Abstract
Small cell lung carcinoma (SCLC) is a neuroendocrine subtype of lung cancer. While SCLC patients often initially respond to therapy, tumors nearly always recur, resulting in a 5-year survival rate of less than 10%. A mouse model has been developed based on the fact that the RB and p53 tumor suppressor genes are mutated in more than 90% of human SCLCs. Emerging evidence in patients and mouse models suggests that p130, a gene related to RB, may act as a tumor suppressor in SCLC cells. To test this idea, we used conditional mutant mice to delete p130 in combination with Rb and p53 in adult lung epithelial cells. We found that loss of p130 resulted in increased proliferation and significant acceleration of SCLC development in this triple knockout mouse model. The histopathological features of the triple mutant mouse tumors closely resembled that of human SCLC. Genome-wide expression profiling experiments further showed that Rb/p53/p130 mutant mouse tumors were similar to human SCLC. These findings indicate that p130 plays a key tumor suppressor role in SCLC. Rb/p53/p130 mutant mice provide a novel pre-clinical mouse model to identify novel therapeutic targets against SCLC.
Keywords: p53, Rb, p130, mouse model, small cell lung carcinoma
INTRODUCTION
The prognosis for patients suffering from small cell lung carcinoma (SCLC) has not improved in the past 25 years. Despite advances in chemotherapy and radiation therapy regimens, 5-year survival rates for SCLC remain only 5-10%. At the time of diagnosis, SCLC has often already metastasized, and patients almost always relapse following treatment (1).
Based on the observation that the RB and p53 tumor suppressor genes are both mutated in more than 90% of human SCLC, a mouse model for human SCLC has been generated. In this model, Cre-mediated deletion of Rb and p53 conditional alleles in the lungs of adult mice results in the development of tumors that share many characteristics with human SCLC, including their histopathology, the expression of neuroendocrine markers, and their ability to metastasize (2). This mouse model provides a system to identify genetic and epigenetic changes that may contribute to the development of SCLC in addition to loss of Rb and p53 (2, 3). In particular, emerging evidence suggests that p130, a cell cycle inhibitor related to RB (4), may normally suppress SCLC development. The first report describing loss of p130 in a human cancer was in SCLC (5). In addition, low levels of the p130 protein have been associated with a higher histological grade, increased proliferation, and a trend toward poorer patient survival (6-8). Recent evidence further indicates that p130 mRNA levels may be downregulated in lung cancer cells by members of the miR-17-92 microRNA cluster, which is often overexpressed in human SCLC and may play a role in the expansion of neuroendocrine cells (9, 10). The possibility that p130 is a bona fide tumor suppressor has gained further support in mice where loss of p130 cooperates with loss of Rb in retinoblastoma development (11) and in the development of small neuroendocrine lung lesions (12). Similarly, loss of E2F4, a major partner for p130, partially suppresses lung neuroendocrine hyperplasias in Rb mutant mice (13). Loss of p130 also results in the acceleration of tumorigenesis in a mouse model for human lung adenocarcinoma (14). Interestingly, while p130 share many characteristics with p107, the third member of the RB family, there is no evidence that p107 may be involved in the suppression of SCLC (5, 12). Here we tested the tumor suppressor role of p130 in SCLC using a mouse genetics approach. We found that loss of p130 accelerates the development of SCLC in Rb/p53 mutant mice and that Rb/p53/p130 mutant mice provide an improved mouse model of SCLC.
MATERIAL AND METHODS
Mice and adenoviral infections
Conditional mutant Rb and p53 mice were bred to mice with a conditional allele for p130 (2, 15) (Supplementary Fig. S1 and Supplementary Methods). Adenoviral infections were performed as described (14). Mice were maintained at the Stanford's Research Animal Facility accredited by the AAALAC.
DNA, RNA, and microarray analysis
Genomic DNA was prepared using lysis buffer containing Proteinase K (Sigma Aldrich). RNA was isolated from tumors or control lungs using TRIzol (Invitrogen). The Dynamo cDNA Synthesis Kit (New England Biolabs) was used to prepare cDNA from RNA. Real-time quantitative PCR was performed on an ABI Prism 7900HT Sequence Detection System with the SYBR GreenER qPCR mix (Invitrogen). The Ct value of each sample was normalized using the values for the TATA Binding Protein gene (TBP). Primer sequences are listed in Supplementary Tables S1 and S2. Affymetrix 430-2.0 arrays were hybridized and analyzed at the Stanford Microarray facility as described (15) (Supplementary Methods).
Immunoblot analysis and immunostaining
For immunoblot analysis (14), the antibodies used were: p130 (BD Biosciences, 610261), p107 (Santa Cruz, SC-318), Synaptophysin (Neomarkers, RB-1461-P1), Karyopherin beta 1 (Santa Cruz, SC-1919), and α-Tubulin (Sigma, T9026).
Paraffin sections were rehydrated in Trilogy reagent (Cell Marque). Sections were mounted in ProLong® Gold Antifade reagent (Invitrogen) after immunostaining (14). The primary antibodies used were: Phospho-Histone 3 Ser10 (PH3) (Millipore, 06-570), BrdU (Becton-Dickinson, 347580), Ki67 (BD Biosciences, 55069), MCM6 (Santa Cruz Biotechnology, sc-9843), PCNA (Santa Cruz Biotechnology, sc-56), Surfactant protein C (SP-C) (Dr. Jeff Whitsett, University of Cincinnati), Clara cell secretory protein (CCSP) (Santa Cruz Biotechnology, sc-9772), Synaptophysin (SYP) (Neomarkers, RB-1461-P1). Alexa Fluor® secondary antibodies (Invitrogen) were used for antibody detection. Quantification was performed using the Bio-Quant image analysis software.
Statistics
Statistical significance was assayed by the Student's t-test with the Prism Graphpad software.
RESULTS AND DISCUSSION
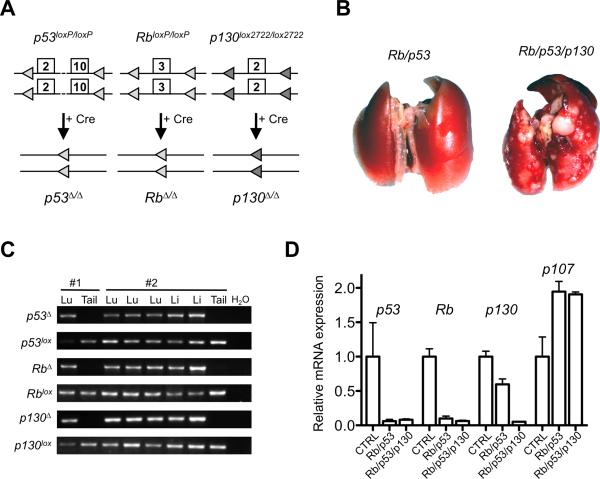
To test the possibility that p130 may act as a tumor suppressor gene in SCLC, we performed intranasal instillation of adenovirus expressing the Cre recombinase (Ad-Cre) in Rb/p53/p130 triple conditional mutant mice (Fig. 1A). After six months, primary lung tumors and liver metastases were detected at the surface of these organs; no other tumor types were identified in the mutant mice at this time point (Fig. 1B and data not shown, see below). PCR analysis on genomic DNA showed deletion of the three genes in the tumor cells but not in tail DNA in all cases examined (Fig. 1C). Quantitative RT-PCR analysis confirmed the decreased expression of these three genes in the triple mutant tumors (Fig. 1D). Finally, immunoblot analysis showed depletion of the p130 protein in all the triple mutant tumors analyzed (Supplementary Fig. S2). p107 levels increased similarly in double and triple mutant tumors compared to wild-type lungs (Fig. 1D and Supplementary Fig. S2), indicating that loss of p130 does not induce p107 expression in SCLC cells. Together, these experiments suggested that loss of p130 was not counter-selected during tumorigenesis, allowing us to test the role of p130 in SCLC development. Recent observations indicate that the combined loss of Rb and p130 is not sufficient for SCLC development in mice (16).The fact that we did not find tumors that were only p53/p130 mutant is also suggestive that loss of p130 does not alleviate the need for loss of Rb in SCLC, but this idea remains to be directly tested in p53/p130 mutant mice.
Figure 1. Deletion of p53, Rb, and p130 in the lungs of adult mice leads to the development of lung tumors.
A, schematic representation of the conditional mutant alleles for p53, Rb, and p130. B, representative photographs of the lungs from double and triple mutant mice 6 months after Ad-Cre infection. C, PCR analysis for the deletion of the conditional alleles on genomic DNA obtained from lung tumors (Lu), liver metastasis (Li), and the tails of two representative mice (#1 and #2). D, RT-qPCR analysis of p53, Rb, p130, and p107 expression levels in control lungs (CTRL) (n=3) and double (n=6) and triple mutant tumors (n=3).
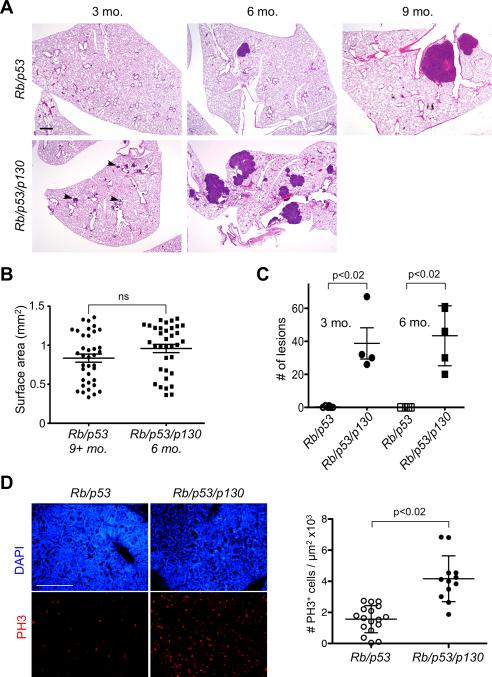
To determine the functional role of p130 in SCLC development, we compared tumorigenesis in the lungs of Rb/p53 and Rb/p53/p130 mutant mice. We found that six months after Ad-Cre infection, few tumors were present at the surface of the lungs of Rb/p53 mutant mice, as reported (2). In contrast, a number of tumors were visible in the lungs of Rb/p53/p130 mutant mice (Fig. 1B). Histological examination confirmed that tumors were rarely found at this time point in Rb/p53 mutant lung while tumors of various sizes were present in Rb/p53/p130 mutant lungs (Fig. 2A). Even three months after Ad-Cre infection, numerous lesions were readily identified in sections of Rb/p53/p130 mutant lungs while similar size lesions were observed six months after infection of Rb/p53 mutant mice (Fig. 2A). Tumor burden was measured in the lungs of Rb/p53/p130 mutant mice 6 months after Ad-Cre infection and was similar to that of Rb/p53 mutant mice 9-12 months after infection (Fig. 2B). Not only did the triple mutant mice developed tumors more rapidly, they also developed significantly more tumors than the double mutant animals (Fig. 2C). We found that very few double mutant mice died before 28 weeks (data not shown), similar to what was described before (2). In contrast, one triple mutant mouse died at 17 weeks, another at 19 weeks, and two at 26 weeks; in addition, all the Rb/p53/p130 mutant mice analyzed at the 6 months time point were moribund. While we have not performed a complete survival analysis, these observations suggest that Rb/p53/p130 mutant mice may die faster than Rb/p53 mutant mice with the dose of Ad-Cre used in these studies. Thus, p130 is a potent suppressor of lung cancer initiated by loss of Rb and p53 function in mice.
Figure 2. Loss of p130 results in accelerated lung cancer development in Rb/p53 mutant mice.
A, hematoxylin and eosin (H&E) staining of sections from the lungs of double and triple mutant mice at three time points post-Ad-Cre injection – no triple mutant mice were alive at the 9 months time point. Bar: 400 μm. Arrowheads show small lesions. B, quantification of tumor surface area in triple mutant mice 6 months after Ad-Cre injection compared to double mutant mice 9-12 months after Ad-Cre infection. C, quantification of tumor numbers in double and triple mutant mice 3 and 6 months after Ad-Cre injection. D, analysis of proliferation in double (~9 months) and triple (~6 months) mutant tumors of similar size by immunostaining for phospho-histone 3 (PH3). Left: representative immunostaining (PH3, red – DAPI, blue). Right: quantification. ns, not significant. Bar: 50 μm.
p130 is a known regulator of cell cycle progression (4), which led us to examine the proliferative status of double and triple mutant tumor cells in vivo. Immunostaining for Ki67, a marker of cycling cells, and PCNA and MCM6, two markers of S phase, showed that both double and triple mutant cells were actively proliferating (Supplementary Fig. S3). Quantitative analysis of the mitotic index of tumors using immunostaining for phospho-histone 3 (PH3) further indicated that triple mutant tumors had more cells in G2/M than double mutant tumors of similar sizes (Fig. 2D). These data suggest that one mechanism of tumor suppression by p130 in SCLC is to limit the proliferation of Rb/p53 mutant cells (see below), without excluding the possibility that p130 loss may accelerate SCLC development by other mechanisms. In particular, p130 loss could change the type of lung cancer developing in the mutant mice. While histopathological analysis showed that a few lesions had some characteristic of lung adenocarcinomas, adenocarcinomas with neuroendocrine features, large cell neuroendocrine carcinomas, and large cell carcinomas, all these were present in mice from both genotypes. Furthermore, the large majority of tumors developing in Rb/p53 and Rb/p53/p130 mutant mice had clear architectural, cytological, and clinical features of human SCLC (Fig. 2A, Supplementary Fig. S4, and data not shown). Furthermore, although the number of mice analyzed thus far remains limited, another striking phenotype of the Rb/p53/p130 mutant model is a high incidence of metastasis to the liver, one of the primary organs to which SCLC is known to metastasize in patients. Three out of five Rb/p53/p130 mice had several liver metastases 6 months after Ad-Cre infection, while no lesions were identified in Rb/p53 mice at the same time point and a lower incidence of liver metastasis was observed or previously reported at 9 months or later time points (Supplementary Fig. S4 and data not shown)(2). In addition, Rb/p53p130 mice developed metastases to pulmonary lymph nodes and kidney, two common sites of metastasis of human SCLC (Supplementary Fig. S4).
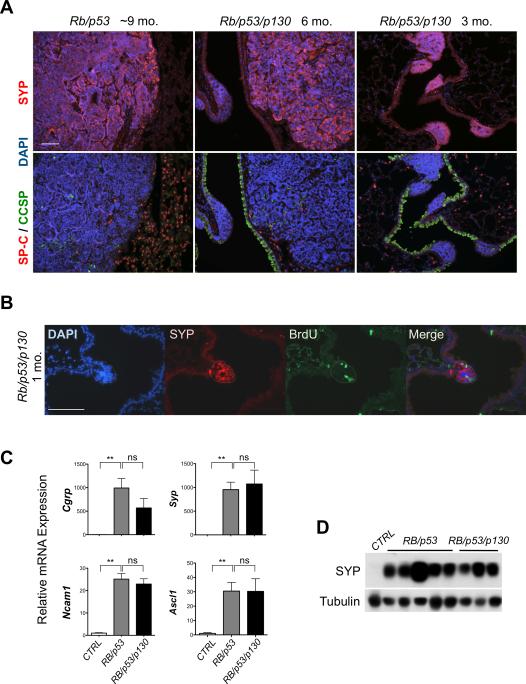
We next examined the expression of lung cancer markers in Rb/p53/p130 mutant tumors. Immunostaining analysis showed that triple mutant lesions expressed Synaptophysin (SYP), a synaptic vesicle protein used as a marker for human SCLC, but were largely negative for SP-C and CCSP, two markers of lung alveolar and bronchiolar epithelial cells, respectively, similar to Rb/p53 mutant tumors (Fig. 3A). Even one month after Ad-Cre infection, at a time when no lesions are visible in Rb/p53 mutant mice, small triple mutant lesions were evident. Cells in these lesions were dividing as assessed by BrdU incorporation and expressed SYP (Fig. 3B). Moreover, quantitative RT-PCR showed that all Rb/p53/p130 mutant tumors examined expressed typical markers of human SCLC such as Cgrp, Syp, Ncam1, and Ascl1 (17) (Fig. 3C). Immunoblot analysis further indicated that the triple mutant tumor cells expressed the SYP protein (Fig. 3D). Cell lines derived from triple and double knock-out mutant tumors grew in floating clusters, similar to human SCLC cells (18); the mouse tumor cells also retained the expression of SYP, but, as expected, Rb/p53/p130 mutant tumor cells did not express p130 (Supplementary Fig. S5).
Figure 3. Rb/p53/p130 triple mutant tumors have features of neuroendocrine SCLC.
A-B, immunostaining analysis of SCLC at different time points (1, 3, 6, and 9 months) for the expression of SYP (neuroendocrine), CCSP (bronchiolar), and SP-C (alveolar) and the incorporation of BrdU (S phase). DAPI stains the DNA in blue. The small lesion in a triple mutant mouse in B is circled by a dotted line. Bar: 50 μm. C, RT-qPCR analysis for the expression of four neuroendocrine genes in micro-dissected double (n=5) and triple (n=3) mutant tumors compared to control lungs (n=4). **, p<0.005. D, immunoblot analysis for SYP in double and triple mutant tumors compared to a control lung. Tubulin serves as a loading control.
To compare genome-wide expression profiles in Rb/p53 and Rb/p53/p130 mutant tumors, we performed a microarray analysis with double mutant tumors 9 months after Ad-Cre infection and triple mutant tumors 6 months after Ad-Cre infection. While this difference in time could affect tumor evolution in the two models, the histology and the size of double and triple mutant tumors at these time points are similar (Figs. 2A and 2B). Significant Analysis of Microarrays (SAM) showed that only 151 genes were differentially expressed between the two genotypes out of total 14712 genes with a minimal 1.5 fold change of expression level and a false discovery rate of 10% or lower (Supplementary Fig. S6). This observation strongly indicates that tumors from the two genotypes are very similar at the level of global gene expression. Using DAVID bioinformatics tools, we found that a few of the differentially expressed genes were implicated in the control of cell cycle progression (Supplementary Fig. S6A).Quantitative RT-PCR analysis confirmed increased levels of the tyrosine kinase gene c-Yes and the DNA repair gene Rad54l in Rb/p53/p130 mutant tumors compared to Rb/p53 mutant tumors; in contrast, the expression of classical E2F targets such as genes coding for B-Myb, Cyclin E and PCNA was not significantly different (Supplementary Figure S7). The functional role of the genes whose expression is significantly different in triple knock-out tumors and the mechanisms underlying the increased proliferation of triple mutant cells versus double mutant cells will be investigated in future experiments.
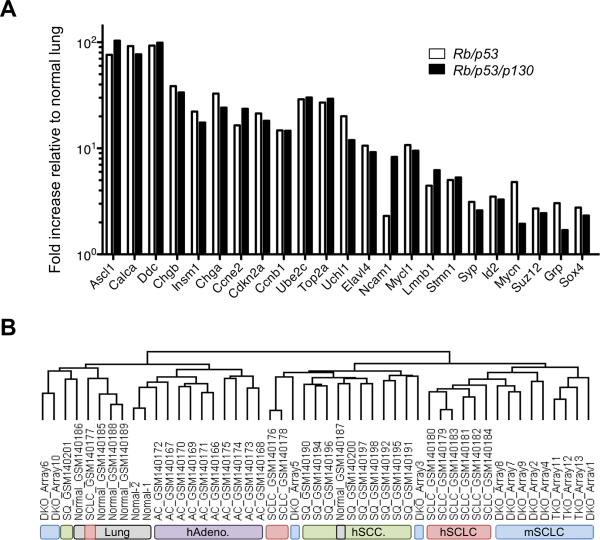
We next compared the datasets from the mouse models to datasets from human SCLC and NSCLC (Non-Small Cell Lung Cancer); because SCLC patients rarely undergo surgery, less than 50 genome-wide transcription profiling microarray data for primary human SCLC samples are currently publicly available(19-21); the 13 datasets from double and triple mutant mouse tumors increase the number of primary tumors analyzed by more than 25%. A number of genes overexpressed in human SCLC were readily identified in the list of overexpressed genes in Rb/p53/p130 and Rb/p53 mutant tumors (Fig. 4A). Gene Set Enrichment Analysis (GSEA) further showed that the genes overexpressed in human SCLC were significantly enriched in Rb/p53/p130 mutant tumors (Supplementary Fig. S8). Importantly, cross-specific clustering analysis showed that the two mouse models clustered with human SCLC rather than with normal lung or NSCLC (Fig. 4B). Taken together, these findings extend the similarity between the Rb/p53 and the Rb/p53/p130 mutant lung tumors and human SCLC from the histopathological level to global gene expression, further validating the Rb/p53/p130 mutant mouse model.
Figure 4. Rb/p53/p130 triple mutant mice provide a novel mouse model for human SCLC.
A, Expression of selected genes previously found to be overexpressed and/or implicated in human SCLC is graphed as fold increase relative to control lungs. The values were obtained from SAM analysis of gene expression profiles from 3 Rb/p53/p130 mutant tumors (dissected 6 months after Ad-Cre), 10 Rb/p53 mutant tumors 9 months after Ad-Cre), and 5 control lungs. Noticeably, expression levels for most of the well-known genes are similar in both mouse models. The q-value for all the genes listed above is less than 0.05%. B, Expression profiles of genes in Rb/p53/p130 and Rb/p53 were compared to those of orthologous genes in human array datasets of lung cancers using hierarchical clustering. hAdeno: human adenocarcinoma; hSCC: human squamous cell carcinoma; hSCLC: human SCLC; mSCLC: mouse SCLC.
In conclusion, the ablation of p130 in the context of Rb and p53 loss accelerates tumor development while maintaining the overall histopathological and molecular features of SCLC. Our data directly demonstrate that p130 serves a potent tumor suppression function in murine SCLC. Loss of p130 function results in increased proliferation, providing an explanation for why reduced p130 levels may be selected in human tumors. Compared to Rb/p53 mutant mice, Rb/p53/p130 mutant mice develop more tumors at a faster rate and will be instrumental in exploring the mechanisms of SCLC initiation, progression, and metastasis. Future studies will employ different doses of virus to change tumor burden and explore the long-term survival and phenotypes of the triple mutant mice. This novel mouse model for human SCLC will also serve to identify novel diagnostic markers and therapeutic targets against this deadly cancer.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. J. Kissil for critical reading of the manuscript and Drs. J. Whitsett and A. Berns for the generous gift of SP-C antibodies and p53lox mice, respectively. We apologize to our colleagues whose important work is not cited because of space considerations.
Financial support: The American Lung Association (J.S. and K.P.), the Parker B. Francis Fellowship Program (K. P.), the California Tobacco-Related Disease Research Program (J. C.), the California Breast Cancer Research Program Dissertation Fellowship (D.B.), the Damon Runyon Cancer Research Foundation (J.S.), and the American Cancer Society (J.S.).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1.Rudin CM, Hann CL, Peacock CD, Watkins DN. Novel systemic therapies for small cell lung cancer. J Natl Compr Canc Netw. 2008;6:315–22. doi: 10.6004/jnccn.2008.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 3.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 4.Claudio PP, Tonini T, Giordano A. The retinoblastoma family: twins or distant cousins? Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-reviews3012. reviews3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, et al. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci U S A. 1997;94:6933–8. doi: 10.1073/pnas.94.13.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cinti C, Macaluso M, Giordano A. Tumor-specific exon 1 mutations could be the ‘hit event’ predisposing Rb2/p130 gene to epigenetic silencing in lung cancer. Oncogene. 2005;24:5821–6. doi: 10.1038/sj.onc.1208880. [DOI] [PubMed] [Google Scholar]
- 7.Caputi M, Groeger AM, Esposito V, De Luca A, Masciullo V, Mancini A, et al. Loss of pRb2/p130 expression is associated with unfavorable clinical outcome in lung cancer. Clin Cancer Res. 2002;8:3850–6. [PubMed] [Google Scholar]
- 8.Campioni M, Ambrogi V, Pompeo E, Citro G, Castelli M, Spugnini EP, et al. Identification of genes down-regulated during lung cancer progression: a cDNA array study. J Exp Clin Cancer Res. 2008;27:38. doi: 10.1186/1756-9966-27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macpherson D. Insights from mouse models into human retinoblastoma. Cell Div. 2008;3:9. doi: 10.1186/1747-1028-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–62. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parisi T, Bronson RT, Lees JA. Inhibition of pituitary tumors in Rb mutant chimeras through E2f4 loss reveals a key suppressive role for the pRB/E2F pathway in urothelium and ganglionic carcinogenesis. Oncogene. 2009;28:500–8. doi: 10.1038/onc.2008.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho VM, Schaffer BE, Karnezis AN, Park KS, Sage J. The retinoblastoma gene Rb and its family member p130 suppress lung adenocarcinoma induced by oncogenic K-Ras. Oncogene. 2009 doi: 10.1038/onc.2008.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, et al. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–28. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson DS, Mason-Richie NA, Gettler CA, Wikenheiser-Brokamp KA. Retinoblastoma Family Proteins Have Distinct Functions in Pulmonary Epithelial Cells In vivo Critical for Suppressing Cell Growth and Tumorigenesis. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taneja TK, Sharma SK. Markers of small cell lung cancer. World J Surg Oncol. 2004;2:10. doi: 10.1186/1477-7819-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calbo J, Meuwissen R, van Montfort E, van Tellingen O, Berns A. Genotype-phenotype relationships in a mouse model for human small-cell lung cancer. Cold Spring Harb Symp Quant Biol. 2005;70:225–32. doi: 10.1101/sqb.2005.70.026. [DOI] [PubMed] [Google Scholar]
- 19.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–9. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrbeck A, Neukirchen J, Rosskopf M, Pardillos GG, Geddert H, Schwalen A, et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.