Abstract
RNA folding occurs via a series of transitions between metastable intermediate states for Mg2+ concentrations below those needed to fold the native structure. In general, these folding intermediates are considerably less compact than their respective native states. Our previous work demonstrates that the major equilibrium intermediate of the 154 residue specificity domain (S-domain) of the B. subtilis RNase P RNA is more extended than its native structure. We now investigate two models with falsifiable predictions regarding the origins of the extended intermediate structures in the S-domains of the B. subtilis and the E. coli RNase P RNA that belong to different classes P RNA and have distinct native structures. The first model explores the contribution of electrostatic repulsion, while the second model probes specific interactions in the core of the folding intermediate. Using small-angle X-ray scattering (SAXS) and Langevin Dynamics (LD) simulations, we show that electrostatics only plays a minor role, whereas specific interactions largely accounts for the extended nature of the intermediate. Structural contacts in the core, including a non-native base-pair, help to stabilize the intermediate conformation. We conclude that RNA folding intermediates adopt extended conformations due to short-range, non-native interactions rather than generic electrostatic repulsion of helical domains. These principles apply to other ribozymes and riboswitches that undergo functionally relevant conformational changes.
Keywords: Langevin dynamics, P RNA, S-domain
Introduction
A major challenge in RNA folding is to identify the physiochemical interactions in folding intermediates and their role in the folding to the native state. Folding to this compact, functional state generally occurs via a series of transitions in which local elements form early and long-range structures occur later. Populated intermediate states have been characterized by a variety of biophysical and biochemical methods.1; 2; 3; 4; 5; 6; 7; 8 Compared to native states, the intermediates generally are less compact, have more solvent exposed residues, and contain fewer tertiary interactions, enabling these intermediates to act as structural checkpoints and contain a majority of the native secondary structure and some tertiary structure. The strength of base-pairing and stacking interactions can result in the formation of non-native structures whose disruption can be rate-limiting in the over-all folding process.3; 8; 9
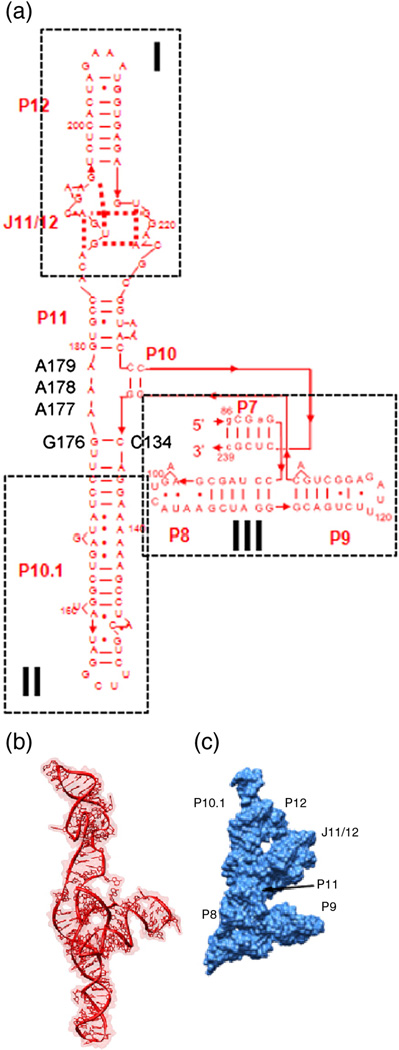
The native structure of the 154 residue specificity domain (S-domain) of B. subtilis RNase P RNA has been solved by X-ray crystallography,10 and its folding behavior in solution has been studied extensively.8; 11 The addition of Mg2+ ions induces folding of the S-domain along an equilibrium pathway with two transitions between three populated states, Unfolded-to-Intermediate-to-Native. The dominant equilibrium intermediate also is a highly populated kinetic folding intermediate11 whose structure has been modeled using experimental data that probe the global (small-angle X-ray scattering, SAXS) and local (chemical and enzymatic probing) conformation11 (Fig. 1A, 1B). The intermediate structure is composed of three helical domains that conjoin in a “core” region. In the intermediate, the helical domains I and II are splayed out with a ~180° angle between them. In the native structure, helical domains I and II rearrange to have a ~60° relative angle, enabling them to interact with each other through the conserved, long-range tetraloop-receptor interaction (Fig. 1C). Helical domain III is located perpendicular to the extended stack of helical domains I and II; its orientation in the intermediate state is only slightly perturbed relative to the native structure.
Figure 1. B. subtilis S-domain structure.
A) Secondary structure presentation of the S-domain folding intermediate. The structure is composed of three helical domains (boxed) that meet in a core. B) Ribbon diagram of the structural model of the folding intermediate.11 C) Native structure of the S-domain (1nbs.pdb)10 which is much more compact.
Electrostatic repulsion of the backbone exerts a substantial influence on the RNA folding thermodynamics and kinetics,12; 13; 14; 15; 16 as demonstrated by the considerable dependence of RNA folding on ionic conditions, with Mg2+-Mg2+ ion correlations inducing more contracted conformational ensembles of helices lacking fixed tertiary interactions.17; 18; 19 However, the relative contribution of these terms compared to other interactions is unclear. Using a “tightly bound ion” model, Chen and coworkers concluded that electrostatic repulsion is insufficient to account for the extended nature of intermediates, and other interactions, e.g., within the junctions and the helix stems, could limit the conformational diversity.18
For the S-domain intermediate, two models are proposed to probe the origin of the extended helical domains in the folding intermediate. The first model assumes the extension is due to the helices’ electrostatic repulsion that is not fully compensated under low ionic conditions where the intermediate is populated. The second model posits that specific interactions in the core are responsible for maintaining the extended geometry of the intermediate. Here, we investigate both models using SAXS, mutational analysis and Langevin Dynamics (LD) simulations and thereby determine that the extended structure of S-domain folding intermediates is stabilized by specific interactions in the core and not from generic electrostatic repulsion of helical domains.
Results and Discussion
Compaction of the folding intermediate of the B. subtilis S-domain
The lack of long-range tertiary interactions in the B. subtilis S-domain intermediate produces an extended conformation in which helical domains I and II are splayed out in opposite directions.11 Whether this intermediate represents a discrete structure or heterogeneous ensembles, e.g., with the angle between helical domains I and II varying by ±30°, remains an open question. Regardless, their extended geometry raises the possibility that the presence considerable electrostatic repulsion between negatively charged backbones of helices precludes their additional collapse. This model makes a strong prediction regarding the compactness of the intermediate as a function of ionic condition because a reduction in the electrostatic repulsion should lead to more compact structures.
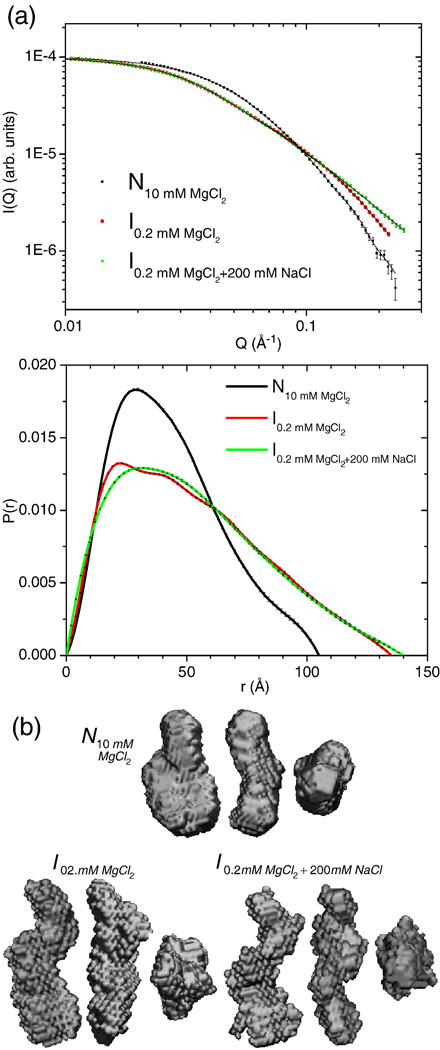
We have tested this prediction by measuring the compactness of the intermediate using SAXS under different monovalent salt conditions with two RNA constructs of the S-domain (Table 1, Fig. 2A,B). The wild-type S-domain intermediate exhibits a radius of gyration (Rg) of 42.2. ± 0.1 Å, as obtained from P(r), the pair distribution function calculated from the scattering data using the indirect Fourier transform procedure GNOM.20; 21 Upon addition of 0.2 M NaCl, no further collapse is observed (Rg = 41.8 ± 0.3 Å). The lack of shape change also is apparent from the invariance in P(r) distributions and the maximum distance, Dmax. Hence, the >100-fold greater ionic strength provided by this high monovalent salt concentration is insufficient to alter the global structure of the intermediate. In particular, helical domains I and II must remain extended, although the conformational diversity may change somewhat.
Table 1.
Compaction of S-domain RNA measured by SAXS.
| Construct | Conditiona | Rg (Å) | Dmax (Å) |
|---|---|---|---|
| B. subtilis (B-type) | |||
| Wild-type | 10 mM MgCl2 | 32.4 ± 0.1 | 100 |
| 0.4 mM MgCl2 | 42.2 ± 0.03 | 130 | |
| 0.4 mM MgCl2+0.2 M NaCl | 42.2 ± 0.3 | 140 | |
| C134U | 0.4 mM MgCl2 | 43.4 ± 0.2 | 150 |
| 0.4 mM MgCl2+0.2 M NaCl | 38.8 ± 0.1 | 150 | |
| L12 GAAA->UUCG | 10 mM MgCl2 | 44.6 ± 0.04 | 150 |
| E. coli (A-type) | |||
| Wild-type | 10 mM MgCl2 | 33.6 ± 0.1 | 105 |
| 0.2 mM MgCl2 | 42.3 ± 0.1 | 135 | |
| 0.2 mM MgCl2+0.2 M NaCl | 42.1 ± 0.1 | 140 | |
All contain 20 mM trisHCl, pH 8, 37 °C, 6 µM RNA.
Figure 2. SAXS data and reconstruction for B. subtilis S-domain and DTL mutant.
A) Scattering data and P(r) at 37 °C are shown. I(0) values are normalized to the same value while P(r) distributions are unit normalized (∫P(r) dr = 1). B) Shown are three orthogonal views of the molecular envelopes derived from the scattering data along with the S-domain crystal structure10 and model of the intermediate.11 The intermediate remains extended in the background of 200 mM NaCl while the removal of the tetraloop interaction (ΔTL) renders the molecule unable to fold to the native state, remaining in an extended conformation similar to the wild-type intermediate. The scale for the reconstructions and models is the same to allow for direct comparison.
We further investigate the lack of compaction of the S-domain using a mutant with a significantly impaired long-range tetraloop-receptor interaction (ΔTL, Fig. 2A, 2B). In this mutant, the L12 tetraloop (G205AAA) is substituted with UUCG which disrupted the tetraloop-receptor interaction critical for the native structure.10; 22; 23; 24 Hence, the ΔTL mutation generates an RNA whose folding is unable to progress past the I state. This arrest affords us the opportunity to study the effects of elevated Mg2+ concentration on the structure of the I state under conditions which otherwise result in the folding to the collapsed native state. This additional measurement further probes whether electrostatic repulsion underlies the extended nature of the intermediate because divalent cations interact much stronger with RNAs than monovalent ions. If this repulsion is significant, the arrested ΔTL mutant at high Mg2+ concentrations might collapse relative to the intermediate conformation, akin to near-native collapsed intermediates found in other RNA folding studies.25; 26; 27
However, we find that the ΔTL mutant does not collapse. SAXS measurements, even at 10 mM Mg2+, show that the ΔTL mutant does not become more compact (Rg = 44.6 ± 0.4 Å), as compared to the wild-type intermediate in 0.4 mM Mg2+ (the RgΔTL is ~ 3 Å larger, which may be due to a slight amount of native species in the wild-type measurement). This Mg2+ concentration is more than ten-fold higher than is required to fold the wild-type S-domain to the native state. Furthermore, folding of the wild-type S-domain is very sensitive to [Mg], with a highly cooperative I-to-N transition having a Hill constant of ~7.11 In similar SAXS study on the P4–P6 region of the Group 1 intron, a mutant also lacking long-range tertiary interactions likewise failed to collapsed in the presence 10 mM Mg2+.28 These results indicate that the extended structure of both the S-domain and the Group 1 intermediate cannot be explained by simple electrostatic repulsion between their helical domains.
To visualize the conformational states, 3D reconstructions are obtained using the programs DAMMIF/DAMMAVER/DAMFILT29 along with the SITUS suite of SAXS analysis programs.30 The envelopes for the wild-type intermediates under low and high ionic conditions are very similar to each other and to the ΔTL conformation populated in 10 mM MgCl2 (Fig. 2). These three states are more extended than the native state. Additionally, the envelopes for the native state and three extended states have very similar aspect ratios and dimensions as do the native crystal structure and our model for the intermediate.
Evaluation of specific interactions in the folding intermediate by LD simulation
Because electrostatic repulsion is insufficient to account for the extended conformation of the S-domain intermediate, we investigate the alternative that specific interactions play an important role. However, most RNA folding intermediates lack long-range, peripheral tertiary interactions, as in the case of the S-domain. This necessitates the investigation of local structural elements in the core of the molecule. For example, in the folding intermediate, helical domains I, II, and III are joined in a three-way junction (Fig. 1A) whose conformation may be important in maintaining the extended structure of the intermediate.
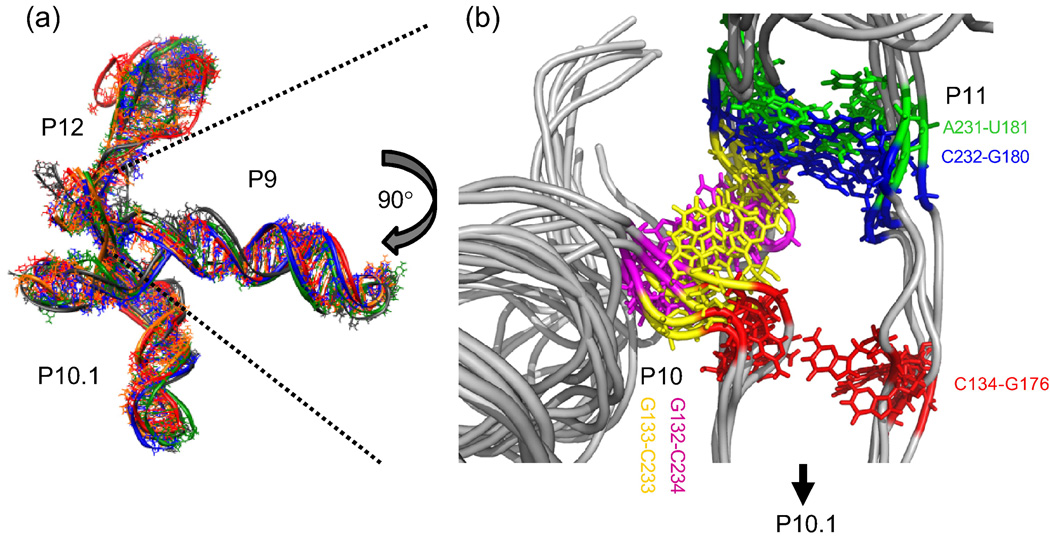
A comparison between the intermediate with a co-linear P12 (helical domain I) and P10.1 (helical domain II) helices and the native structure reveals that the loss of long-range tertiary interactions is concomitant with a major rearrangement of a five nucleotide junction between the P10.1 and P11 helices (J10.1/11, residues 175–179 in Fig. 1A). This junction exists in the core of the molecule and mediates the connection between helical domains I and II. In the native structure, the J10.1/11 residues form a U-turn capped by a reverse Watson-Crick U175-A179 base pair. In addition, the junction participates in tertiary structure stability; A177 and A178 form an A-minor motif with two G-C base pairs in the stacked P7 and P10 helices. In the intermediate, the entire J10.1/11 region is proposed to be extended, and this extension results in the intermediate in the reorientation of the P10.1 helix (helical domain I), which points in the opposite direction to that observed in the native structure.11
All-atom molecular dynamics simulations are a powerful complement for probing atomic-level dynamics.31 To investigate the possible interactions between residues in the three-way junction at the core of the S-domain, we have applied all-atom implicit solvent LD simulations using the Amber94 force field with a new distance-dependent dielectric constant modified to account for the monovalent ion concentrations.32; 33 Initial configurations begin with the structure shown in Fig. 3A. Stable trajectories on the 5 nsec time-scale require the placement of one fully hydrated Mg2+ ion at each of four native-like regions having a high negative electrostatic potential. Due to the electrostatic strong interactions, these ions remain in their initial positions over the course of the simulation. The placement of these Mg2+ ions is derived from the crystal structure of the native S-domain where five Mg2+ have been localized in the tertiary core.10
Figure 3. Characterization of the B. subtilis intermediate structure by LD simulation.
A) All-atom LD simulations of the intermediate with an extended P9 region. Overlay of five different simulation trajectories at 2 ns. The overall conformation is globally very similar, but locally diverse. Simulation was performed using an S-domain with extended P9 helix to better visualize the global similarities and differences of the five runs. B) Close-up view of the intermediate core. Though variability in H-bonds exists, the main shape of the core and the whole molecule changes little.
During each of the five LD trajectories, greater than half of the core residues form base-pair and stacking within the three-way junction, although the details of the pairings vary within each trajectory (Fig. 3B). Nevertheless, the three helical domains remain at essentially the same orientation, partially due to the triangular geometry, the multiple base-pairing interactions within the core, and the finite length of the single-stranded regions.
One of the base pairs in the core (C134-G176) exhibits a non-native base-pairing (Fig. 3B) that can be considered to be short range because it extends the native, P10.1 helix (Fig. 1A). We have previously proposed this pairing based upon the proximity of these two bases within the core.11 This non-native base pair is of particular interest because the phosphate backbone undergoes a dramatic change of direction at C134 in the folding intermediate, suggesting that its interaction with G176 stabilizes C134 in the intermediate conformation. In the native conformation, C134 is located far from G176. We therefore propose that this non-native base pair C134-G176 is partially responsible for the orientation of helical domain II relative to the core in the intermediate.
Compaction of a core mutant
To further investigate the importance of the C134-G176 non-native base pair in stabilizing the intermediate’s extended structure, we examine the compaction and folding of a C134U mutant in which the non-native base pair is changed from G-C to G-U (Fig. 4). Mutating C134 to G or A is not possible since this would disrupt the interaction involving residue 134 in the native structure. To maintain all interactions in the native structure, we also have incorporated compensatory mutations (A231-U181 to G231-C181) to allow for an isosteric U134xG231-C181 base triple in the native structure. The C134U mutant exhibits a very similar folding behavior as the wild-type S-domain as measured by CD (Fig. 4A).
Figure 4. Characterization of the B. subtilis C134 U core mutant by SAXS.
A) Mg2+-dependent folding of the wild-type S-domain and C134U mutant monitored by CD spectroscopy. B) Scattering data and P(r) distributions for the mutant at low and high ionic conditions. I(0) values are normalized to the same value while P(r) distributions are unit normalized (∫P(r) dr = 1). C) Three orthogonal views of the molecular envelopes derived from the scattering data are shown. Under high salt conditions, the Rg of the C134U intermediate is smaller by 2 Å relative to its low salt counterpart as well as the wild-type intermediate at low and high salt conditions. The scale for two reconstructions is the same to allow for direct comparison.
In the absence of monovalent ions, the C134U mutant also compacts in the same manner as the wild-type S-domain as measured by the Rg values and P(r) distribution (Fig. 4B, Table 1). However, the addition of 0.2 M NaCl induces a substantial compaction of the C134U mutant as evidenced by a significant reduction in Rg by 4.6Å, a shortened P(r) profile, and a slightly more compact molecular envelope (Fig. 4, Table 1), behavior consistent with a stabilizing C134 interaction in the folding intermediate. Weakening this interaction upon C134U mutation produces an intermediate structure that is more readily collapsed at higher monovalent cation concentrations.
Compaction of the folding intermediate of the E. coli S-domain
To determine whether electrostatic repulsion also exerts a minor role in forming the extended structure of other folding intermediates, we have performed SAXS studies of the S-domain from E. coli RNase P RNA34 (Fig. 5), which belongs to the A-type family of bacterial RNase P RNAs. The A and B-type S-domain native structures share a common core, but have a different number and arrangement of peripheral structures. As described previously, folding of the E. coli S-domain also displays an intermediate state populated at submillimolar Mg2+ concentrations from which it folds cooperatively to the native state.34.
Figure 5. SAXS analysis and reconstructions for the E. coli S-domain.
A) Scattering data and P(r) distributions for the native state and the intermediate under low and high ionic conditions at 37 °C are shown. I(0) values are normalized to the same value while P(r) distributions are unit normalized (∫P(r) dr = 1). B) Three orthogonal views of the molecular envelopes derived from the scattering data are shown. The scale for the reconstructions is the same to allow for direct comparison.
The folding intermediate of E. coli S-domain also has a significantly extended intermediate structure (Rg = 42.1 ± 0.1 Å) as compared to the native (Rg = 33.6 ± 0.1 Å). This compaction is readily observed in the P(r) functions and molecular envelopes where the Dmax decreases from 140 to 105 Å (Fig. 5). As with the B-type S-domain, the intermediate remains extended even upon the addition of 200 mM NaCl (Rg = 42.3 ± 0.1 Å) with a nearly unchanged P(r) distribution, Dmax and molecular envelope. These findings indicate that electrostatic repulsion does not play a significant role in the formation of the extended structure of this folding intermediate, as well as the intermediate for the B-type S-domain.
Conclusion
Based on the SAXS measurements, mutations, and all-atom simulations, we conclude that the global conformation for both the A- and B-type S-domain folding intermediates are derived from the specific sequence and structure of the S-domain and not from generic electrostatic repulsion of the RNA backbone. In the case of the B-type RNA, we demonstrate that the intermediate contains a non-native core stabilized by local interactions. The extended structure of the S-domain intermediate arises both from native and non-native interactions in the core of the molecule, consistent with predictions using the “tightly bound ion” model.18 The lack of long-range tertiary interactions allows for this alternative conformation to be stably populated.
This folding behavior is similar to other reported cases of non-native RNA conformations. In the S-domain, for example, the stabilization of an alternative core conformation (C134-G176) in the absence of long-range tertiary interactions is analogous to the various conformations observed for the hammerhead ribozyme core where a minimal construct lacking peripheral tertiary interactions is stably populated and even crystallizable in a conformation having non-native interactions.35 However, the full-length ribozyme construct includes additional long-range tertiary contacts that help stabilize native interactions in the active site core.36 Consequently, the ribozyme undergoes a large-scale rearrangement which is similar to the rearrangement the B-type S-domain core adopts upon folding from the intermediate to native state. Similarly, in the case of several riboswitches, an extended conformation is stabilized in the absence of cognate metabolite. Only upon addition of metabolite does the riboswitch reorganize and form the long-range tertiary interactions.37; 38 Hence, different conformational states stabilized by different, specific interactions are common in both RNA folding and function.
Materials and Methods
Solution analysis
All RNA constructs are in vitro transcripts prepared by standard T7 RNA polymerase transcription using plasmid DNA templates. Transcripts are purified on denaturing polyacrylamide gels and stored in water at −20 °C. The unfolded state (U) is produced upon heating the RNA in 20 mM Tris–HCl (pH 8.1) at 85 °C for 2 minutes followed by 5 minutes at room temperature.
The native structure contains C134 which forms a base triple with the A231-U181 base pair. In order to probe the consequences of non-native C134-G176 base pair in the intermediate, we have incorporated compensatory mutations of A231-U181 to G231-C181 to allow the isosteric U134xG231-C181 triple in the native structure. The RNA folding is monitored spectroscopically by adding aliquots of concentrated Mg2+ solutions to RNA (0.3 µM) in the U state, with CD and absorbance readings taken after the addition of each aliquot. MgCl2 solution is added using a Hamilton titrator connected to a Jasco J715 spectrometer as described previously.39
SAXS experiments are performed at the BioCAT beamline at the Advanced Photon Source at the Argonne National Laboratories as described previously.40 The S-domain constructs (all at 0.3mg/ml or ~6 µM) are prepared in 20 mM Tris, pH 8.1 and heated for 2 minutes at 85 °C. Wild-type and C134U S-domains are incubated in 0.4 mM MgCl2 for 5 minutes at 50°C, followed by incubation at room temperature for 5 minutes. The particular [MgCl2] is determined that maximally populates the intermediate according to CD titrations performed under identical conditions. NaCl is then added to the solution while maintaining a constant 0.4 mM Mg2+. The L12 mutant is prepared as above except 10 mM MgCl2 is added to the sample. Each sample is loaded into a capillary tube controlled by a Hamilton titrator with the sample holder thermostatically controlled at 37 °C. Buffer data are collected immediately prior to taking sample data under continuous-flow conditions to minimize radiation damage.
To visualize the conformational states, 3D reconstructions have been obtained using the program DAMMIF.29 Ten such reconstructions enable generating a consensus space-filling bead model reconstruction using the DAMMAVER/DAMFILT programs. This model is converted using the PDB2VOL program, a part of the SITUS suite of SAXS analysis programs,30 to create a final molecular envelope for the RNA. Molecular envelopes are visualized with the program VMD.41
LD simulations
All-atom simulations are performed using an implicit solvent model,42 a modified version of the TINKER dynamics package, and the Amber94 force field.43 The model incorporates a nonlinear, distance and ionic strength dependent dielectric permittivity44 with the solute-solvent interaction energy described by the Ooi-Scheraga solvent accessible surface area (SASA) potential 45 and the atomic friction coefficients calculated with the Pastor-Karplus scheme 46. After an initial energy minimization step, trajectories are calculated for approximately 5 ns, with a structure being saved every 5 ps. Mg2+ ions are placed at four native-like regions having high negative potential (< -8 kT/e in the presence of 0.1 M monovalent ions) determined using a non-linear Poisson-Boltzmann calculation.16
ACKNOWLEDGMENTS
This work was supported by NIH grants (GM57880 to TP and TRS). We thank L. Guo for assistance with the SAXS measurements. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center RR-08630. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Center for Research Resources or the National Institutes of Health.
Abbreviations
- Dmax
maximum distance
- LD
Langevin Dynamics
- Rg
radius of gyration
- SAXS
small-angle X-ray scattering
- TL
tetra-loop
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Ann. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 2.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 3.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 4.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 5.Woodson SA. Folding mechanisms of group I ribozymes: role of stability and contact order. Biochem Soc Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 6.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird NJ, Fang XW, Srividya N, Pan T, Sosnick TR. Folding of a universal ribozyme: the ribonuclease P RNA. Q. Rev. Biophys. 2007;40:113–161. doi: 10.1017/S0033583507004623. [DOI] [PubMed] [Google Scholar]
- 9.Sosnick TR, Pan T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 10.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 11.Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J. Mol. Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 13.Chen SJ. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu VB, Bai Y, Lipfert J, Herschlag D, Doniach S. A repulsive field: advances in the electrostatics of the ion atmosphere. Curr. Opin. Chem. Biol. 2008;12:619–625. doi: 10.1016/j.cbpa.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin K, Sharp KA, Honig B, Pyle AM. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nature Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 17.Tan ZJ, Chen SJ. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J Chem Phys. 2005;122:44903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Tan ZJ, Chen SJ. Salt-dependent folding energy landscape of RNA three-way junction. Biophys. J. 2010;98:111–120. doi: 10.1016/j.bpj.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y, Chu VB, Lipfert J, Pande VS, Herschlag D, Doniach S. Critical assessment of nucleic acid electrostatics via experimental and computational investigation of an unfolded state ensemble. J. Am. Chem. Soc. 2008;130:12334–12341. doi: 10.1021/ja800854u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svergun D, Semenyuk A. GNOM Small-angle scattering data processing by means of the regularization technique. 2000 http://www.srs.dl.ac.uk/ncd/computing/manual.gnom.html.
- 21.Svergun DI. Determination of the Regularization Parameter in Indirect- Transform Methods Using Perceptual Criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 22.Qin H, Sosnick TR, Pan T. Modular construction of a tertiary RNA structure: the specificity domain of the Bacillus subtilis RNase P RNA. Biochemistry. 2001;40:11202–11210. doi: 10.1021/bi010076n. [DOI] [PubMed] [Google Scholar]
- 23.Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 24.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 25.Fang X, Littrell K, Yang X, Henderson SJ, Siefert S, Thiyagarajan P, Pan T, Sosnick TR. Mg(2+)-dependent compaction and folding of yeast tRNA(Phe) and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle Xray scattering. Biochemistry. 2000;39:11107–11113. doi: 10.1021/bi000724n. [DOI] [PubMed] [Google Scholar]
- 26.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42:13869–13878. doi: 10.1021/bi035476k. [DOI] [PubMed] [Google Scholar]
- 28.Schlatterer JC, Kwok LW, Lamb JS, Park HY, Andresen K, Brenowit M, Pollack L. Hinge stiffness is a barrier to RNA folding. J. Mol. Biol. 2008;379:859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wriggers W, Chacón P. Using Situs for the Registration of Protein Structures with Low-Resolution Bead Models from X-ray Solution Scattering. J. Appl. Cryst. 2001;34:773–776. [Google Scholar]
- 31.Li W, Frank J. Transfer RNA in the hybrid P/E state: correlating molecular dynamics simulations with cryo-EM data. Proc. Natl. Acad. Sci. U S A. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramstein J, Lavery R. Energetic coupling between DNA bending and base pair opening. Proc. Natl. Acad. Sci. U S A. 1988;85:7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen MY, Freed KF. Long time dynamics of met-enkephalin: Comparison of explicit and implicit solvent models. Biophys. J. 2002;82:1791–1808. doi: 10.1016/s0006-3495(02)75530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baird NJ, Srividya N, Krasilnikov AS, Mondragon A, Sosnick TR, Pan T. Structural basis for altering the stability of homologous RNAs from a mesophilic and a thermophilic bacterium. RNA. 2006;12:598–606. doi: 10.1261/rna.2186506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 36.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J. Mol. Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulshina N, Baird NJ, Ferre-D'Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nature Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sosnick TR, Fang X, Shelton VM. Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 40.Pan T, Sosnick TR. Structural analysis of RNA and RNA-protein complexes by small-angle X-ray scattering. In: Hartmann R, B A, Schon A, Westhof E, editors. Handbook of RNA biochemistry. Weinheim: Wiley-VCH Verlag; 2005. pp. 385–397. [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 42.Shen MY, Freed KF. All-atom fast protein folding simulations: the villin headpiece. Proteins. 2002;49:439–445. doi: 10.1002/prot.10230. [DOI] [PubMed] [Google Scholar]
- 43.Pearlman D, Case DA, Caldwell JW, Ross WS, Cheatam ITE, Ferguson DM, Singh UC, Weiner P, Kollman P. Amber 4.1. 1995 [Google Scholar]
- 44.Jha AK, Freed KF. Solvation effect on conformations of 1,2:dimethoxyethane: charge-dependent nonlinear response in implicit solvent models. J. Chem. Phys. 2008;128:034501. doi: 10.1063/1.2815764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ooi T, Oobatake M, Nemethy G, Scheraga HA. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc. Natl. Acad. Sci. U S A. 1987;84:3086–3090. doi: 10.1073/pnas.84.10.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastor RW, Karplus M. Parametrization of the Friction Constant for Stochastic Simulations of Polymers. J. Phys. Chem. 1988;92:2636–2641. [Google Scholar]