Abstract
β-barrel membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts are assembled by highly conserved multi-protein complexes. The mechanism by which these molecular machines fold and insert their substrates is poorly understood. It has not been possible to dissect the folding and insertion pathway because the process has not been reproduced in a biochemical system. We purified the components that fold and insert E. coli outer membrane proteins and reconstituted β-barrel protein assembly in proteoliposomes using the enzymatic activity of a protein substrate to report on its folding state. The assembly of this protein occurred without an energy source but required a soluble chaperone in addition to the multi-protein assembly complex.
The outer membranes of Gram-negative bacteria and the mitochondria and chloroplasts of higher eukaryotes contain proteins with β-barrel structure, which are assembled in their respective membranes by multi-protein machines (1-6). The folding and insertion of these β-barrels must be coordinated because they would have many unsatisfied hydrogen bonds in the membrane if they were inserted in an unfolded state, but, conversely, they would be “inside out” if they folded first in the aqueous environment and were subsequently inserted. In order to understand how β-barrel proteins assemble into membranes, we purified the proteins comprising the E. coli outer membrane protein (OMP) folding machinery and established a reconstituted system to monitor the activity of this machinery.
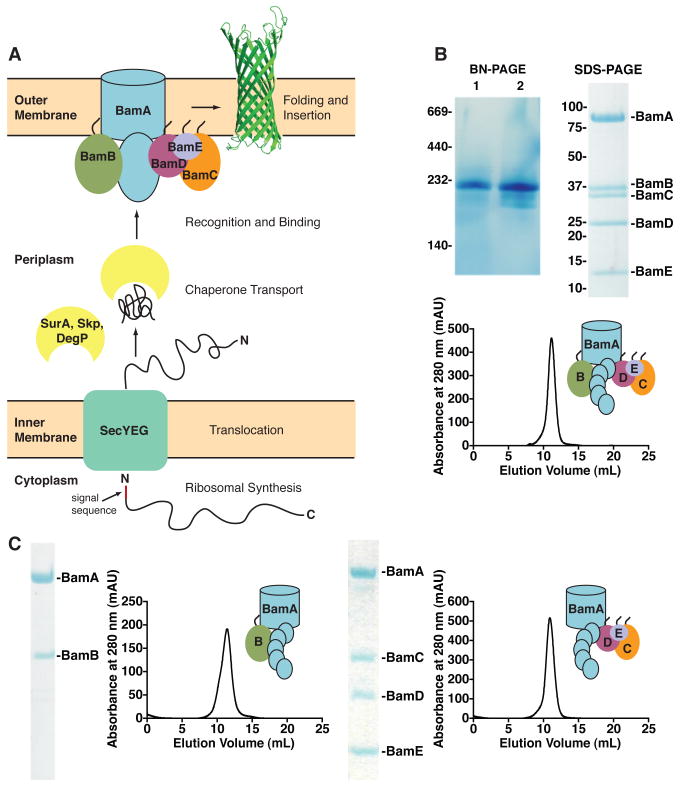
The β-barrel assembly machine (Bam) in E. coli consists of an integral β-barrel protein, BamA (formerly YaeT), and four lipoproteins, BamB, C, D, and E (formerly YfgL, NlpB, YfiO, and SmpA, respectively). Only BamA and BamD are essential for cell survival, but deleting or depleting any member of the complex causes defects in OMP assembly (6-9). BamA has homologs in prokaryotes and eukaryotes and contains five periplasmic polypeptide transport associated (POTRA) domains, which scaffold the Bam lipoproteins (2-5, 10). Unfolded OMPs are delivered to this complex following their synthesis in the cytoplasm and translocation across the inner membrane by the secretion machinery (SecYEG) (Figure 1A) (11). The chaperones, SurA, Skp, and DegP, prevent unfolded OMPs that are released from the Sec machine from aggregating and misfolding as they transit the periplasm. These chaperones are thought to transport OMPs in two parallel, but separate pathways—one that relies on SurA and one that involves both Skp and DegP (12, 13). Most OMPs can be handled by either pathway, but SurA delivers the bulk of OMPs to the OM (14).
Figure 1.
The outer membrane protein assembly pathway and the purified Bam complex and sub-complexes. A. The E. coli OMP biogenesis pathway. B. Blue native gel analysis, SDS-PAGE, and gel filtration chromatogram of the purified Bam complex. The complex isolated from cells expressing the complex at basal levels (lane 1) and the over-produced and reconstructed complex (lane 2). C. SDS-PAGE and gel filtration chromatograms of the purified two- and four-protein Bam sub-complexes.
The process of OMP assembly can be monitored in isolated mitochondria (15) and in vivo in E. coli (16). We sought to develop an in vitro system to study the function of the Bam proteins. Expressing all five bam genes in a single strain produced a mixture of complexes and sub-complexes that could not be easily separated. Previous lipoprotein and POTRA domain deletion experiments indicated that BamA binds BamB independently from BamCDE (8, 10); thus, we expressed the sub-complexes BamAB and BamCDE separately and reconstructed the full complex in vitro (17). By blue native polyacrylamide gel electrophoresis (BN-PAGE), the reconstructed complex was identical to the native complex (Figure 1B). The purified Bam complex had an apparent molecular weight that is too small to accommodate more than one copy of BamA and the ratio of BamA:B:C:D was 1:1:1:1 (Figure S1). Either one or two copies of BamE may have been present; its small size prevents a definitive conclusion. The Bam complex may exist as a higher order oligomer in the membrane in vivo, but the relative stoichiometry is expected to be as we have determined.
Purified Bam complex was incorporated into liposomes, as others have done to reconstitute complexes that handle membrane proteins (18-22). To follow OMP assembly in the proteoliposomes, we used a substrate OMP that possesses enzymatic activity, OmpT. OmpT is a β-barrel outer membrane protease that cleaves peptides between two consecutive basic residues, and its activity can be monitored using a fluorogenic peptide (23). Urea-denatured OmpT was incubated with the periplasmic chaperone SurA, and this chaperone-OmpT complex was diluted into solutions containing the proteoliposomes, the fluorogenic peptide, and lipopolysaccharide (LPS), which is required for OmpT activity (17). OmpT assembly was observed as an increase in the rate of fluorescence production.
OmpT activity increased with the concentration of SurA and in the presence of the Bam complex (Figures 2A, S2, and S4). OmpT activity saturated at the same SurA concentration in the presence or absence of the complex, which suggests that SurA plays a general role in delivering the unfolded protein in a state that can be folded and that the Bam complex handles OmpT bound to SurA (Figure 2B and C). The concentrations of SurA at which we saw a significant improvement in OmpT assembly (above 20 μM) are comparable to the reported binding constants for SurA and model peptides (1-14 μM) (24, 25). OmpT contains several putative SurA binding sites (26, 27), and the sigmoidal relationship may indicate that multiple SurA molecules are involved in generating a SurAn-OmpT complex that can be folded efficiently.
Figure 2.
SurA and the Bam complex facilitate OmpT assembly in proteoliposomes. A. Preincubated solutions of urea-denatured OmpT with SurA were diluted (at t = 0 min) into liposomes (left panel) or proteoliposomes that contain the Bam complex (right panel). OmpT was present in each reaction at a final concentration of 10 μM, and the concentration of SurA was varied from 0-100 μM. B. The amount of active OmpT (reflected in the slope from 15-30 minutes) in the reactions in A as a function of SurA concentration in the presence (black triangles) or absence (red squares) of the Bam complex. C. Schematic of the reconstitution reaction pathway.
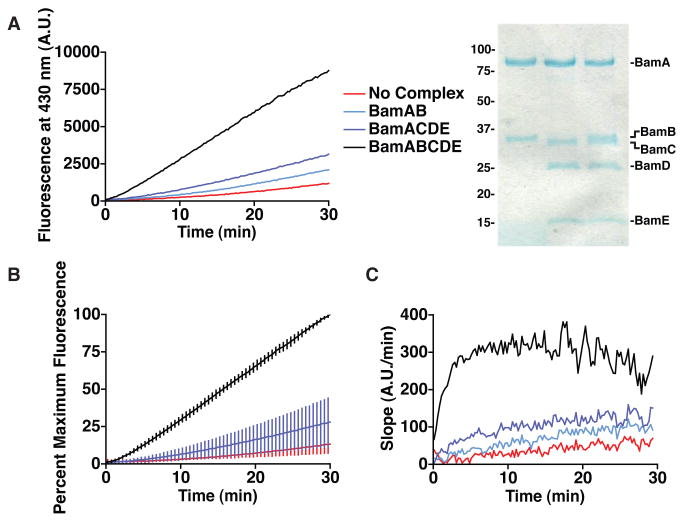
It was important to determine whether the assembly observed in the presence of the Bam complex reflected the function of the complex and not a non-specific property of the proteoliposomes (28, 29). We compared the activity of the five-protein Bam complex to sub-complexes containing two (BamAB) and four proteins (BamACDE), which had comparable stability as judged by their behavior on gel filtration chromatography (Figure 1C). The components of these sub-complexes were incorporated into proteoliposomes in equal proportion to those of the five-protein complex (Figure 3A). Cleavage of the fluorogenic substrate was greatly reduced in proteoliposomes containing the sub-complexes compared to those containing the five-protein complex (Figures 3A and S3). The activity of the four-protein complex was always less than that of the five-protein complex (Figure 3B). Importantly, in the first five minutes of the experiment, OmpT assembly was significantly faster in the presence of the five-protein complex (Figure 3C). These large differences in activity at early time points are relevant given that in vivo pulse-chase experiments indicate that OMP assembly occurs in the cell on a similar time scale of 30 seconds to several minutes (30, 31). The four-protein complex appeared to have some activity on this time scale, which BamB dramatically improved. While there was a background rate of OmpT folding in the absence of the Bam complex, the Bam proteins significantly improved the kinetics of OmpT assembly.
Figure 3.
OmpT assembly requires specific components of the Bam complex. A. Fluorescence produced in the presence of the Bam complex and sub-complexes (left panel). OmpT and SurA are diluted to final concentrations of 10 μM and 100 μM, respectively. SDS-PAGE of these proteoliposomes indicates that they contain equal amounts of the complexes (right panel). B. Eight different experiments were normalized to their maximum fluorescence values and then averaged. The error bars represent the standard deviation among these experiments. C. The gradient of the fluorescence data in A.
To verify that increased fluorescence correlated with increased amounts of folded OmpT, we ran the products of our folding reactions on SDS-PAGE gels without and with prior heat denaturation. β-barrel proteins remain folded on SDS-PAGE if they are not boiled and consequently run faster than their unfolded forms. Purified 35S-labeled OmpT was incubated with SurA and then diluted into proteoliposomes as in the fluorescence experiments. After 30 minutes, the reaction solutions were centrifuged and the pellets were run on SDS-PAGE (Figures 4 and S5). Much more folded OmpT was observed in the reaction of SurAn-OmpT with the full Bam complex than in the reactions with the sub-complexes. The yield of folded protein represented by the faster migrating band was about 7% as determined by five separate experiments. The folded OmpT was resistant to extraction from the pellet by incubation in 100 mM sodium carbonate for 30 minutes, suggesting that the folded protein had been integrated into the membrane.
Figure 4.

OmpT assembled by the Bam complex is folded on SDS-PAGE and resistant to membrane extraction. Pellets of the reactions of SurA and 35S-labeled OmpT with proteoliposomes containing the Bam complexes. 35S-labeled OmpT and SurA were diluted to final concentrations of 0.4 μM and 100 μM, respectively. Extraction in 100 mM sodium carbonate results in a three-fold enrichment of the folded material relative to the unfolded material.
The non-essential lipoprotein BamB is important in the assembly of OMP substrates delivered by SurA; the five-protein complex had significantly higher activity than the four-protein complex, BamACDE. Thus, some or all of the BamCDE proteins are required for OmpT assembly, but BamB appears to have a specific function that, when coordinated with the other components, increases the activity of the complex. The functional importance of both SurA and BamB is consistent with in vivo observations that these proteins play related, but not redundant roles in OMP biogenesis. Deleting surA or bamB produces identical phenotypes with respect to the kinetics of conversion of unfolded, mature LamB to folded monomers (31). In addition, simultaneous deletion of the surA and bamB genes results in a phenotype that is severely defective in the assembly of OMPs and much sicker than either single deletion strain (32, 33). Our results also suggest that SurA and BamB both affect the efficiency of OMP assembly. Our highly simplified system thus appears to recapitulate elements of the cellular process and clearly demonstrates that a non-essential protein can alter the activity of essential proteins to regulate the efficiency of the process they catalyze.
We do not know if the Bam complex in proteoliposomes can process multiple substrate molecules, but clearly the Bam complex can assemble a β-barrel protein in the membrane without an input of energy. The inner membrane secretion machinery, by contrast, couples energy-driven translocation to insertion (11). While it remains possible that β-barrel assembly may be coupled to an energy-driven process in the cell, it is remarkable that these six proteins (the Bam complex and a chaperone) are sufficient to perform the folding and insertion process. Together they perform a chemical transformation that we do not understand, and that they are able to do so without energy suggests that their structure inherently facilitates OMP assembly.
Supplementary Material
Acknowledgments
The Biophysics Resource of the Keck Facility at Yale University performed the size exclusion-light scattering analysis and amino acid analysis. This work is supported by NIH Grant AI081059. C.L.H. is supported by a NSF Graduate Research Fellowship and a Kellogg fellowship from Amherst College.
Footnotes
Supporting Online Material: www.sciencemag.org
Materials and Methods
References
- 1.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 2.Reumann S, Davila-Aponte J, Keegstra K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc Natl Acad Sci USA. 1999;96:784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 4.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 5.Paschen SA, et al. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 6.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Eggert US, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 8.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 9.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 11.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 12.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–2443. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutik S, et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 16.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Information on materials and methods is available on Science Online.
- 18.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 19.Nicchitta CV, Blobel G. Assembly of translocation-competent proteoliposomes from detergent-solubilized rough microsomes. Cell. 1990;60:259–269. doi: 10.1016/0092-8674(90)90741-v. [DOI] [PubMed] [Google Scholar]
- 20.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 22.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 23.Kramer RA, Zandwijken D, Egmond MR, Dekker N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease OmpT. Eur J Biochem. 2000;267:885–893. doi: 10.1046/j.1432-1327.2000.01073.x. [DOI] [PubMed] [Google Scholar]
- 24.Bitto E, McKay DB. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- 25.Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem. 2005;280:23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Wang S, Hu YX, McKay DB. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J Mol Biol. 2007;373:367–381. doi: 10.1016/j.jmb.2007.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 28.Kleinschmidt JH. Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers. Chem Phys Lipids. 2006;141:30–47. doi: 10.1016/j.chemphyslip.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen C, Heutink M, Tommassen J, de Cock H. The assembly pathway of outer membrane protein PhoE of Escherichia coli. Eur J Biochem. 2000;267:3792–3800. doi: 10.1046/j.1432-1327.2000.01417.x. [DOI] [PubMed] [Google Scholar]
- 31.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.