Int. J. Oncol. 2009;35:33–40. [PubMed] [Google Scholar]
- 18.Shintani S, Hamakawa H, Ueyama Y, Hatori M, Toyoshima T. Identification of a truncated cystatin SA-I as a saliva biomarker for oral squamous cell carcinoma using the SELDI ProteinChip platform. Int. J. Oral Maxillofac. Surg. 2009 doi: 10.1016/j.ijom.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Rudney JD, Staikov RK, Johnson JD. Potential biomarkers of human salivary function: a modified proteomic approach. Arch. Oral Biol. 2009;54:91–100. doi: 10.1016/j.archoralbio.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR, 3rd, Fisher SJ. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitelegge JP, Gundersen CB, Faull KF. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zabrouskov V, Whitelegge JP. Increased coverage in the transmembrane domain with activated-ion electron capture dissociation for top-down Fourier-transform mass spectrometry of integral membrane proteins. J. Proteome Res. 2007;6:2205–2210. doi: 10.1021/pr0607031. [DOI] [PubMed] [Google Scholar]
- 23.Whitelegge JP, Zhang H, Aguilera R, Taylor RM, Cramer WA. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: cytochrome b(6)f complex from spinach and the cyanobacterium Mastigocladus laminosus. Mol. Cell. Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Veerman EC, van den Keybus PA, Vissink A, Amerongen A. V. Nieuw. Human glandular salivas: their separate collection and analysis. Eur. J. Oral Sci. 1996;104:346–352. doi: 10.1111/j.1600-0722.1996.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 25.Messana I, Cabras T, Pisano E, Sanna MT, Olianas A, Manconi B, Pellegrini M, Paludetti G, Scarano E, Fiorita A, Agostino S, Contucci AM, Calo L, Picciotti PM, Manni A, Bennick A, Vitali A, Fanali C, Inzitari R, Castagnola M. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides: a proteomics approach. Mol. Cell. Proteomics. 2008;7:911–926. doi: 10.1074/mcp.M700501-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Whitelegge JP, Zabrouskov V, Halgand F, Souda P, Bassilian S, Yan W, Wolinsky L, Loo JA, Wong DT, Faull KF. Protein-Sequence Polymorphisms and Post-translational Modifications in Proteins from Human Saliva using Top-Down Fourier-transform Ion Cyclotron Resonance Mass Spectrometry. Int. J. Mass Spectrom. 2007;268:190–197. doi: 10.1016/j.ijms.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez SM, Nishio JN, Faull KF, Whitelegge JP. The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell. Proteomics. 2002;1:46–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 28.Isemura S, Saitoh E, Sanada K, Minakata K. Identification of full-sized forms of salivary (S-type) cystatins (cystatin SN, cystatin SA, cystatin S, and two phosphorylated forms of cystatin S) in human whole saliva and determination of phosphorylation sites of cystatin S. J. Biochem. 1991;110:648–654. doi: 10.1093/oxfordjournals.jbchem.a123634. [DOI] [PubMed] [Google Scholar]
- 29.Johnsson M, Richardson CF, Bergey EJ, Levine MJ, Nancollas GH. The effects of human salivary cystatins and statherin on hydroxyapatite crystallization. Arch. Oral Biol. 1991;36:631–636. doi: 10.1016/0003-9969(91)90014-l. [DOI] [PubMed] [Google Scholar]
- 30.Ramasubbu N, Reddy MS, Bergey EJ, Haraszthy GG, Soni SD, Levine MJ. Large-scale purification and characterization of the major phosphoproteins and mucins of human submandibular-sublingual saliva. Biochem. J. 1991;280:341–352. doi: 10.1042/bj2800341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grubb A, Loefberg H, Barrett AJ. The disulphide bridges of human cystatin C (gamma-trace) and chicken cystatin. FEBS Lett. 1984;170:370–374. [Google Scholar]
- 32.Hu S, Denny P, Xie Y, Loo JA, Wolinsky LE, Li Y, McBride J, Loo R. R. Ogorzalek, Navazesh M, Wong DT. Differentially expressed protein markers in human submandibular and sublingual secretions. Int. J. Oncol. 2004;25:1423–1430. [PubMed] [Google Scholar]
- 33.Messana I, Inzitari R, Fanali C, Cabras T, Castagnola M. Facts and artifacts in proteomics of body fluids. What proteomics of saliva is telling us? J. Sep. Sci. 2008;31:1948–1963. doi: 10.1002/jssc.200800100. [DOI] [PubMed] [Google Scholar]
- 34.Cabras T, Pisano E, Boi R, Olianas A, Manconi B, Inzitari R, Fanali C, Giardina B, Castagnola M, Messana I. Age-dependent modifications of the human salivary secretory protein complex. J. Proteome Res. 2009;8:4126–4134. doi: 10.1021/pr900212u. [DOI] [PubMed] [Google Scholar]
- 35.Castagnola M, Messana I, Inzitari R, Fanali C, Cabras T, Morelli A, Pecoraro AM, Neri G, Torrioli MG, Gurrieri F. Hypo-Phosphorylation of Salivary Peptidome as a Clue to the Molecular Pathogenesis of Autism Spectrum Disorders. J. Proteome Res. 2008 doi: 10.1021/pr8004088. [DOI] [PubMed] [Google Scholar]
- 36.Higgins DG. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 1994;25:307–318. doi: 10.1385/0-89603-276-0:307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.