Abstract
Colorectal cancer (CRC) is a leading cause of morbidity and mortality and alterations in mismatch repair (MMR) genes, leading to absent protein (negative) expression, are responsible for approximately 20% of CRC cases. Immunohistochemistry is a tool for prescreening of MMR protein expression in CRC but the literature on its use on Hispanics is scarce. However, Hispanics represent the second leading ethnicity in the United States (US) and CRC is a public health burden in this group. Our objectives were to determine the frequency of MMR protein-negative CRC and to evaluate its association with clinical and pathological characteristics among Hispanics from Puerto Rico, for the first time to our knowledge. A retrospective observational study of unselected CRC patients from the Puerto Rico Medical Center from 2001 to 2005 was done. MLH1 and MSH2, the most commonly altered MMR genes, protein expression was evaluated using immunohistochemistry, with microsatellite instability (MSI) and BRAF gene analyses in the absence of MLH1 protein expression. One-hundred sixty-four CRC patients were evaluated: the overall MMR protein-negative frequency was 4.3%, with 0.6% frequency of co-occurrence of MLH1-protein negative expression, MSI-high, and normal BRAF gene. MMR protein-negative expression was associated with proximal colon location (p = 0.02) and poor histological tumor differentiation (p = 0.001), but not with other characteristics. The frequency of MMR protein-negative CRC in Hispanics from Puerto Rico was lower than reported in other populations. This finding may explain the lower CRC incidence rate among US Hispanics as compared to US non-Hispanic whites and blacks.
Keywords: colorectal cancer, genetics, Hispanics, immunohistochemistry, mismatch repair
Introduction
Colorectal cancer (CRC) is a preventable and highly curable disease if detected in the early stages of tumorigenesis. However, it is the third most common cancer and the fourth most frequent cause of cancer death worldwide [1]. Furthermore, CRC will be the third most common cancer in incidence and cause of death for both sexes in the United States (US) in 2009 [2]. CRC arising as a result of microsatellite instability (MSI) have distinct clinical and pathological features that distinguish them from those with microsatellite stability [3]. MSI occurs when a germline repeated nucleotide sequence (microsatellite) allele has gained or lost repeated units, and has, thus, undergone a somatic change in length [4]. MSI is the result of inactivation of both alleles of a DNA nucleotide mismatch repair (MMR) gene: MLH1, MSH2, MSH6, PMS1, or PMS2. These genes are implicated in post-replication repair, DNA damage signaling, and apoptosis when repair is overwhelmed by DNA damage [5–7].
A germline mutation in any of the MMR genes is associated with hereditary nonpolyposis colorectal cancer (HNPCC) or Lynch syndrome [8–13]. HNPCC comprises 2–4% of all CRC [8, 14, 15], is the most common form of hereditary CRC [16], and probably the most frequent cause of hereditary cancer [17]. More than 90% of patients with HNPCC have MMR defects causing MSI [18, 19], with MLH1 and MSH2 germline mutations accounting for more than 90% of [20, 21] or most [14, 15, 22] HNPCC cases.
MSI associated with MMR gene alterations account for 20% of all CRC cases [23], 15–20% of sporadic CRC [15, 19, 24], and 85% of hereditary CRC [25]. Specifically, MMR germline mutation is associated with a 70–80% lifetime risk of developing CRC [26, 27], compared to 5–6% in the general population [20]. Tumors with a MMR gene defect show absence of MMR protein expression [28], which may be secondary to either a germline mutation (3–5% of all CRC cases, expressed as HNPCC) or a somatic mutation, usually hypermethylation of MLH1 promoter (15–20% of all CRC cases) [29]. Additionally, BRAF (gene involved in growth factor signaling [30]) V600E activating (BRAFV600E) mutation is evaluated in MLH1-protein negative expression because it is often present when the MLH1 promoter is methylated [14].
MSI or immunohistochemical testing (with or without BRAFV600E mutation testing) of the tumor tissue are strategies to select patients for subsequent diagnostic testing [14, 31–33]. Immunohistochemistry is a simple, accessible, rapid, and relatively inexpensive prescreening method [22, 34–36] to evaluate tumor tissue for MMR protein expression as compared to polymerase chain reaction (PCR)-based MSI assays. Absence of protein expression also indicates the MMR gene most appropriate for DNA analysis [15, 34, 35, 37, 38]. For screening of HNPCC among patients with CRC, immunohistochemistry is almost equally sensitive as MSI [34, 39]. If restricted to experienced pathologists [35], immunohistochemistry is a valid tool to identify patients at risk for HNPCC and patients with sporadic microsatellite instable CRC [40]. For these reasons, immunohistochemistry of these colorectal tumors has gained popularity as the first step in checkingfor MMR gene mutations, even in community hospitals [15].
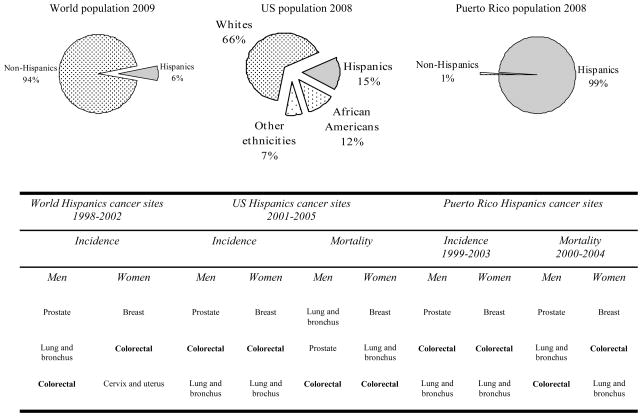
Hispanics or Latinos are individuals of Mexican, Puerto Rican, Cuban, Central or South American, or other Spanish culture or origin, regardless of race [41]. Hispanics represent a notable fraction of the world population [42], the second leading ethnicity in the US [43], and almost all the population of Puerto Rico [44]. Indeed, CRC basically has the second highest incidence rate and the third highest mortality rate among cancer sites in Hispanics [2, 45, 46] (Figure 1). These are reasons why it is important to study this highly preventable but deadly disease in this ethnicity.
Fig. 1.
Percentage of Hispanics within the world [42], United States [43], and Puerto Rico [44] populations and their corresponding leading cancer sites in incidence and mortality rates, for men and women [2, 45, 46] in decreasing order. World population 2009 [42]: it is still an underestimation of the actual Hispanic population for not considering those living in non-Hispanic countries, including United States. World Hispanic cancer sites 1998–2002 [45]: based on good quality data provided by United States and Hispanic countries, registries, and populations.
Studies of MMR protein expression in unselected CRC patients have been performed in various countries or ethnicities [37, 47–58]. However, literature in Hispanic patients is scarce, showing a frequency of MMR protein-negative expression of approximately 7% with no documentation on its association with clinical and pathological characteristics [57, 58]. Furthermore, most of the literature about MMR protein expression in CRC is from selected high risk cancer clinics and registries, including HNPCC cohorts [59–66], or sporadic colorectal tumors excluding HNPCC patients [67]. Hence, we designed a study of unselected patients with the objective of evaluating the frequency of MMR protein-negative CRC and the association with clinical and pathological characteristics in Hispanics from Puerto Rico. Moreover, to our knowledge and considering that immunohistochemistry is a relatively inexpensive prescreening method, this is the first study evaluating MMR protein expression by immunohistochemistry in Hispanic CRC patients from Puerto Rico.
Material and methods
Study design and population
A retrospective observational study of unselected patients that visited the Puerto Rico Medical Center (PRMC, San Juan, Puerto Rico), which includes the Hospital Oncologico Isaac Gonzalez-Martinez (HOIGM) between 2001 and 2005 was conducted. These hospitals provide tertiary and supra-tertiary medical care and are the main referral centers for patients in Puerto Rico in need of specialized health care. Hispanic patients with the diagnosis of CRC who were 21 years of age or older and who had pathological tissue from biopsy or surgery at the PRMC or HOIGM were included. Individuals with familial adenomatous polyposis phenotype based on surgical or pathological report were excluded because it is a genetically different disease [68–70]. Patients were identified originally by evaluation of pathology reports and the HOIGM cancer registry for the diagnosis of colorectal carcinoma in situ or adenocarcinoma. The study was approved by the University of Puerto Rico Medical Sciences Campus and the HOIGM institutional review boards.
Hispanics comprised 98.5% of the population in Puerto Rico for 2005 [71]. CRC was diagnosed in approximately 7000 patients in Puerto Rico in 1999–2003: 53% men and 47% women, with more than 90% of patients being older than 50 years old. Additionally, CRC was the cause of death in 2907 patients in Puerto Rico in 2000–2004: 54% men and 46% women [46].
Tumor MLH1 and MSH2 protein expression
Tumor tissue blocks were retrieved from the departments of pathology of both institutions. Hematoxylin and eosin slides from each block were evaluated by a pathologist (C.G.K.) to assess adequacy of the sample and confirm the diagnosis. Coded slides were prepared from each block for blind assessment.
Formalin-fixed, paraffin-embedded tumor tissues were sectioned at 4 μm and affixed to Fisherbrand Colorfrost / PLUS microscope slides (No. 12-550-19, Fisher Scientific, Pittsburgh, PA) and heated at 60°C for at least 1 hour. The slides were deparaffinized and rehydrated with xylene, descending graded alcohols, and distilled water. Endogenous peroxidase activity was quenched using 3% hydrogen peroxide in methanol solution for 5 minutes. The slides underwent antigen retrieval with 10X Tris-ethylenediamine tetraacetic acid buffer, pH 7.5 (Code MB-006, Rockland Immunochemicals, Inc., Gilbertsville, PA) for 45 minutes on boiling water in a Flavor Scenter Steamer Plus (SKU HS900, Black and Decker, Hunt Valley, MD). The sections were cooled for 20 minutes and transferred to DakoCytomation wash buffer (Product S3006, DakoCytomation, Inc., Carpinteria, CA). The subsequent immunohistochemical staining protocol was performed on an automated immunostainer (Dako Autostainer Plus Universal Staining System, Dako North America, Inc., Carpinteria, CA). The sections were incubated with mouse monoclonal antibodies against MLH1 (Clone G168-728, 1:60; Catalog No. 554073, BD Pharmingen, San Jose, CA) or MSH2 (Clone FE11, 1:30; Catalog No. NA27, Calbiochem, La Jolla, CA) for 60 minutes. The antibodies were diluted with antibody diluent with background reducing components (Product S3022, DakoCytomation, Inc., Carpinteria, CA). Wash buffer (Product S3006, DakoCytomation, Inc., Carpinteria, CA) was used for rinsing. Immunoreactivity was detected using the DakoCytomation EnVision+ System-HRP kit (Code K4007, DakoCytomation, Inc., Carpinteria, CA). Mayer’s hematoxylin was used as counterstain for 8 minutes. The sections were dehydrated through ascending graded alcohols, cleared in xylene, and mounted.
Two independent pathologists (C.G.K. and S.R.H.) evaluated the tissue slide staining. Protein expression was scored as positive if both the tumor and internal control (normal germinal centers and basal crypt epithelium) showed nuclear staining; negative if the tissue lacked staining in tumor while the internal control was stained, or uninterpretable if no immunostaining of tumor or internal control could be shown.
Microsatellite instability (MSI)
To evaluate for suggestion of germline mutation or hypermethylation of the MLH1 promoter, any patient with MLH1-protein negative expression was evaluated for MSI by fluorescently labeled PCR reaction amplification with five microsatellite markers from the panel described by the National Cancer Institute conference on MSI: BAT-26, BAT-40, D2S123, D5S346, and D17S250 [72]. MSI-high (MSI-H) was defined by shifts of bands compared with control DNA in ≥ 30% of evaluable markers and / or in a mononucleotide repeat, MSI-low was defined by shifts in one dinucleotide marker representing < 30% of evaluable markers, and microsatellite-stable was defined by absence of shifts in any marker.
Sequencing of BRAF gene
In the case of MLH1-protein negative expression, BRAFV600E mutation was evaluated by amplification of BRAF gene exon 15 by genomic PCR using intronic primers and a commercial DNA sequencing kit according to the manufacturer’s instructions (BigDye Terminator v1.1 Cycle Sequencing kit, Part 4337449, Applied Biosystems, Foster City, CA). The PCR products were analyzed with an automated sequencer (3730 DNA Analyzer, Applied Biosystems, Foster City, CA) using forward and reverse primers. All mutations were confirmed by an independent PCR amplification and sequencing.
Medical record review and independent covariates
Medical records were examined for the following clinical and pathological variables: sex, age at diagnosis, follow up time, status on follow up (alive no vs. yes), personal history of any other cancer, family history of any cancer, tumor location, tumor size (greatest dimension), history of surgery for the primary tumor, history of chemotherapy, history of radiotherapy, and tumor recurrence. The tumor location was classified as proximal (from cecum to transverse colon, also known as right-sided) or distal (from splenic flexure to rectum, also known as left-sided).
Tumor histological differentiation was classified as well / moderately differentiated (more than 50% gland formation) or poorly differentiated (less than 50% gland formation). The tumor was classified according to the American Joint Committee on Cancer staging (I vs. II vs. III vs. IV vs. not available or not applies) [73]. Stages were I (tumor confined within the muscularis propria of the wall of the large bowel), II (tumor penetrates through muscularis propria of the bowel wall), III (tumor has lymph node involvement), and IV (tumor spread to systemic organs).
Statistics
The primary outcome variable for MMR protein expression was either positive or negative. Hypothesis testing for the association of MMR protein-negative expression with clinical and pathological characteristics was done using Pearson Χ2 test, two-sample t test with equal variances, or Fisher’s exact test as appropriate for small cell sizes. The follow up time was determined from date of diagnosis until May 31, 2007, when patients’ survival was sought in the Social Security Death Index at http://www.rootsweb.com if unknown otherwise. Statistical analysis was performed using STATA 9.0 software (Stata Corporation, College Station, TX). A significance value less than 0.05 was considered statistically significant.
Results
Patients
Two-hundred twenty-six potential patients were identified during the study period. Of these, 39 patients were excluded because the block did not include tumor tissue, 20 due to lack of availability of tissue blocks, and 3 due to failure to meet eligibility criteria. Thus, 164 patients were eligible and were included in the study. Table 1 shows the clinical and pathological features of these patients. Our study population included 94 (57.3%) women, had a mean age at diagnosis of 62.72 years, and a mean tumor size of 4.4 cm. At a mean follow up time of 39.2 months, 73.8% of patients were alive. Most of the patients had no personal history of any other cancer (90.2%), had tumor localized in distal colon (70.4%), had well / moderate tumor histological differentiation (81.1%), were stage II (40.2%), had surgery (92.1%), received chemotherapy (56.1%), did not receive radiotherapy (70.7%), and did not have tumor recurrence (90.0%). There were no statistically significant differences between the final cohort evaluated (n = 164) and those excluded (n = 62) with regards to basic demographic and clinical and pathological characteristics (data not shown).
Table 1.
Clinical and pathological characteristics of study population
| Characteristic | |
|---|---|
| Patients, n | 164 |
| Sex, n (%) | |
| Men | 70 (42.7) |
| Women | 94 (57.3) |
| Age at diagnosis (years), mean ± SD (range) | 62.72 ± 13.54 (23-9) |
| Follow up time (months), mean ± SD (range) | 39.2 ± 22 (0.67–154.1) |
| Alive on follow up, n (%) | |
| No | 43 (26.2) |
| Yes | 121 (73.8) |
| Personal history of any other cancer, n (%) | |
| No | 148 (90.2) |
| Yes | 16 (9.8) |
| Family history of any cancer if available, n (%) | |
| No | 51 (41.8) |
| Yes | 71 (58.2) |
| Tumor location if available, n (%) | |
| Proximal | 47 (29.6) |
| Distal | 112 (70.4) |
| Tumor size if available (cm), mean ± SD (range) | 4.42 ± 2.34 (1–13.2) |
| Tumor histological differentiation, n (%) | |
| Well / moderate | 133 (81.1) |
| Poor | 31 (18.9) |
| Tumor stage, n (%) | |
| I | 9 (5.5) |
| II | 66 (40.2) |
| III | 46 (28.1) |
| IV | 14 (8.5) |
| Not available or not applies | 29 (17.7) |
| Surgery received, n (%) | |
| No | 13 (7.9) |
| Yes | 151 (92.1) |
| Chemotherapy received, n (%) | |
| No | 72 (43.9) |
| Yes | 92 (56.1) |
| Radiotherapy received, n (%) | |
| No | 116 (70.7) |
| Yes | 48 (29.3) |
| Tumor recurrence, n (%) | |
| No | 149 (90.9) |
| Yes | 15 (9.2) |
SD = Standard deviation, cm = Centimeters
Tumor MLH1 and MSH2 protein expression
Figure 2 shows representative examples of immunohistochemistry for MLH1 and MSH2 protein expression. MLH1 protein was expressed in all but one tumor (0.6%). MSH2 protein was expressed in all but 6 tumors (3.7%). One patient had uninterpretable results. Overall, 7 of 164 tumors evaluated for MMR protein expression showed absence of expression (4.3%).
Fig. 2.
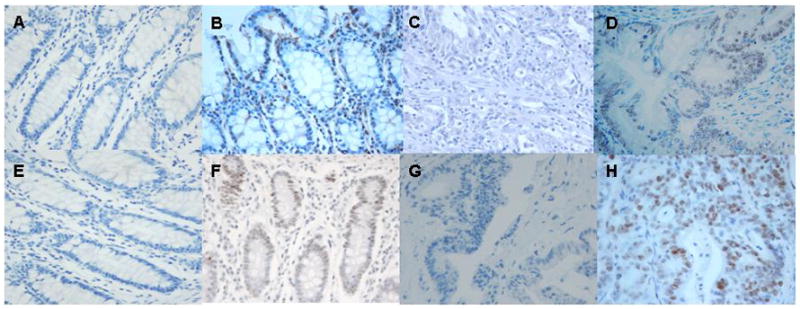
Immunohistochemistry for MLH1 and MSH2 protein expression (hematoxylin, 400x). (a) Normal colorectal tissue with no antibody staining for MLH1 (negative control). (b) Normal colorectal tissue with MLH1 antibody staining (positive control). (c) Tumor with absence of MLH1 protein expression. (d) Tumor with MLH1 protein expression. (e) Normal colorectal tissue with no antibody staining for MSH2 (negative control). (f) Normal colorectal tissue with MSH2 antibody staining (positive control). (g) Tumor with absence of MSH2 protein expression. (h) Tumor with MSH2 protein expression.
MLH1 evaluation for MSI and sequencing for BRAFV600E mutation
The only patient who showed absence of MLH1 protein expression was evaluated for both MSI and BRAFV600E mutation. Two of 5 (40%) microsatellite markers were altered in the tumor, thus classifying the tumor as MSI-H. BRAF exon 15 sequencing did not reveal V600E activating mutation. Thus, the BRAF gene was considered to be wild type or normal.
Clinical and pathological characteristics
Tumor location and histological differentiation were the only statistically significant clinical and pathological differences observed between colorectal tumors based on MMR protein expression (Table 2). Colorectal tumors with absent MMR protein expression were more commonly located in the proximal colon compared to tumors that expressed both MMR proteins, which were more commonly located in the distal colon (Fisher’s exact p = 0.024). Colorectal tumors with MMR protein expression were more commonly well or moderately differentiated compared to those with absence of protein expression, which more commonly were poorly differentiated (Pearson X2 p = 0.001). There was no significant difference according to MMR protein status with regards to sex, age at diagnosis of cancer, personal history of any other cancer, family history of any cancer, mean tumor size, tumor stage or tumor recurrence.
Table 2.
Association of mismatch repair protein expression with clinical and pathological characteristics
| Characteristic | Mismatch repair protein expression |
p-value | |
|---|---|---|---|
| negative | positive | ||
| Sex, n (%) | |||
| Men | 3 (42.9) | 67 (42.7) | 0.99a |
| Women | 4 (57.1) | 90 (57.3) | |
| Age at diagnosis (years), mean ± SD (95% confidence interval) | 59.9 ± 21.7 (39.8–79.9) | 62.85 ± 13.2 (60.8–64.9) | 0.57b |
| Personal history of any other cancer, n (%) | |||
| No | 6 (85.7) | 142 (90.4) | 0.68a |
| Yes | 1 (14.3) | 15 (9.6) | |
| Family history of any cancer if available, n(%) | |||
| No | 1 (25) | 50 (42.4) | 0.49a |
| Yes | 3 (75) | 68 (57.6) | |
| Not available | 42 | ||
| Tumor location if available, n (%) | |||
| Proximal | 5 (71.4) | 42 (27.6) | 0.024c |
| Distal | 2 (28.6) | 110 (72.4) | |
| Not available | 5 | ||
| Tumor size (cm), mean ± SD (95% CI) | 6.0 ± 2.0 (3.9–8.1) | 4.4 ± 2.33 (4.0–4.8) | 0.09b |
| Tumor histological differentiation, n (%) | |||
| Well / moderate | 2 (28.6) | 131 (83.4) | 0.001a |
| Poor | 5 (71.4) | 26 (16.6) | |
| Tumor stage, n (%) | |||
| I | 0 (0) | 9 (5.7) | 0.63a |
| II | 3 (42.9) | 63 (40.1) | |
| III | 3 (42.9) | 43 (27.4) | |
| IV | 1 (14.2) | 13 (8.3) | |
| Not available | 0 (0) | 29 (18.5) | |
| Tumor recurrence, n (%) | |||
| No | 7 (100) | 142 (90.4) | 0.39a |
| Yes | 0 (0) | 15 (9.6) | |
SD = Standard deviation, cm = Centimeters
p-value determined by
Pearson Χ2 test,
two-sample t test with equal variances, and
Fisher’s exact test
Discussion
CRC is a deadly illness of great concern in Hispanic populations and the island of Puerto Rico is a great site to study this ethnicity. During 1998–2002, in both sexes, Puerto Ricans had CRC incidence and mortality rates similar to those for US Hispanics, but their rates were lower than those for non-Hispanic whites and non-Hispanic blacks. However, Puerto Rican men and women with ages 40–59 years had a greater risk of incidence and mortality than their US Hispanic counterparts [74].
Evaluation of colon cancers for MMR defects may have significant therapeutic implications. In patients with stage II or III CRC, adjuvant 5-fluorouracil chemotherapy provides survival improvement in patients with MMR-competent tumors while the survival of patients with MMR-defective tumors does not improve with the adjuvant chemotherapy [75–77]. Also, loss of tumor MMR function may predict improved outcome in stage III colon cancer patients postoperatively treated with a weekly bolus irinotecan, fluorouracil, and leucovorin regimen as compared with those receiving weekly bolus of fluorouracil and leucovorin [78].
HNPCC is characterized by young onset CRC [8, 9, 15], an increased risk for gynecologic, urinary tract, and gastrointestinal cancers [9, 27, 79–88], and most commonly occurs in men [89]. In HNPCC, a single mutation is inherited in the germline, and MSI occurs only after inactivation of the other allele [90]. Most sporadic MSI-H cancers are caused by methylation and silencing of the MLH1 gene [91, 92], most probably develop in women, and have their origin within serrated polyps [89]. Colorectal tumors with MSI are associated with location proximal to the splenic flexure and poor histological differentiation [67, 93, 94].
MSI occurs in approximately 20% of CRC cases [23]. These tumors typically fail to express MMR protein as seen on immunohistochemistry [28]. In the present study, only 4.3% of 164 patients lacked protein MMR expression. Lack of protein expression as a surrogate for MSI is clinically important to patients. A patient with absence of MMR protein expression may be a member of a Lynch syndrome kindred, hence results may have implications for themselves and other family members. Furthermore, individuals belonging to a HNPCC-kindred have a higher risk of developing other non-colorectal cancers such as endometrial, ovarian, and gastric [9, 27, 79–88]. Of the seven tumors in our study that lacked MMR protein expression, six tumors lacked expression of MSH2, which usually indicates a germline mutation in the MSH2 gene, and one patient had absence of MLH1 with MSI-H.
The tumor with absence of MLH1 protein expression with MSI-H may have a either a germline mutation or hypermethylation of the MLH1 promoter. Additionally, this patient had normal BRAF gene, which may be suggestive of HNPCC-associated MLH1 germline mutation. Virtually 100% of individuals with HNPCC do not carry the BRAFV600E mutation [14, 15], whereas 68% of those without HNPCC do [14]. Moreover, BRAFV600E mutation is associated with sporadic CRC with MSI [30, 95]. MLH1 promoter hypermethylation leads to gene inactivation [96] and, if present, may be secondary to CpG island methylation phenotype (CIMP), progressing via the MSI-H pathway. CpG islands are regions rich in cytosine-guanosine dinucleotide repeats at the 5′ region of approximately half of all human genes. Methylation of cytosine residues within CpG islands of promoters and proximal exons is associated with loss of gene expression [97]. CIMP tumors have similarities to tumors with MSI-H, such as right-sided location and poor histological differentiation [98]. However, BRAF mutation is frequently seen in sporadic MSI-H CRC, associated with DNA methylation secondary to CIMP-high (3–4 methylated in tumor markers are methylated) status [99]. For these reasons, it is unlikely that a MLH1 promoter hypermethylation secondary to CIMP may have occurred. However, none of the patients underwent germline genetic testing.
In our study of Hispanics from Puerto Rico, the frequency of MMR protein-negative tumors was far lower than in other studies of unselected CRC patients in other countries or ethnicities, especially regarding MLH1-protein negative expression (Table 3) [37, 47–58]. Moreover, the frequency of MMR protein-negative tumors in our study was lower than in Spain, including MLH1-protein negative expression [57, 58].
Table 3.
Comparison of our study with others in other countries or ethnicities for mismatch repair protein expression in unselected colorectal cancer patients
| Authors | Country or ethnic group | Evaluated patients, n | MLH1-protein negative expression, n (%) | MSH2-protein negative expression, n (%) |
|---|---|---|---|---|
| Lindor et al [22]a | Minnesota, US | 255 | 48 (18.8) | 3 (1.2) |
| De Jesus-Monge et al | Hispanics from Puerto Rico | 164 | 1 (0.6) | 6 (3.7) |
| Brim et al [47] | Iran | 25 | 10 (40) | 1 (4) |
| Ashktorab et al [48] | African | b | 16 of 34 (47.1)c | 12 of 31 (47.1)c |
| Americans | 3 of 34 (8.8)d | 1 of 31 (8.8)d | ||
| Pandey et al [49] | India | 46 | 7 (15.2) | 1 (2.2) |
| Ashktorab et al [50] | Oman | 49 | 5 (10.2)e | 3 (6.1)e |
| Brim et al [47] | Oman | 61 | 19 (31.1) | 5 (8.2) |
| Erdamar et al [51] | Turkey | 74 | 29 (39.2) | 7 (9.5) |
| Brim et al [47] | African Americans | b | 26 of 76 (34.2) | 8 of 74 (10.8) |
| Jin et al [52] | China | 146 | 17 (11.6) | 9 (6.2) |
| Jensen et al [53] | Denmark | 262 | 37 (14.1) | 3 (1.1) |
| Boardman et al [54] | Alaska Natives | 329 | 42 (12.8) | 3 (0.9) |
| Molaei et al [37] | Iran | 343 | 19 (5.5) | 24 (7) |
| Wright et al [55] | New Zealand | 458 | 80 (17.5) | 9 (2) |
| Coggins et al [56] | England | 732 | 52 (7.1) | 5 (0.7) |
| Xicola et al [57] | Spain | 1058 | 59 (5.6) | 22 (2.1) |
| Piñol et al [58] | Spain | 1222 | 60 (4.9) | 21 (1.7) |
Representative sample for United States
See columns to the right to see the number of evaluated patients for each protein
Partially negative expression
Completely negative expression
Partially or completely negative expression
As seen in other studies [4, 15, 37, 56, 67, 93, 94, 100–102], colorectal tumors with lack of MMR protein expression, suggesting MMR deficiency, were associated with a proximal location in the colon and poor histological differentiation. However, opposing previous publications [4, 17, 19, 37, 55, 72, 79, 101–104], we did not observe associations of MMR protein-negative tumors with gender, earlier age at diagnosis of cancer, positive personal and family history of cancer, or earlier tumor stage, when compared to tumors with MMR protein-positive expression.
We acknowledge that our study has several limitations. First, the study was retrospective, which limited the information to that present in the medical record. This limited our ability to accurately evaluate family history of cancer and other factors, like environmental exposure. Second, we were only able to include patients that had tumor blocks available for analysis. However, the percentage of patients excluded from pathological analysis was small and did not statistically differ with regards to basic demographic characteristics from the patients included in this investigation. As such, we believe our results are representative of the referral population seen at our Center. Third, the sample size was relatively small. The sample size may have limited the power of the study to identify important differences according to MMR protein expression status. This is also a pilot retrospective study and a larger prospective study is undergone to verify our findings. Fourth, only MLH1 and MSH2 protein expression were evaluated. However, over 90% [20, 21] or most [14, 15, 22] of genetically characterized HNPCC cases are accounted for by germline mutations in any of them.
Additionally, a fifth limitation is that all patients were not subsequently evaluated for MSI to confirm the suggestive findings from immunohistochemistry. However, for screening of HNPCC among patients with CRC, immunohistochemistry is almost equally sensitive as MSI [33, 34, 39]. Indeed, MSI testing is not available in the majority of routine pathology service laboratories, and these rely on immunohistochemistry to detect loss of MMR protein expression as a surrogate marker for the presence of MSI [105]. In consequence, immunohistochemistry is a valid tool to identify patients at risk for HNPCC and patients with sporadic microsatellite instable CRC [40] when evaluated by experienced pathologists [35] like the authors. Additionally, many consider immunohistochemistry and MSI testing as complementary. In a sequential approach, immunohistochemistry is done first and, if informative, may result in cost savings. In the other hand, if immunohistochemistry is not informative, MSI testing can then be performed [15]. Nonetheless, this investigation provides insightful information about the frequency and clinical and pathological characteristics associated with MMR protein deficiency in Hispanics from Puerto Rico, which may be ethnically related.
To our knowledge, the present study is the first one to evaluate the frequency of MMR protein-negative expression in unselected Hispanic CRC patients from Puerto Rico using the relatively inexpensive [35] prescreening method of immunohistochemistry and its association with clinical and pathological characteristics.
In conclusion, this investigation of unselected Hispanic CRC patients from Puerto Rico revealed a low frequency of MMR protein-negative tumors. Similar to other populations, Hispanic MMR protein-negative tumors were proximally located and exhibited poor histological differentiation. The results may be relevant to the evaluation and management of Hispanic patients with CRC from Puerto Rico in immigrant as well as native populations. The relatively low frequency of MMR protein-negative CRC in unselected Hispanic patients from Puerto Rico may be a reflection of, or explain, the lower CRC incidence rate among US Hispanics as compared to non-Hispanic whites and blacks. Our study is significant on shedding light on the scarce literature on MMR protein expression in CRC.in Hispanic populations.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) National Center for Research Resources, grants R25 RR 017589, P20 RR 011126-13, and G12 RR 003051-22; and NIH National Cancer Institute, grants K07 CA 092445, K22 CA 115913-02, and U54 CA 096297. S.R.H. is the Frederick F. Becker Distinguished University Chair in Cancer Research from The University of Texas. We thank the Department of Pathology of the PRMC and the HOIGM for tissue access; the Department of Medical Records of the PRMC and the HOIGM and the HOIGM cancer registry for data access; Mr. Nelson Santiago and Mrs. Janet E. Quiñones for laboratory technical support; Mr. Alexis A. Florian-Ayala and Mr. Alejandro Lopez-Araujo for data collection and laboratory technical support; Kerry Sieger for MSI evaluation and BRAF gene sequencing; and Dr. Sara S. Strom, Dr. Francis M. Giardiello, Dr. Elena Martinez-Stoffel, Prof. Vivianna M. De Jesus-Monge, and the faculty and scholars of the University of Puerto Rico Master of Science in Clinical Research Program for critical feedback. This work was done as fulfillment for the studies for a graduate degree from the University of Puerto Rico Medical Sciences Campus by Wilfredo E. De Jesus-Monge.
Abbreviations
- CIMP
CpG island methylation phenotype
- CRC
Colorectal cancer
- HNPCC
Hereditary nonpolyposis colorectal cancer
- HOIGM
Hospital Oncologico Isaac Gonzalez-Martinez
- MMR
Mismatch repair
- MSI
Microsatellite instability
- MSI-H
Microsatellite instability-high
- PCR
Polymerase chain reaction
- PRMC
Puerto Rico Medical Center
- US
United States
Contributor Information
Wilfredo E. De Jesus-Monge, Master of Science in Clinical Research Program, University of Puerto Rico, San Juan, Puerto Rico; Program in Gene Function and Expression, University of Massachusetts Medical School, Worcester, Massachusetts; Department of Medicine, University of Puerto Rico, San Juan, Puerto Rico; Clinical Research Center, Medical Sciences Campus, P.O. Box 365067, San Juan, Puerto Rico 00936-5067
Carmen Gonzalez-Keelan, Department of Pathology and Laboratory Medicine, University of Puerto Rico, San Juan, Puerto Rico.
Ronghua Zhao, Comprehensive Cancer Center, University of Puerto Rico, San Juan, Puerto Rico; Department of Surgery, University of Saskatchewan College of Medicine, Saskatoon, SK, Canada.
Stanley R. Hamilton, Division of Pathology and Laboratory Medicine, The University of Texas M.D. Anderson Cancer Center, Houston, Texas
Miguel Rodriguez-Bigas, Department of Surgical Oncology, The University of Texas M.D. Anderson Cancer Center, Houston, Texas.
Marcia Cruz-Correa, Department of Medicine, University of Puerto Rico, San Juan, Puerto Rico; Clinical Research Center, Medical Sciences Campus, P.O. Box 365067, San Juan, Puerto Rico 00936-5067 telephone: (787) 759-0306, fax: (787) 759-0305, marcia.cruz1@upr.edu; Comprehensive Cancer Center, University of Puerto Rico, San Juan, Puerto Rico; Department of Surgical Oncology, The University of Texas M.D. Anderson Cancer Center, Houston, Texas; Department of Biochemistry, University of Puerto Rico, San Juan, Puerto Rico.
References
- 1.Steward BW, Kleihues P, editors. World cancer report. IARC Press; Lyon: 2003. Colorectal cancer. [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Raut CP, Pawlik TM, Rodriguez-Bigas MA. Clinicopathologic features in colorectal cancer patients with microsatellite instability. Mutat Res. 2004;568:275–282. doi: 10.1016/j.mrfmmm.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 4.Soreide K, Janssen EA, Soiland H, et al. Microsatellite instability in colorectal cancer. Br J Surg. 2006;93:395–406. doi: 10.1002/bjs.5328. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Gu L, Wang H, et al. Mismatch repair processing of carcinogen-DNA adducts triggers apoptosis. Mol Cell Biol. 1999;19:8292–8301. doi: 10.1128/mcb.19.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci U S A. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koessler T, Azzato EM, Perkins B, et al. Common germline variation in mismatch repair genes and survival after a diagnosis of colorectal cancer. Int J Cancer. 2009;124:1887–1891. doi: 10.1002/ijc.24120. [DOI] [PubMed] [Google Scholar]
- 8.Wijnen JT, Brohet RM, van Eijk R, et al. Chromosome 8q23.3 and 11q23.1 variants modify colorectal cancer risk in Lynch syndrome. Gastroenterology. 2009;136:131–137. doi: 10.1053/j.gastro.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Lynch HT, Smyrk T. Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer. 1996;78:1149–1167. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1149::AID-CNCR1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Boland CR, Koi M, Chang DK, et al. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer. 2008;7:41–52. doi: 10.1007/s10689-007-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raevaara TE, Korhonen MK, Lohi H, et al. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129:537–549. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Hampel H, Stephens JA, Pukkala E, et al. Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology. 2005;129:415–421. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Shin YK, Heo SC, Shin JH, et al. Germline mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis colorectal cancer families. Hum Mutat. 2004;24:351. doi: 10.1002/humu.9277. [DOI] [PubMed] [Google Scholar]
- 14.Recommendations from the EGAPP Working Group. genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch HT, Boland CR, Rodriguez-Bigas MA, et al. Who should be sent for genetic testing in hereditary colorectal cancer syndromes? J Clin Oncol. 2007;25:3534–3542. doi: 10.1200/JCO.2006.10.3119. [DOI] [PubMed] [Google Scholar]
- 16.Lynch HT, Boland CR, Gong G, et al. Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet. 2006;14:390–402. doi: 10.1038/sj.ejhg.5201584. [DOI] [PubMed] [Google Scholar]
- 17.Ricciardiello L, Boland CR. Lynch syndrome (hereditary non-polyposis colorectal cancer): current concepts and approaches to management. Curr Gastroenterol Rep. 2005;7:412–420. doi: 10.1007/s11894-005-0012-2. [DOI] [PubMed] [Google Scholar]
- 18.De Jong AE, Morreau H, Van Puijenbroek M, et al. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology. 2004;126:42–48. doi: 10.1053/j.gastro.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 19.Soreide K. Molecular testing for microsatellite instability and DNA mismatch repair defects in hereditary and sporadic colorectal cancers--ready for prime time? Tumour Biol. 2007;28:290–300. doi: 10.1159/000110427. [DOI] [PubMed] [Google Scholar]
- 20.Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med. 2003;138:560–570. doi: 10.7326/0003-4819-138-7-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 21.Rouleau E, Lefol C, Bourdon V, et al. Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: application to MLH1 germline mutations in Lynch syndrome. Hum Mutat. 2009;30:867–875. doi: 10.1002/humu.20947. [DOI] [PubMed] [Google Scholar]
- 22.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 23.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 24.Pawlik TM, Raut CP, Rodriguez-Bigas MA. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers. 2004;20:199–206. doi: 10.1155/2004/368680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Correa M, Giardiello FM. Diagnosis and management of hereditary colon cancer. Gastroenterol Clin North Am. 2002;31:537–549. x. doi: 10.1016/s0889-8553(02)00009-2. [DOI] [PubMed] [Google Scholar]
- 26.Bleiker EM, Menko FH, Taal BG, et al. Screening behavior of individuals at high risk for colorectal cancer. Gastroenterology. 2005;128:280–287. doi: 10.1053/j.gastro.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Guillem JG, Wood WC, Moley JF, et al. ASCO/SSO review of current role of risk-reducing surgery in common hereditary cancer syndromes. J Clin Oncol. 2006;24:4642–4660. doi: 10.1200/JCO.2005.04.5260. [DOI] [PubMed] [Google Scholar]
- 28.Balmana J, Balaguer F, Castellvi-Bell S, et al. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008;45:557–563. doi: 10.1136/jmg.2008.059311. [DOI] [PubMed] [Google Scholar]
- 29.Sidelnikov E, Bostick RM, Flanders WD, et al. MutL-Homolog 1 expression and risk of incident, sporadic colorectal adenoma: search for prospective biomarkers of risk for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1599–1609. doi: 10.1158/1055-9965.EPI-08-0800. [DOI] [PubMed] [Google Scholar]
- 30.De Jesus-Monge WE, Gonzalez-Keelan C, Cruz-Correa M. Serrated adenomas. Current Gastroenterology Reports. 2009;11:420–427. doi: 10.1007/s11894-009-0063-x. [DOI] [PubMed] [Google Scholar]
- 31.Vasen HF, Moslein G, Alonso A, et al. Recommendations to improve identification of hereditary and familial colorectal cancer in Europe. Fam Cancer. 2009 doi: 10.1007/s10689-009-9291-3. Available via INTERNET. http://www.springerlink.com/content/u165m5k773817528/Cited 28 Oct 2009. [DOI] [PubMed]
- 32.Ponz de Leon M, Bertario L, Genuardi M, et al. Identification and classification of hereditary nonpolyposis colorectal cancer (Lynch syndrome): adapting old concepts to recent advancements. Report from the Italian Association for the Study of Hereditary Colorectal Tumors Consensus Group. Dis Colon Rectum. 2007;50:2126–2134. doi: 10.1007/s10350-007-9071-9. [DOI] [PubMed] [Google Scholar]
- 33.Lynch HT, Fusaro RM, Lynch PM. Sebaceous skin lesions as clues to hereditary non-polyposis colorectal cancer. J Invest Dermatol. 2006;126:2158–2159. doi: 10.1038/sj.jid.5700534. [DOI] [PubMed] [Google Scholar]
- 34.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de La Chapelle A. Microsatellite instability phenotype of tumors: genotyping or immunohistochemistry? The jury is still out. J Clin Oncol. 2002;20:897–899. doi: 10.1200/JCO.2002.20.4.897. [DOI] [PubMed] [Google Scholar]
- 36.Morales-Burgos A, Sanchez JL, Figueroa LD, et al. MSH-2 and MLH-1 protein expression in Muir Torre syndrome-related and sporadic sebaceous neoplasms. P R Health Sci J. 2008;27:322–327. [PMC free article] [PubMed] [Google Scholar]
- 37.Molaei M, Mansoori BK, Ghiasi S, et al. Colorectal cancer in Iran: immunohistochemical profiles of four mismatch repair proteins. Int J Colorectal Dis. 2009 doi: 10.1007/s00384-009-0784-1. Available via INTERNET. http://www.springerlink.com/content/932045231028jj4k/Cited 28 Oct 2009. [DOI] [PubMed]
- 38.Evans DG, Walsh S, Hill J, et al. Strategies for identifying hereditary nonpolyposis colon cancer. Semin Oncol. 2007;34:411–417. doi: 10.1053/j.seminoncol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek LI, Ligtenberg MJ, Willems RW, et al. Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am J Surg Pathol. 2008;32:1246–1251. doi: 10.1097/pas.0b013e31816401bb. [DOI] [PubMed] [Google Scholar]
- 41.Recommendations from the Interagency Committee for the Review of the Racial and Ethnic Standards to the Office of Management and Budget concerning changes to the standards for the classification of federal data on race and ethnicity. US Office of Management and Budget. 1997 Available via INTERNET. http://www.census.gov/population/www/socdemo/race/Directive_15.htmlCited 17 Oct 2009.
- 42.International Data Base. US Census Bureau, Population Division; Washington. : 2009. http://www.census.gov/ipc/www/idb/region.phpCited 18 Oct 2009. [Google Scholar]
- 43.US American Community Survey demographic and housing estimates: 2008. US Census Bureau, American Community Survey; Washington: 2008. http://factfinder.census.gov/servlet/ADPTable?_bm=y&-geo_id=01000US&-qr_name=ACS_2008_1YR_G00_DP5&-ds_name=ACS_2008_1YR_G00_&-_lang=en&-_caller=geoselect&-state=adp&-format=Cited 16 Oct 2009. [Google Scholar]
- 44.Puerto Rico American Community Survey demographic and housing estimates: 2008. US Census Bureau, American Community Survey; Washington: 2008. http://factfinder.census.gov/servlet/ADPTable?_bm=y&-state=adp&-context=adp&-qr_name=ACS_2008_1YR_G00_DP5&-ds_name=ACS_2008_1YR_G00_&-tree_id=308&-_caller=geoselect&-geo_id=04000US72&-format=&-_lang=enCited 17 Oct 2009. [Google Scholar]
- 45.Curado MP, Edwards B, Shin HR, et al. Cancer incidence in five continents. International Agency for Research on Cancer; Lyon: 2007. http://www-dep.iarc.fr/Cited 20 Oct 2009. [Google Scholar]
- 46.Figueroa-Valles NR. Boletin del Registro de Cancer. Puerto Rico Cancer Central Registry. 2008 Available via INTERNET. http://www.salud.gov.pr/RCancer/Documents/Vol.%201%20No.%202.pdfCited 17 Oct 2009.
- 47.Brim H, Mokarram P, Naghibalhossaini F, et al. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashktorab H, Smoot DT, Farzanmehr H, et al. Clinicopathological features and microsatellite instability (MSI) in colorectal cancers from African Americans. Int J Cancer. 2005;116:914–919. doi: 10.1002/ijc.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandey V, Prabhu JS, Payal K, et al. Assessment of microsatellite instability in colorectal carcinoma at an Indian center. Int J Colorectal Dis. 2007;22:777–782. doi: 10.1007/s00384-006-0241-3. [DOI] [PubMed] [Google Scholar]
- 50.Ashktorab H, Brim H, Al-Riyami M, et al. Sporadic colon cancer: mismatch repair immunohistochemistry and microsatellite instability in Omani subjects. Dig Dis Sci. 2008;53:2723–2731. doi: 10.1007/s10620-007-0189-3. [DOI] [PubMed] [Google Scholar]
- 51.Erdamar S, Ucaryilmaz E, Demir G, et al. Importance of MutL homologue MLH1 and MutS homologue MSH2 expression in Turkish patients with sporadic colorectal cancer. World J Gastroenterol. 2007;13:4437–4444. doi: 10.3748/wjg.v13.i33.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin HY, Liu X, Lin VK, et al. Detection of mismatch repair gene germline mutation carrier among Chinese population with colorectal cancer. BMC Cancer. 2008;8:44. doi: 10.1186/1471-2407-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen LH, Lindebjerg J, Byriel L, et al. Strategy in clinical practice for classification of unselected colorectal tumours based on mismatch repair deficiency. Colorectal Dis. 2008;10:490–497. doi: 10.1111/j.1463-1318.2007.01378.x. [DOI] [PubMed] [Google Scholar]
- 54.Boardman LA, Lanier AP, French AJ, et al. Frequency of defective DNA mismatch repair in colorectal cancer among the Alaska Native people. Cancer Epidemiol Biomarkers Prev. 2007;16:2344–2350. doi: 10.1158/1055-9965.EPI-07-0577. [DOI] [PubMed] [Google Scholar]
- 55.Wright CL, Stewart ID. Histopathology and mismatch repair status of 458 consecutive colorectal carcinomas. Am J Surg Pathol. 2003;27:1393–1406. doi: 10.1097/00000478-200311000-00001. [DOI] [PubMed] [Google Scholar]
- 56.Coggins RP, Cawkwell L, Bell SM, et al. Association between family history and mismatch repair in colorectal cancer. Gut. 2005;54:636–642. doi: 10.1136/gut.2003.017517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xicola RM, Llor X, Pons E, et al. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99:244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 58.Piñol V, Castells A, Andreu M, et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 59.Terdiman JP, Gum JR, Jr, Conrad PG, et al. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology. 2001;120:21–30. doi: 10.1053/gast.2001.20874. [DOI] [PubMed] [Google Scholar]
- 60.Christensen M, Katballe N, Wikman F, et al. Antibody-based screening for hereditary nonpolyposis colorectal carcinoma compared with microsatellite analysis and sequencing. Cancer. 2002;95:2422–2430. doi: 10.1002/cncr.10979. [DOI] [PubMed] [Google Scholar]
- 61.Wahlberg SS, Schmeits J, Thomas G, et al. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 62.Park JG, Vasen HF, Park YJ, et al. Suspected HNPCC and Amsterdam criteria II: evaluation of mutation detection rate, an international collaborative study. Int J Colorectal Dis. 2002;17:109–114. doi: 10.1007/s003840100348. [DOI] [PubMed] [Google Scholar]
- 63.Hitchins M, Williams R, Cheong K, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 64.Bermejo JL, Eng C, Hemminki K. Cancer characteristics in Swedish families fulfilling criteria for hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1889–1899. doi: 10.1053/j.gastro.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Woerner SM, Kloor M, Mueller A, et al. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005;24:2525–2535. doi: 10.1038/sj.onc.1208456. [DOI] [PubMed] [Google Scholar]
- 66.Plaschke J, Engel C, Kruger S, et al. Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J Clin Oncol. 2004;22:4486–4494. doi: 10.1200/JCO.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 67.Lim SB, Jeong SY, Lee MR, et al. Prognostic significance of microsatellite instability in sporadic colorectal cancer. Int J Colorectal Dis. 2004;19:533–537. doi: 10.1007/s00384-004-0596-2. [DOI] [PubMed] [Google Scholar]
- 68.Cruz-Correa M, Giardiello FM. Familial adenomatous polyposis. Gastrointestinal Endoscopy. 2003;58:885–894. doi: 10.1016/s0016-5107(03)02336-8. [DOI] [PubMed] [Google Scholar]
- 69.Half ED, Bercovich D, Rozen P. Familial adenomatous polyposis. Orphanet J Rare Dis. 2009;4:22. doi: 10.1186/1750-1172-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rozen P, Macrae F. Familial adenomatous polyposis: The practical applications of clinical and molecular screening. Fam Cancer. 2006;5:227–235. doi: 10.1007/s10689-005-5674-2. [DOI] [PubMed] [Google Scholar]
- 71.Puerto Rico general demographic characteristics: 2005. US Census Bureau, American Community Survey; Washington: 2005. http://factfinder.census.gov/servlet/ADPTable?_bm=y&-state=adp&-context=adp&-qr_name=ACS_2005_EST_G00_DP1&-ds_name=ACS_2005_EST_G00_&-tree_id=305&-redoLog=true&-all_geo_types=N&-_caller=geoselect&-geo_id=04000US72&-format=&-_lang=enCited 16 Oct 2009. [Google Scholar]
- 72.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fleming ID, Cooper JS, Henson DE, et al., editors. AJCC cancer staging manual. 5. Lippincott-Raven; Philadelphia: 1997. [Google Scholar]
- 74.Soto-Salgado M, Suarez E, Calo W, et al. Incidence and mortality rates for colorectal cancer in Puerto Rico and among Hispanics, non-Hispanic whites, and non-Hispanic blacks in the United States, 1998–2002. Cancer. 2009;115:3016–3023. doi: 10.1002/cncr.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jover R, Zapater P, Castells A, et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 76.Carethers JM, Smith EJ, Behling CA, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 77.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grover S, Syngal S. Genetic testing in gastroenterology: Lynch syndrome. Best Pract Res Clin Gastroenterol. 2009;23:185–196. doi: 10.1016/j.bpg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 80.Beiner ME, Rosen B, Fyles A, et al. Endometrial cancer risk is associated with variants of the mismatch repair genes MLH1 and MSH2. Cancer Epidemiol Biomarkers Prev. 2006;15:1636–1640. doi: 10.1158/1055-9965.EPI-06-0257. [DOI] [PubMed] [Google Scholar]
- 81.Yoon SN, Ku JL, Shin YK, et al. Hereditary nonpolyposis colorectal cancer in endometrial cancer patients. Int J Cancer. 2008;122:1077–1081. doi: 10.1002/ijc.22986. [DOI] [PubMed] [Google Scholar]
- 82.Park JG, Kim DW, Hong CW, et al. Germ line mutations of mismatch repair genes in hereditary nonpolyposis colorectal cancer patients with small bowel cancer: International Society for Gastrointestinal Hereditary Tumours Collaborative Study. Clin Cancer Res. 2006;12:3389–3393. doi: 10.1158/1078-0432.CCR-05-2452. [DOI] [PubMed] [Google Scholar]
- 83.Scaife CL, Rodriguez-Bigas MA. Lynch syndrome: implications for the surgeon. Clin Colorectal Cancer. 2003;3:92–98. doi: 10.3816/CCC.2003.n.015. [DOI] [PubMed] [Google Scholar]
- 84.Watson P, Riley B. The tumor spectrum in the Lynch syndrome. Fam Cancer. 2005;4:245–248. doi: 10.1007/s10689-004-7994-z. [DOI] [PubMed] [Google Scholar]
- 85.Watson P, Lynch HT. Cancer risk in mismatch repair gene mutation carriers. Fam Cancer. 2001;1:57–60. doi: 10.1023/a:1011590617833. [DOI] [PubMed] [Google Scholar]
- 86.Manchanda R, Menon U, Michaelson-Cohen R, et al. Hereditary non-polyposis colorectal cancer or Lynch syndrome: the gynaecological perspective. Curr Opin Obstet Gynecol. 2009;21:31–38. doi: 10.1097/GCO.0b013e32831c844d. [DOI] [PubMed] [Google Scholar]
- 87.Brown GJ, St John DJ, Macrae FA, et al. Cancer risk in young women at risk of hereditary nonpolyposis colorectal cancer: implications for gynecologic surveillance. Gynecol Oncol. 2001;80:346–349. doi: 10.1006/gyno.2000.6065. [DOI] [PubMed] [Google Scholar]
- 88.Schmeler KM, Lu KH. Gynecologic cancers associated with Lynch syndrome/HNPCC. Clin Transl Oncol. 2008;10:313–317. doi: 10.1007/s12094-008-0206-9. [DOI] [PubMed] [Google Scholar]
- 89.Dionigi G, Bianchi V, Villa F, et al. Differences between familial and sporadic forms of colorectal cancer with DNA microsatellite instability. Surg Oncol. 2007;16:S37–S42. doi: 10.1016/j.suronc.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 90.Aaltonen LA, Peltomaki P, Mecklin JP, et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54:1645–1648. [PubMed] [Google Scholar]
- 91.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuismanen SA, Holmberg MT, Salovaara R, et al. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol. 2000;156:1773–1779. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gryfe R, Gallinger S. Microsatellite instability, mismatch repair deficiency, and colorectal cancer. Surgery. 2001;130:17–20. doi: 10.1067/msy.2001.112738. [DOI] [PubMed] [Google Scholar]
- 94.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 95.O’Brien MJ. Hyperplastic and serrated polyps of the colorectum. Gastroenterol Clin North Am. 2007;36:947–968. viii. doi: 10.1016/j.gtc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 96.Esteller M, Fraga MF, Guo M, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 97.Chirieac LR, Chen L, Catalano PJ, et al. Phenotype of microsatellite-stable colorectal carcinomas with CpG island methylation. Am J Surg Pathol. 2005;29:429–436. doi: 10.1097/01.pas.0000155144.53047.7d. [DOI] [PubMed] [Google Scholar]
- 98.van Rijnsoever M, Grieu F, Elsaleh H, et al. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kambara T, Simms LA, Whitehall VL, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Al-Kuraya KS, Bavi PP, Ezzat AA, et al. Colorectal carcinoma from Saudi Arabia. Analysis of MLH-1, MSH-2 and p53 genes by immunohistochemistry and tissue microarray analysis. Saudi Med J. 2006;27:323–328. [PubMed] [Google Scholar]
- 101.Samowitz WS, Curtin K, Ma KN, et al. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10:917–923. [PubMed] [Google Scholar]
- 102.Chen JR, Chiang JM, Changchien CR, et al. Mismatch repair protein expression in Amsterdam II criteria-positive patients in Taiwan. Br J Surg. 2008;95:102–110. doi: 10.1002/bjs.5786. [DOI] [PubMed] [Google Scholar]
- 103.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 104.Mecklin JP, Jarvinen HJ. Clinical features of colorectal carcinoma in cancer family syndrome. Dis Colon Rectum. 1986;29:160–164. doi: 10.1007/BF02555012. [DOI] [PubMed] [Google Scholar]
- 105.Watson N, Grieu F, Morris M, et al. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J Mol Diagn. 2007;9:472–478. doi: 10.2353/jmoldx.2007.060162. [DOI] [PMC free article] [PubMed] [Google Scholar]