Abstract
Cell fates are instructed by signals emitted from specialized cell populations called organizers. The study of epidermal patterning in Drosophila is contributing novel insights concerning the establishment and action of such organizers. Juxtaposed rows of cells express either the wingless or hedgehog signaling molecules and thereby act as organizers of segment pattern. These signals mediate a mutually re-enforcing interaction between the two rows of cells to sustain organizer function. In a distinct and subsequent phase, wingless and hedgehog act to specify the fates of cells.
Introduction
One of the early successes of experimental embryology was the demonstration of the importance of induction to embryonic development. In this process, organizing centers emit signals that direct the choice of fate in surrounding cells [1]; for example, a signaling center in the posterior portion of the limb bud, the zone of polarizing activity (ZPA), organizes the pattern of digits across the entire limb [2]. Such organizers also operate within the insect epidermis, in which signaling centers near the borders of each segment specify cell fate [3,4]. Although the organisms favored by early experimental embryologists were well suited to the transplantation experiments that defined organizers, genetically tractable organisms are more suited to investigating the mechanisms by which organizers act.
In Drosophila, genetic screens have identified mutations in many of the genes involved in patterning the epidermis [5,6]. Genetic and molecular analyses of these segment polarity genes have revealed that two signaling molecules, encoded by wingless (wg) and hedgehog (hh), specify most of the epidermal cell fates. The wg product is a member of the evolutionarily conserved Wnt family of signaling proteins [7,8], and the hh product defines a novel class of conserved developmental signaling molecules [9–12,13•–16•]. Recent experiments have demonstrated that not only are the molecules conserved across species, but their function is also conserved. This observation has stimulated broad interest in the action of these two proteins, as well as other segment polarity genes. In this review, we outline a new framework for thinking about the action of these patterning genes. We focus on the establishment and action of the wg- and hh-expressing cells as organizers of pattern. The other segment polarity genes fit into this framework as components of signal transduction pathways, or as factors required to maintain and accurately position the signaling centers.
Steps in segmental patterning
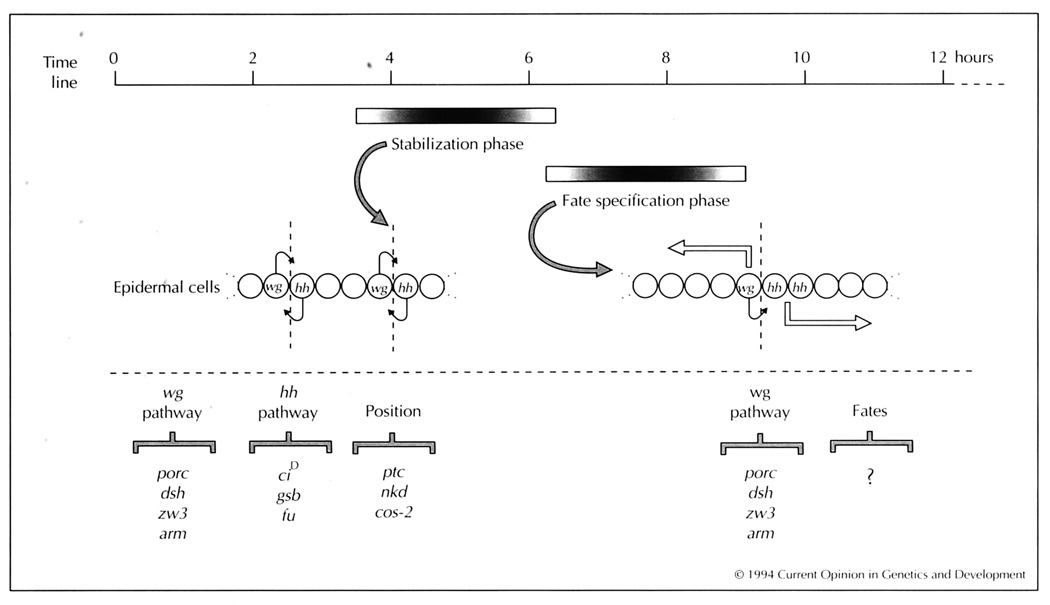
Early in development, a cascade of regulatory genes generates stripes of localized transcription factors that define repeating units along the body axis, called parasegments [17]. Within each parasegment, the transcription factors initiate the expression of wg in one row of cells, and the expression of both the secreted protein Hh [9–12] and the transcription factor Engrailed (En) in the adjacent, posterior row [18–21] (Fig. 1). These two rows of cells flank the boundary between adjacent parasegments and have been identified as sources of signaling. Each row of cells signals at two different times during segmentation, with distinct outcomes [22–26,27•,28••].
Fig. 1.
The stabilization and fate specification phases of epidermal patterning. Cells first form at two hours after fertilization, and the epidermal cells differentiate at twelve hours (time line). Shaded bars indicate the approximate period during which signals stabilize wingless (wg) and hedgehog (hh) expression, or specify cell fates (shading reflects uncertainty in timing). Line of circles represent a short antero-posterior strip of epidermal cells. During the early period, wg- and hh-expressing cells signal to one another (short arrows) across the parasegment boundary (vertical dashed line). During the late period, wg specifies fates anteriorly (leftward open arrow) and the fate of the adjacent hh-expressing cell [which co-expresses engrailed (en)]. hh function specifies cell fates posteriorly (rightward open arrow). The other segment polarity genes are grouped below according to their postulated roles in either the wg or hh signaling pathways, or in restricting the position of the signaling cells. porc—porcupine, dsh—dishevelled, zw3—zeste-white3, arm—armadillo, CiD—cubitus interruptusD, gsb—gooseberry, fu—fused, ptc—patched, nkd—naked, and cos-2—costal-2. The mechanism by which pair-rule segmentation genes first establish wg and en/hh expression is reviewed in [60].
The early phase: stabilization of the signaling centers
The wg- and hh-expressing cells signal to each other, reinforcing gene expression in each cell (Fig. 1; for a comprehensive review, see [29]). It has been demonstrated that the secreted glycoprotein Wg is the ligand required for continued expression of en and hh in the neighboring cells [22–24,30••]. Reciprocally, it has been proposed that the secreted protein Hh is the signal that maintains wg expression [31,32].
Several segment polarity genes act in the signal transduction pathways that operate during the stabilization phase. Although no Wg receptor has yet been identified, the genes porcupine, dishevelled, zeste-white3 and armadillo have been implicated in the sending or transduction of the wg signal (Fig. 1; [33,34•,35•,36••,37,38•–40•]; review in preparation, J Klingensmith, R Nusse, personal communication). One target of this transduction cascade may be en autoactivation, and en, in turn, positively regulates hh expression [25,41].
Less is known about transduction of the putative hh signal, although some genetic evidence [42,43••] implicates the gooseberry and cubitus interruptusD transcription factors [44,45] and the fused serine/threonine kinase [46] in this pathway (Fig. 1).
At this stage in patterning, both wg and hh signaling appear to act over only short distances [30••,32]. Locally restricted signaling ensures that the domain of cells expressing either wg or hh remains narrow during development, even though the width of the parasegment grows threefold; for instance, as cell division and movements occur, some en/hh-expressing cells are displaced from the interface with wg-expressing cells and the expression of en is shut off in cells furthest away from the sustaining wg influence [47••]. Such refinement in the domains of en and wg expression is crucial for patterning, as several studies have demonstrated that widened domains of either wg or en/hh expression cause severe mis-specification of cell fates [22,23,32,48,49].
The late phase: signaling centers specify fates
The cell signaling events executed during the stabilization phase do not specify the final fate of cells, as alterations in the expression of either signal at later times dramatically affect cell fates [24,26,28••,32,48,50,51]. After the positions of the signaling centers have stabilized, however, the two signals then act to specify the distinct cell types across the parasegment [24,26,28••,50,51] (Fig. 1).
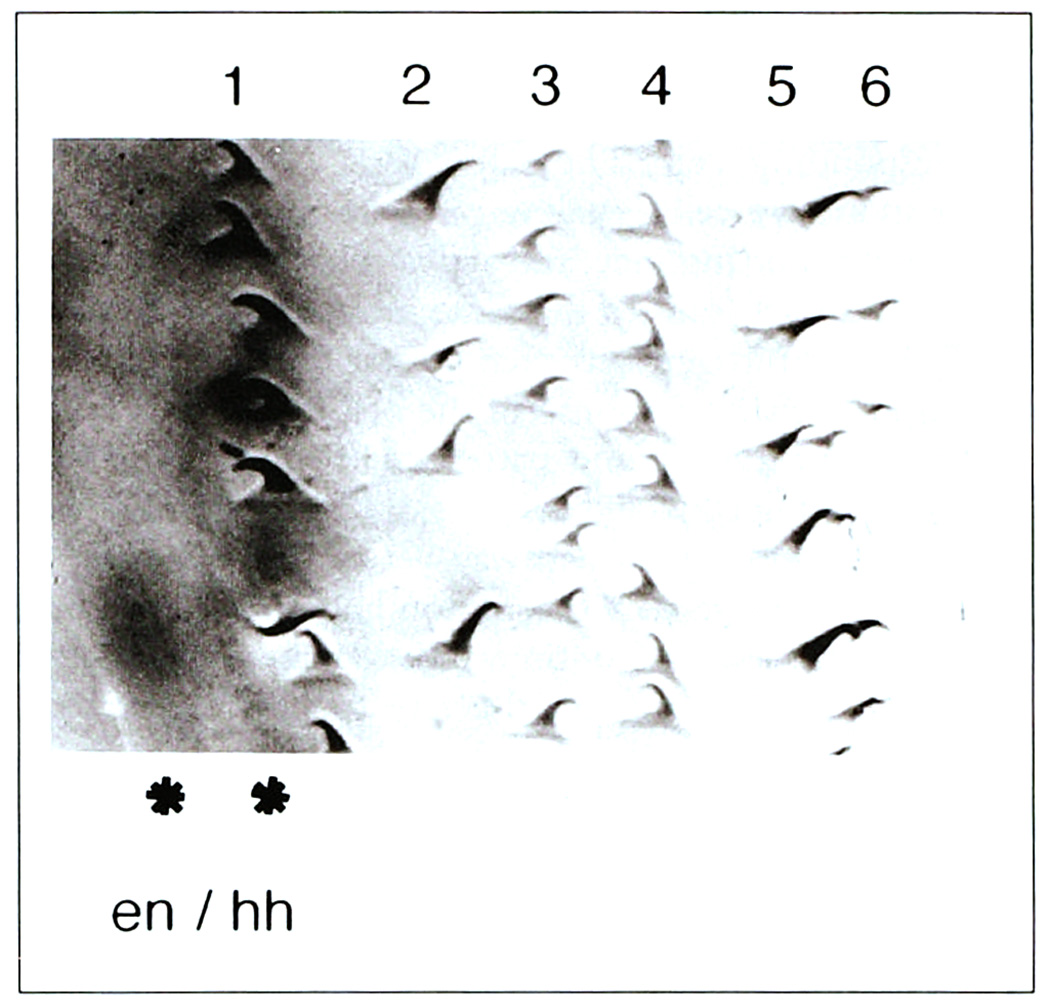
Roughly ten diverse epidermal cell types are generated within each parasegment. The final fate adopted by a cell is visualized at differentiation when the cytoskeleton distorts cells into distinct shapes (Fig. 2) [52]. Each cell then secretes a cuticular covering that indelibly reflects its shape change; therefore, cellular identity is easily visualized in the stereotyped pattern of segmentally repeated cuticular features [53].
Fig. 2.
Epidermal cell fate. The photograph shows a portion of the ventral epidermis within a parasegment (anterior to the left, posterior to the right). Cell bodies are below the plane of focus. Six rows of cuticular protrusions, called denticles, are indicated, and each row exhibits unique characteristics of size and orientation that reflect the distinct positional identity of the row of underlying epidermal cells. Two rows of cells at the left express en/hh as revealed by an en–lacZ reporter gene (*; dark stain). Note that the posterior row adopts a denticle row #1 fate, whereas the anterior row adopts a smooth cell fate. This smooth fate is instructed by late wg input [26], as are other smooth cell fates further to the anterior in the segment [24] (not shown).
The wg input is necessary for several rows of cells anterior to the wg-expressing cells to adopt their normal smooth cell fate [24,50]. The wg product also signals in the posterior direction, but in this case its effect is local. Two rows of cells posterior to the wg-expressing cells express en/hh. The most posterior row of en/hh-expressing cells adopts a denticle fate. However, wg input to the more anterior row, instructs those cells to adopt the smooth fate (Fig. 2) [26].
The hh gene appears to signal many of the remaining cell fates across the parasegment [28••]. In the dorsal epidermis, Hh acts as a morphogen in executing this role, whereas ventrally, Hh may cooperate with an unidentified signal from the en-expressing cells ([54••]; S DiNardo, unpublished data).
Uncoupling stabilization from fate specification
The mutual dependence of wg and hh expression during the stabilization phase initially masked their later, separate roles in fate specification. The standard genetic approach by which to uncover a role for a gene is to remove gene function and analyze the consequences on cell fate specification. If this gene is required for the expression of another signaling molecule, however, it is difficult to determine which ligand is responsible for which fate changes; for example, any of the changes in cell fate observed in a wg mutant could be attributed to loss of direct action of Wg, to subsequent loss of Hh activity, or to combined loss of both Hh and Wg activity. Two kinds of experiment have enabled researchers to distinguish between these possibilities and have led to the above proposal that wg and hh signaling centers operate as the two organizers of segmental pattern.
First, a temperature-sensitive allele of wg made it possible to inactivate wg at various times during development [24,28••,50,55]. Loss of Wg activity during the later, fate specification phase no longer affects the continued expression of en or of the hh signal [24,25]. This has provided a way in which to analyze the contribution of wg to fate specification without affecting the fates specified by hh signaling. Second, a key role for hh has been uncovered through experiments that bypass the stabilization phase, maintaining the expression of hh in the absence of any wg input. In this manner, we have discovered that hh can organize substantial pattern in the dorsal epidermis independently of wg signaling [28••].
The difficulty in identifying the separate early and late roles for wg and hh illustrates a general problem concerning all segment polarity genes. Does a given gene act early, during stabilization, or does it act both early and later, during fate specification?
Components of the wg signal transduction pathway act both early and late. These genes were first identified through their action during the stabilization phase, in which the target is en gene expression. However, the same signal transduction cassette mediates wg signaling during limb and wing patterning [35•,37,38•–40•], even though the target is not en expression. Thus, it is likely that porcupine, dishevelled, zeste-white3, and armadillo mediate wg function during fate specification also (Fig. 1).
In contrast to this, the segment polarity genes implicated in transduction of the hh signal act only early. In embryos lacking gooseberry, cubitus interruptusD or fused, wg expression is lost, but hh-dependent cell types are still specified ([42,56]; J Heemskerk, S DiNardo, PH O’Farrell, unpublished data). Therefore, these three genes act only during early hh signaling, when hh is needed for the maintenance of wg expression, and are not required in the hh pathway for signaling cell fates.
Particular segment polarity genes position the signaling centers
Segment polarity mutants that result in mis-specification of some cell fates were first thought to define genes involved directly in the specification of the affected cell types. Genes in this class include naked, patched, and costal-2. We argue that mutations in these genes affect cell fates only indirectly and do so because they change the distribution of the important signaling molecules Wg and Hh. The changes in Wg and Hh expression precede cell fate specification [22,23], and the ultimate changes in cell fate can be explained by the altered positions of the two signaling centers, or the distance over which they now act ([26]; J Heemskerk, S DiNardo, PH O’Farrell, unpublished data). One test of this hypothesis is to determine whether the cell fates missing in a mutant background can be restored by manipulating organizer function without restoring the missing gene product; for example, naked mutants mis-express wg and lack several cell types [22,26], but if wg function is inactivated after its mis-expression, but prior to final fate specification, the missing cell types are restored [26]. Therefore, naked activity is not required for the fates of these cells, but, rather, for the control of where wg is expressed. Analogous experiments have not yet been carried out for patched and costal-2. Nevertheless, as mutations in these genes also change the position of the organizers, we postulate that these genes do not act directly in establishing cell fate, but, rather, constitute a genetic circuit that assures the accurate positioning of the signaling centers during the stabilization phase (Fig. 2). The patched and naked genes may execute this role by modulating the transduction of either the wg or hh signal during stabilization, perhaps by encoding components of the signal transduction apparatus itself [31,32,54••]. It is presently less clear how costal-2 acts to modify patterns of wg and hh expression. The view that these genes do not specify cell fate directly contrasts with a recent proposal by Bejsovec and Wieschaus [54••].
Conclusions
The wg and hh genes act as organizers of epidermal pattern. Most other segment polarity genes fit into this framework as components of the signal transduction apparatus, or as factors required to maintain or accurately position these signaling centers. Thus, few if any of the other segment polarity genes act specifically in signaling final cell fate; instead, most act in the feedback between the adjacent signaling centers.
Reinforcement is a general property of organizers
The mutual reinforcing signals that occur during the stabilization phase may be a general feature of organizers. In vertebrates, cell signaling interactions appear to sustain organizers; for instance, in the limb bud, feedback from the apical ectodermal ridge is required for the maintenance of the ZPA [57]. Recent analysis strongly suggests that the signaling molecule in the apical ectodermal ridge is fibroblast growth factor 4 [57,58], whereas the vertebrate hh homolog (vhh) encodes a ZPA signal [13•]. Although such feedback will maintain an organizer, it may also serve a larger purpose. Neighboring organizers that rely on mutually reinforcing signals would remain highly localized during growth and proliferation. This would constrain each organizer from inappropriately extending its influence and thereby disrupting overall pattern.
Distinct responses to the same signal
Early wg input stabilizes en and hh expression, whereas the later input specifies the smooth cell fate. It appears that the same components transduce the wg signal at both times. At present, we do not understand how the same transduction pathway leads to different read-outs from the responding cell. The same issue is unresolved in other inductive cell signaling processes used during development. For instance, the activation of most receptor tyrosine kinases leads to the same intracellular cascade of Ras→Raf→mitogen-activated protein (MAP) kinase interactions, yet the response of the cell differs depending on the tissue type being patterned (reviewed in [59]). In the fly epidermis, either there are novel components in the wg transduction pathway yet to be identified, or the available targets in the responding en/hh cell must be different at the two times. Perhaps a solution will be found by focusing on the fate specification phase, in which more components the wg pathway need to be identified.
Acknowledgements
Work in the lab of S DiNardo is supported by ACS grant #DB-51 and NIH grant #GM45747. Work in the lab of PH O’Farrell is supported by the NSF. S Dougan is supported by NIH pre-doctoral training grant #GM07982.
Apologies to PA Lawrence for the partially borrowed title.
Abbreviations
- en
engrailed
- hh
hedgehog
- wg
wingless
- ZPA
zone of polarizing activity
Contributor Information
Stephen DiNardo, The Rockefeller University, 1230 York Avenue, New York, New York 10021-6399, USA..
Jill Heemskerk, Department of Physiology, Columbia University College of Physicians and Surgeons, 630 West 168th Street, New York, New York 10032, USA..
Scott Dougan, The Rockefeller University, 1230 York Avenue, New York, New York 10021-6399, USA..
Patrick H O’Farrell, Department of Biochemistry and Biophysics, University of California at San Francisco, San Francisco 94143, USA..
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
• of outstanding interest
- 1.Spemann H. Embryonic Development and Induction. New Haven: Yale University Press; 1993; reprinted. In: Moore J, editor. Great Books in Experimental Biology. vol 10. New York, London: Garland Publishing; 1988. [Google Scholar]
- 2.Tickle C, Summerbell O, Wolpert I. Positional Signalling and Specification of Digits in Chick limb Morphogenesis. Nature. 1975;254:199–202. doi: 10.1038/254199a0. [DOI] [PubMed] [Google Scholar]
- 3.Locke M. The Cuticular Pattern in an Insect, Rhodnius prolixus. J Exp Biol. 1959;36:459–477. [Google Scholar]
- 4.Nubler-Jung K. Pattern Stability in the Insect Segment. I. Pattern Reconstitution by Intercalary Regeneration and Cell Sorting in Dysdercus intermedius. Roux Arch Dev Biol. 1977;183:17–40. doi: 10.1007/BF00849032. [DOI] [PubMed] [Google Scholar]
- 5.Nusslein-Volhard C, Wieschaus E. Mutations Affecting Segment Number and Polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 6.Perrimon N, Engstrom L, Mahowald AP. Zygotic Lethal with Specific Maternal Effect Phenotypes in Drosophila melanogaster. I. Loci on the X Chromosome. Genetics. 1989;121:333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rijsewijk F, Scheurmann M, Wagenaar E, Parren P, Weigel D, Nusse R. The Drosophila Homolog of the Mouse Mammary Oncogene int-1 is Identical to the Segment Polarity Gene wingless. Cell. 1987;50:649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera CV, Alonso MC, Johnston P, Phillips RG, Lawrence PA. Phenocopies Induced with Antinsense RNA Identify the wingless Gene. Cell. 1987;50:659–663. doi: 10.1016/0092-8674(87)90039-0. [DOI] [PubMed] [Google Scholar]
- 9.Mohler J, Vani K. Molecular Organization and Embryonic Expression of the hedgehog Gene Involved in Cell–Cell Communication in Segmental Patterning of Drosophila. Development. 1992;115:957–971. doi: 10.1242/dev.115.4.957. [DOI] [PubMed] [Google Scholar]
- 10.Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog Gene is Expressed Specifically in Posterior Compartment Cells and is a Target of engrailed Regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- 11.Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and Localized Transcription Suggest a Role in Positional Signaling for Products of the Segmentation Gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 12.Tabata T, Kornberg B. Hedgehog is a Signalling Protein with a Key Role in Patterning Drosophila Imaginal Discs. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 13. Riddle R, Johnson RL, Laufer E, Tabin C. Sonic hedgehog Mediates the Polarizing Activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. The chicken hedgehog homolog acts as the signal from the ZPA that organizes anterior/posterior pattern across the vertebrate limb.
- 14. Echelard Y, Epstein DP, St-Jacques B, Shen L, Mohler J, McMahon IA, McMahon AP. Sonic hedgehog, a Member of a Family of Putative Signaling Molecules is Implicated in the Regulation of CNS Polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. The mouse hedgehog homolog may act in patterning the ventral neural lube, as ectopic expression can induce floor plate markers.
- 15. Krauss S, Concordet J-P, Ingham PW. A Functionally Conserved Homolog of the Drosophila Segment Polarity Gene hedgehog is Expressed in Tissues with Polarising Activity in Zebrafish Embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. The zebrafish hedgehog homolog can induce floor plate markers upon ectopic expression of hedgehog. There is functional homology between Drosophila and fish hedgehog, as ectopically expressed fish and Drosophila genes can cause the same pattern changes in Drosophila.
- 16. Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i, Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell M, Dodd J. Floor Plate and Motor Neuron Induction by vhh-1, a Vertebrate Homolog of hedgehog Expressed by the Notochord. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. The vertebrate hedgehog homolog induces floor plate and motor neurons in neural plate explants, suggesting that hedgehog contributes to the inductive capacity of the notochord.
- 17.Martinez-Arias A, Lawrence PA. Parasegments and Compartments in the Drosophila Embryo. Nature. 1985;313:639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- 18.Kornberg T, Siden I, O’Farrell P, Simon AM. The engrailed locus of Drosophila: In Situ Localization of Transcripts Reveals Compartment-Specific Expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- 19.Fjose A, McGinnis WJ, Gehring WJ. Isolation of a Homoeobox-Containing Gene from the engrailed Region of Drosophila and the Spatial Distribution of its Transcripts. Nature. 1985;313:284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- 20.Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed Locus of Drosophila: Structural Analysis of an Embryonic Transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- 21.Jaynes JB, O’Farrell PH. Activation and Repression of Transcription by Homeodomain-Containing Proteins that Bind a Common Site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Arias A, Baker NE, Ingham PW. Role of Segment Polarity Genes in the Definition and Maintenance of Cell States in the Drosophila Embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- 23.DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O’Farrell PH. Two-Tiered Regulation of Spatially Patterned engrailed Gene Expression during Drosophila Embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bejsovec A, Martinez-Arias A. Roles of wingless in Patterning the Larval Epidermis of Drosophila. Development. 1991;113:471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- 25.Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH. Multiple Modes of engrailed Regulation in the Progression Towards Cell Fate Determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougan ST, DiNardo S. wingless Generates Cell Type Diversity among engrailed Expressing Cells. Nature. 1992;360:347–350. doi: 10.1038/360347a0. [DOI] [PubMed] [Google Scholar]
- 27. Sampedro J, Johnston P, Lawrence PA. A Role for wingless in the Segmental Gradient of Drosophila? Development. 1993;117:677–687. doi: 10.1242/dev.117.2.677. The authors propose that early wingless function is to seal the parasegment borders so that later fate specification can take place.
- 28. Heemskerk J, DiNardo S. Drosophila hedgehog Acts as a Morphogen in Cellular Patterning. Cell. 1994;76:449–460. doi: 10.1016/0092-8674(94)90110-4. This paper uncovered the organizing role of en-expressing cells in patterning the epidermis and showed that hh encoded the signal that specified cell fates, acting as a graded morphogen. This leads to the model for two organizing centers for pattern across the segment.
- 29.Martinez Arias A. Development and Patterning of the Larval Epidermis of Drosophila. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 517–608. [Google Scholar]
- 30. Cumberledge S, Krasnow MA. Intercellular Signalling in Drosophila Segment Formation Reconstructed in Vitro. Nature. 1993;363:549–552. doi: 10.1038/363549a0. The authors separately purified engrailed- or wingless-expressing cells from embryos and demonstrated directly that wingless input to the en cell was necessary and probably sufficient for the maintenance of en gene expression. The wg signaling appears to be contact dependent.
- 31.Ingham P, Taylor A, Nakano Y. Role of the Drosophila patched Gene in Positional Signalling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- 32.Ingham PW. Localized hedgehog Activity Controls Spatial Limits of wingless Transcription in the Drosophila Embryo. Nature. 1993;366:560–562. doi: 10.1038/366560a0. [DOI] [PubMed] [Google Scholar]
- 33.Wieschaus E, Riggleman R. Autonomous Requirements for the Segment Polarity Gene armadillo during Drosophila Embryo-genesis. Cell. 1987;49:177–184. doi: 10.1016/0092-8674(87)90558-7. [DOI] [PubMed] [Google Scholar]
- 34. Siegfried E, Wilder EL, Perrimon N. Components of wingless Signalling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. Through genetic epistasis using multiple mutants, the relationship of several genes that disrupt wingless signaling in the embryo was investigated. The results suggest that porcupine and dishevelled act upstream of the zeste-white3 kinase, whereas armadillo acts downstream.
- 35. Noordermeer J, Klingensmith J, Perrimon N, Nusse R. dishevelled and armadillo Act in the Wingless Signalling Pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. This paper investigates which genes are required to mediate the wingless signal by using ectopic expression of wingless in combination with particular mutants. This demonstrates that wingless acts through dishevelled and armadillo to control en expression and pattern.
- 36. Peifer M, Sweeton D, Casey M, Wieschaus E. wingless Signal and Zeste-White 3 Kinase Trigger Opposing Changes in the Intracellular Distribution of armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. In response to wingless input, cells increase the level of cytoplasmic armadillo (arm) protein relative to the membrane-associated form of arm. zeste-white3 also regulates arm distribution, and epistasis demonstrates that arm acts directly in wg signaling, and downstream of wg input.
- 41.Siegfried E, Chou T-B, Perrimon N. wingless Signaling Acts through zeste-white 3, the Drosophila Homolog of Glycogen Synthase Kinase-3, to Regulate engrailed and Establish Cell Fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 37.Peifer M, Rauskolb C, Williams M, Riggleman B, Wieschaus E. The Segment Polarity Gene armadillo Interacts with the wingless Signalling Pathway in Both Embryonic and Adult Pattern Formation. Development. 1991;111:1029–1043. doi: 10.1242/dev.111.4.1029. [DOI] [PubMed] [Google Scholar]
- 38. Theisen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh JL. dishevelled is Required during wingless Signalling to Establish Both Cell Polarity and Cell Identity. Development. 1994;120:347–360. doi: 10.1242/dev.120.2.347. The dishevelled gene shares homology with junction-associated proteins and is required on the receiving side of wingless signaling in all wg pathways.
- 39. Klingensmith J, Nusse R, Perrimon N. The Drosophila Segment Polarity Gene dishevelled Encodes a Novel Protein Required for Response to the wingless Signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. The segment polarity gene dishevelled shares homology with junction-associated proteins and acts autonomously as a common component to all wg signaling pathways.
- 40. Couso JP, Bishop SA, Martinz-Arias A. The wingless Signalling Pathway and the Patterning of the Wing Margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. This paper describes the role of wg in forming the wing margin. The armadillo, dishevelled and zeste-white3 genes function in this pathway just as they do in the embryo. High levels of wingless are suggested to induce the expression of the homeodomain protein encoded by cut, whereas lower levels induce the expression of the helix-loop-helix protein encoded by Achaete. These transcription factors then contribute to cell type specification.
- 42.Li X, Noll M. Rose of gooseberry Gene in Drosophila Embryos: Maintenance of wingless Expression by a wingless–gooseberry Autoregulatory Loop. EMBO J. 1993;12:4499–4509. doi: 10.1002/j.1460-2075.1993.tb06139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic Analysis of hedgehog Signaling in the Drosophila Embryo. Development. 1993 suppl:115–124. Using genetic epistasis, this paper categorizes interactions among several segment polarity genes, implicating specific members in a hedgehog-dependent pathway for cellular patterning.
- 44.Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. Cloning Characterization of the Segment Polarity Gane cubitus interruptus Dominant of Drosophila. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 45.Eaton S, Kornberg TB. Repression of ci-D in Posterior Compartments of Drosophila by engrailed. Genes Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- 46.Preat T, Therond P, Lamour-Isnard C, Limbourg-Bouchon B, Tricoire H, Erk I, Mariol MC, Busson D. A Putative Serine/Threonine Kinase Encoded by the Segment Polarity fused Gene of Drosophila. Nature. 1990;347:87–89. doi: 10.1038/347087a0. [DOI] [PubMed] [Google Scholar]
- 47. Vincent JP, O’Farrell PH. The State of engrailed Expression is not Clonally Transmitted during Early Drosophila Development. Cell. 1992;68:923–931. doi: 10.1016/0092-8674(92)90035-b. In this important 1992 paper the authors traced the lineage of single cells in the embryo and demonstrated that cells initially expressing engrailed will lose expression as they lose proximity to wingless-expressing cells.
- 48.Noordermeer J, Johnston P, Rijsewik F, Nusse R, Lawrence PA. The Consequences of Ubiquitous Expression of the wingless Gene in the Drosophila Embryo. Development. 1992;116:711–719. doi: 10.1242/dev.116.3.711. [DOI] [PubMed] [Google Scholar]
- 49.Poole SJ, Kornberg TB. Modifying Expression of the engrailed Gene of Drosophila melanogaster. Development. 1988;104 Suppl:85–93. doi: 10.1242/dev.104.Supplement.85. [DOI] [PubMed] [Google Scholar]
- 50.Baker NE. Embryonic and Imaginal Requirements for wg a Segment Polarity Gene in Drosophila. Dev Biol. 1988;125:96–108. doi: 10.1016/0012-1606(88)90062-0. [DOI] [PubMed] [Google Scholar]
- 51.Hidalgo A, Ingham P. Cell Patterning in the Drosophila Segment: Spatial Regulation of the Segment Polarity Gene patched. Development. 1990;110:291–301. doi: 10.1242/dev.110.1.291. [DOI] [PubMed] [Google Scholar]
- 52.Hillman R, Lesnick LH. Cuticle Formation in the Embryo of Drosophila melanogaster. J Morphol. 1970;131:383–396. [Google Scholar]
- 53.Lohs-Schardin M, Cremer C, Nusslein-Volhard C. A Fate Map of the Larval Epidermis of Drosophila melanogaster. Localized Cuticle Defects Following Irradiation of the Blastoderm with an Ultraviolet Laser Beam. Dev Biol. 1979;73:239–255. doi: 10.1016/0012-1606(79)90065-4. [DOI] [PubMed] [Google Scholar]
- 54. Bejsovec A, Wieschaus E. Segment Polarity Gene Interactions Modulate Epidermal Patterning in Drosophila Embryos. Development. 1993;119:501–537. doi: 10.1242/dev.119.2.501. Multiple mutant combinations are examined to investigate the contribution of each of five segment polarity genes to epidermal patterning. Such data lead to a slightly different interpretation of epidermal patterning than is offered in our review. The authors conclude that patched, naked, engrailed, and hedgehog modulate the ability of cells to respond to wingless input, and that they each can affect pattern independently of wg signaling.
- 55.Mohler J. Requirements for hedgehog a Segmental Polarity Gene in Patterning Larval and Adult Cuticle of Drosophila. Genetics. 1988;120:1061–1072. doi: 10.1093/genetics/120.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orenic T, Chidsey J, Holmgren R. Cell and Cubitus interruptus Dominant Two Segment Polarity Genes on the Fourth Chromosome in Drosophila. Dev Biol. 1987;124:50–56. doi: 10.1016/0012-1606(87)90458-1. [DOI] [PubMed] [Google Scholar]
- 57.Vogel A, Tickle C. FGF-4 Maintains Polarizing Activity of Posterior Limb Bud Cells in Vivo and in Vitro. Development. 1993;119:199–206. doi: 10.1242/dev.119.1.199. [DOI] [PubMed] [Google Scholar]
- 58.Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 Replaces the Apical Ectodermal Ridge and Directs Outgrowth and Patterning of the Limb. Cell. 1993;75:579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- 59.Perrimon N. The torso Receptor Protein-Tyrosine Kinase Signaling Pathway: an Endless Story. Cell. 1993;74:219–222. doi: 10.1016/0092-8674(93)90412-j. [DOI] [PubMed] [Google Scholar]
- 60.Ingham PW. The Molecular Genetics of Embryonic Pattern Formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]