Abstract
The NF-κB family of transcription factors has been implicated in the propagation of ovarian cancer, but the significance of constitutive NF-κB signaling in ovarian cancer is unknown. We hypothesized that constitutive NF-κB signaling defines a subset of ovarian cancer susceptible to therapeutic targeting of this pathway. We investigated the biological relevance of NF-κB in ovarian cancer using a small molecule inhibitor of IKKβ, and confirmed with RNA interference towards IKKβ. We developed a gene expression signature of IKKβ signaling in ovarian cancer using both pharmacologic and genetic manipulation of IKKβ. The expression of IKKβ protein itself and the 9-gene ovarian cancer-specific IKKβ signature were related to poor outcome in independently collected sets of primary ovarian cancers (p=0.02). IKKβ signaling in ovarian cancer regulated the transcription of genes involved in a wide range of cellular effects known to increase the aggressive nature of the cells. We functionally validated the effect of IKKβ signaling on proliferation, invasion and adhesion. Downregulating IKKβ activity, either by a small molecule kinase inhibitor or by shRNA depletion of IKKβ, blocked all of these cellular functions, reflecting the negative regulation of the target genes identified. The diversity of functions controlled by IKKβ in ovarian cancer suggest that therapeutic blockade of this pathway could be efficacious if specific IKKβ inhibitor therapy is focused to patients whose tumors express a molecular profile suggestive of dependence on IKKβ activity.
Keywords: NF-κB, IKK, gene expression signature, ovarian cancer, signal transduction
INTRODUCTION
Ovarian cancer is the fifth most common form of cancer in women in the United States, resulting in over 15,000 deaths per year, making it the most lethal gynecological cancer in this country (1). The majority of deaths are attributed to serous carcinoma, most commonly identified at an advanced stage with metastases present beyond the ovaries, which precludes curative treatment. Goals in the field include improved screening and diagnostics, and until they result in early detection, improved therapy of advanced disease at the outset and for recurrence (2, 3). Gene expression profiling studies have identified subsets of patients presenting aggressive disease who respond differently to standard surgery and chemotherapy treatment (4). Attempts were made to identify molecular signatures of patients that correlate with better survival. The Cancer Genome Atlas project has rapidly and comprehensively advanced the molecular profiling of ovarian cancer through large-scale gene expression profiling, comparative genomic hybridization, single nucleotide polymorphism analysis, and gene exon sequencing (cancergenome.nih.gov). Analysis of these efforts has made clear that ovarian cancer is an extremely heterogeneous disease. A single approach to chemotherapy for all ovarian cancer patients is unlikely to achieve similar success across all patients. Therefore, there is pressing need to identify the molecular etiology driving defined subgroups of ovarian cancers, and to develop alternate treatments targeting such pathways in order to improve specific patient survival.
The involvement of NF-κB in cancer dissemination makes it a logical target (5). The NF-κB family of transcription factors is ubiquitously expressed and has been studied extensively in lymphoid development and lymphoid malignancies. These transcription factors have been implicated in the propagation of solid tumors (6). While found in ovarian cancer, the significance and the mechanism of constitutive NF-κB signaling in ovarian cancer is unexplored. We hypothesized that constitutive NF-κB signaling defines a subset of ovarian cancer susceptible to therapeutic targeting of this pathway.
Constitutive NF-κB signaling has been identified in tumors of epithelial origin including breast, colon, lung and ovarian carcinomas (6). A systems biology approach integrating three global screening techniques discovered IKKε, a cytosolic kinase involved in triggering NF-κB signaling, as an oncogene in breast cancer (7). Recent work suggested the importance of these transcription factors in the propagation of ovarian cancer cell lines (8). We recently demonstrated the NF-κB family proteins are expressed and co-regulated in primary ovarian cancer tumors, and overexpression of the NF-kB subunit p50 at diagnosis conveyed poor outcome (Annunziata, et al, Cancer, in press).
Having established the coordinate presence of the NF-κB machinery in ovarian cancers, we sought to modulate its activity. The NF-κB family consists of five subunits that join into active dimers. Homo- or hetero-dimers form the active transcription factor complex. Inhibitors of NF-κB (IκBs) are tagged for degradation through the proteasome upon specific inducible phosphorylation by IκB kinases (IKKs) (9). Therefore, targeted inhibition of IKKs could isolate NF-κB as a mechanism for the growth and survival of ovarian cancers. IKKβ was the focus of the current investigation in ovarian cancer. Here we examined the biological relevance of NF-κB in ovarian cancer using a small molecule inhibitor of IKKβ, ML120b (Millennium Pharmaceuticals) and confirmed with RNA interference using stably expressed short hairpin RNA molecules towards IKKβ.
MATERIALS AND METHODS
Reagents
LPA was purchased from Avanti Polar Lipids (Alabaster, AL). Before use LPA was dissolved in PBS containing 1% fatty acid-free bovine serum albumin. ML120b (N-(6-chloro-7-methoxy-9H-β-carbolin-8-yl)-2-methylnicotinamide), a b-carboline compound, was obtained from Millennium Pharmaceuticals (Cambridge, MA) (10). IKK-2 inhibitor IV (5-(p-Fluorophenyl)-2-ureido]thiophene-3-carboxamide, catalog #401481), a ureidocarboxamido thiophene compound; and IKK-2 inhibitor VI (5-Phenyl-2-ureido)thiophene-3-carboxamide, catalog #401483) a ureido-thiophenecarboxamide compound, were purchased from EMD Biosciences (La Jolla, CA). Each compound is reportedly highly specific to IKKβ kinase inhibition (10, 11). Puromycin, DMSO, MTT, XTT, PMS, Noble agar and mouse monoclonal antibody to β-tubulin (T5201) were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies to IKKα (2682) were from Cell Signaling Technology, Inc (Danvers, MA), IKKβ (32135) from Abcam (Cambridge, MA), and IKKε (07-580) from Millipore (Billerica, MA).
Patients and samples
The Regional Committee for Medical Research Ethics in Norway approved this study. Tissue microarrays (TMAs) containing 2-mm cores (n=270) from 119 ovarian carcinoma specimens (42 primary carcinomas, 77 solid metastases) operated on at the Norwegian Radium Hospital were additionally studied. Tumors were predominantly (>80%) of the serous type. Metastases were to the omentum (n=46), peritoneum (n=15), intestine (n=12), lymph nodes (n=2) or various other sites (n=2). Tumors were from 56 patients, of whom 38 had both primary carcinoma and one or more metastasis for evaluation, 4 had only primary carcinoma and 14 had only one or more metastasis. Tumors underwent microscopic confirmation of diagnosis, histological type and grade by a gynecopathologist (BD). Grading was according to the FIGO system.
Gene expression profiles were obtained from http://www.ncbi.nlm.nih.gov/geo/ (4, 12). The probesets in the IKKβ signature are CLDN1, 218182_s_at; CXCL1, 204470_at; CXCL2, 209774_x_at; IL8, 211506_s_at; INSIG1, 201627_s_at; ITGB6, 208083_s_at; PTGER2, 206631_at; PTGS1, 215813_s_at; SOD2, 215078_at.
Immunohistochemistry
Expression was scored based on the number of positive cells in a sample rather than the intensity of staining, in order to minimize bias due to technical variation across samples. Staining extent was scored on a scale of 0 to 4, as follows: 0 = no staining, 1 = 1%–5%, 2 = 6% –25%, 3 = 26%–75%, 4 = 76%–100% stained tumor cells.
Statistical analysis
Statistical analysis was performed applying the SPSS- PC package (Version 15.0, Chicago IL). Probability of <0.05 was considered statistically significant. Analysis of the association between IKKβ expression by immunohistochemistry and clinicopathologic parameters was undertaken using the two-sided Chi-squared test. Progression-free survival (PFS) and overall survival (OS) were calculated from the date of diagnosis to the date of recurrence/death or last follow-up. Univariate survival analyses of PFS and OS were executed using the Kaplan-Meier method and log-rank test. Expression categories in the latter test were grouped based on staining extent above or below 25%, so as to allow for a sufficient number of cases to be included in each category.
Cell lines and culture
Ovarian cancer cell lines CAOV3, SKOV3 cells were obtained from ATCC, A2780 cell line was a gift from Tito Fojo (National Cancer Institute, Bethesda MD), HeyA8 was a gift from Gordon Mills (MD Anderson Cancer Center, Houston TX), and human vascular endothelial (HMVEC) cells were obtained from Cascade Biologics (Invitrogen). OVCAR5, OVCAR8 and IGROV1 cells were from the NCI-Frederick Developmental Therapeutics Program tumor/cell line repository (Frederick, MD). All lines were cultured in RPMI plus 10% fetal bovine serum (Hyclone, Pittsburg, PA) and standard antibiotics. Cell lines were authenticated in July 2009 at the Johns Hopkins University Fragment Analysis Facility (Baltimore, MD), using Promega PowerPlex 1.2 System to test for 8 STR markers (D16S539, D7S820, D13S317, D5S818, CSF1PO, TPOX, THO1, vWA) and amelogenin for gender determination. Authenticity was confirmed against the ATCC database (www.atcc.org/CulturesandProducts/CellBiology/STRProfileDatabase/tabid/174/Default.aspx), CLIMA database (http://bioinformatics.istge.it/clima/) and NCI-60 database published data (13).
Cell growth assays
Attached ovarian cancer cell growth was assessed using MTT and XTT as described (14). Plates were incubated for up to 10 days, and inhibitors or vehicle replenished every 3–4 days. Anchorage-independent cell growth was assessed using XTT as described (15). Briefly, cells were seeded in 96-well plates pre-coated with 0.5% soft agar in 0.3% soft agar/10% RPMI at a density of 1–2,000 cells/50µl/well and incubated overnight. Cell density in treated wells was expressed as a percent of vehicle-treated control wells. Experiments included duplicate samples and were repeated at least three times.
Cell invasion assays
Ability of ovarian cancer cells to migrate and invade through Matrigel or basement membrane extract was measured using 12-well Biocoat Matrigel Invasion Chambers (BD Biosciences, San Jose, CA) or Cultrex 96-well BME Cell Invasion Assays (Trevigen, Gaithersburg, MD) according to manufacturer specifications. Briefly, cells suspended in serum-free RPMI were plated in coated chambers in the presence or absence of 25µM ML120b, and allowed to migrate toward RPMI containing 10% FCS as a chemoattractant, for 24h. Matrigel chambers were stained and cells were counted. Migrated cell number in duplicate ML120b-treated samples was compared to vehicle-control treated samples. Cells migrated through BME chambers in IKKβshRNA-inhibited/ controlshRNA samples, ML120b-treated and vehicle-treated samples were stained with calcein, solubilized, and numbers assessed by measuring fluorescence. Migrated cell numbers in triplicate samples were reported as percent control.
Heterotypic cell adhesion assays
The attachment of ovarian cancer cells to human vascular endothelial cells was assessed using the Cytoselect Tumor-endothelium Adhesion Assay (Cell Biolabs, Inc., San Diego, CA) according to manufacturer specifications. Briefly, ovarian cells pre-exposed to ML120b or vehicle control for 48–72 hours were labeled with a fluorescent cytotracker dye in serum-free RPMI and allowed to adhere to a monolayer of HMVEC cells in a 96-well plate. Plates were incubated for 90 minutes, followed by gentle serial washing to dislodge unattached cells. Cells were solubilized and supernantant fluorescence measured in a Tecan fluorimeter (Tecan, Research Triangle Park, NC). Cell number from triplicate samples were reported as percent control.
IKKβ shRNA depletion
IKKβ shRNA retroviral constructs have been previously described (16). Two rounds of viral supernatants were applied to ovarian cancer cell lines over the course of 48h, followed by incubation with growth medium for 24h, and selection with 2ug/ml puromycin for 4 days. Selected cells were used as RNA sources for microarray analysis and all other assays.
Western analysis
Protein from nuclei and cytoplasm was extracted from ovarian cell cultures using the Biovision Nuclear/Cytosol Fractionation kit according to manufacturer protocol (Biovision, Mountain View, CA) and concentrations estimated with BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). SDS-Page and Western analysis were performed using, respectively, the NuPage system (Invitrogen, Carlsbad, CA) and the Supersignal Chemiluminescent Substrate system (Thermo Scientific, Rockford, IL).
RNA isolation and microarray analysis
Total RNA was isolated from 6 independent cultures of ovarian cells using Trizol (Invitrogen, Carlsbad, CA) according to manufacturer protocol. Samples were submitted to the Laboratory of Molecular Technology Microarray Core, SAIC/NCI-Frederick and RNA quality checked on an Agilent Bioanalyzer; only samples having a high quality score (RIN>9) were used for microarray analysis. All replicates of experimental samples were prepared, labeled and hybridized to Affymetrix H133 Plus 2.0 gene chips and scanned on an Affymetrix GeneChip scanner 3000 (Affymetrix, Santa Clara, CA). Data analysis was performed using the RMA normalization algorithm of BRB-Array Tools version 3.7(Biometrics Research Branch, NCI, publicly available at http://linus.nci.nih.gov/BRB-ArrayTools.html ). Genes showing minimal variation across the set of arrays were excluded from the analysis. Genes whose expression differed by at least 1.5 fold from the median in at least 10% of the arrays, or within the 25th the percentile of the log-ratio variation were retained. Class comparison was used to identify genes that were differentially expressed among control and test groups using a random variance t-test. Gene expression differences were considered statistically significant if their p-value was less than 0.001. Data sets containing significant genes deregulated in ML120b treated samples and in shRNA induced samples were compared using the Compare function of Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com).
Quantitative real-time PCR
cDNA was synthesized from total RNA using Superscript II RT (Invitrogen, Carlsbad, CA). PCR was performed using SYBR PCR Master Mix (Applied Biosystems, Foster City, CA) on an ABI 7500 thermal cycler. B2M expression was used as an internal control to normalize between samples. Primer sequences are:
IKBKB: 5'- TCCGATGGCACAATCAGGAAAC -3' (forward),
5'-TCCAGGCACCACCGCTCTC -3' (reverse)
ITGB6: 5'- GAAGACTGCCTGCTTATTGGACCTC -3' (forward),
5'- TGCTGGGGTATCACACCTTTCG -3' (reverse)
CLDN1: 5'- CCATCGTCAGCACTGCCCTG -3' (forward),
5'- AGGACATCCACAGCCCCTCG -3' (reverse)
IL8: 5'- TCAGAGACAGCAGAGCACACAAGC -3' (forward),
5'- CACACAGTGAGATGGTTCCTTCCG -3' (reverse)
CXCL2: 5'- TGTCAGTGCTGCTACTCCACCTCTG -3' (forward),
5'- GCTGCCGTGTGAAGCCCAC -3' (reverse)
Primers for B2M and PTGS1 were purchased from RealTimePrimers.com
Cytokine analysis
Cells stably expressing IKKβ shRNA or a control shRNA were seeded in a 96-well format at a density of 1000 cells/well. After 72 hours cells were serum starved for 24 hours in RPMI 1640 0.5% fetal calf serum, and then stimulated with 10µM LPA in medium containing 0.5% fetal calf serum. Supernatants were harvested 18 hours later. Cells treated with IKK-2 Inhibitor IV (2µM) were handled similarly, except treatment was begun 48 hours before supernatant collection. IL-8 concentration was measured using the Quantikine Human CXCL8/IL-8 ELISA (R&D Systems, Minneapolis, MN) as per the manufacturer’s instructions. Values were normalized to cell density as determined by XTT.
RESULTS
IKKβ protein expression is associated with poor overall survival in ovarian cancer
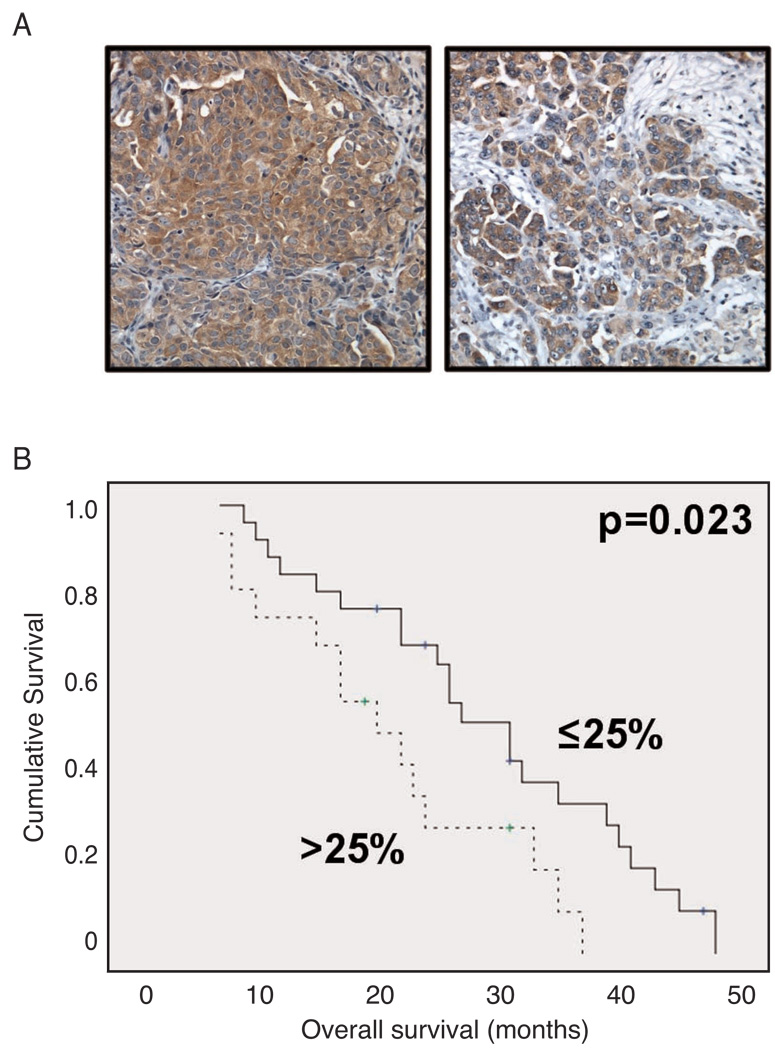
IKKβ expression was explored by immunohistochemistry in 119 tumors comprised of 42 primary carcinomas and 77 solid metastases on tissue microarray. Of the 270 tissue cores, 261 were informative. IKKβ was expressed in the cytoplasmic location in 89 of 119 tumors (Figure 1A), with the following staining extent: score=1: 27 tumors; score=2: 14 tumors; score=3: 23 tumors; score=4: 25 tumors. Expression in primary carcinomas and metastases was comparable (data not shown). High expression of IKKβ, as indicated by greater than 25% of cells expressing IKKβ, was associated with poor overall survival in primary carcinomas (Figure 1B; median survival 20 months versus 31 months, p=0.023). IKKβ was not differentially expressed in tumors based on stage, grade or level of residual disease at the time of initial surgery (Supplemental Table).
Figure 1. IKKβ protein expression is associated with poor overall survival in ovarian cancer.
A – IKKβ staining in two serous carcinomas. All tumor cells are positive. B – Kaplan-Meier survival curve showing the association between IKKβ expression by immunohistochemistry in primary carcinomas and overall survival (OS) for 42 patients. Patients with tumors expressing IKKβ in >25% of cells (n=16, dashed line) had a mean OS of 21 months compared to 29 months for patients with tumors with expression in ≤25% of cells (n=26, solid line; p=0.023).
IKKβ regulates critical target genes in ovarian cancer
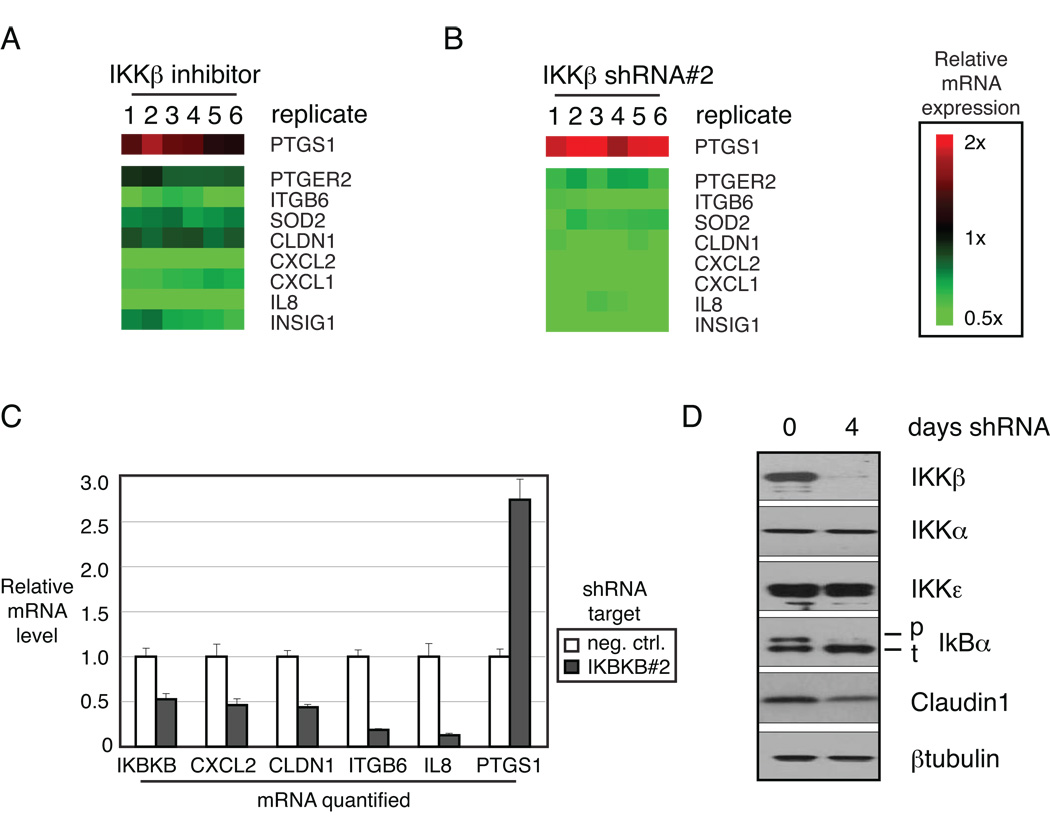
Ovarian cancer cell line CAOV3 expresses a moderate level of IKKβ protein compared to most ovarian cancer cell lines tested (Figure S2A). CAOV3 was treated with a highly specific IKKβ small molecule inhibitor ML120b and gene expression was profiled to measure changes in target genes specifically regulated by IKKβ. Differentially expressed genes were selected by class comparison analysis using BRB Array Tools (http://linus.nci.nih.gov/BRB-rrayTools.html; Figure 2A). The same cells were subjected to RNA interference with two individual short hairpin RNAs (shRNAs) directed against IKKβ, in order to confirm the target gene list (Figure 2B and Figure S1). The overlapping set of 9 genes, 8 down regulated and 1 upregulated, create a novel ovarian cancer-specific signature of IKKβ-regulated genes. The individual genes were validated by quantitative PCR after either small molecule IKKβ inhibition or shRNA-mediated IKKβ depletion (Figure 2C and Figure S1).
Figure 2. IKKβ regulates critical target genes in ovarian cancer.
IKKβ target genes in ovarian cancer: A – CAOV3 cells were treated with the IKKβ inhibitor ML120b for the indicated times and gene expression changes were assessed using Affymetrix U133plus2.0 microarrays and depicted according to the color scale shown. B – IKKβ was depleted in CAOV3 cells by retroviral transduction of targeted shRNA. Regulated genes were identified by class comparison in six replicates compared to control shRNA. C – IKKβ target gene regulation was confirmed with quantitative RT-PCR after shRNA-mediated depletion of IKKβ. D – Specificity of shRNA target was confirmed by western blot demonstrating depleted protein expression of IKKβ, but not IKKα or IKKε. Functionally, IKKβ depletion resulted in decreased phosphorylation of IκBα and lower level of IKKβ target Claudin1.
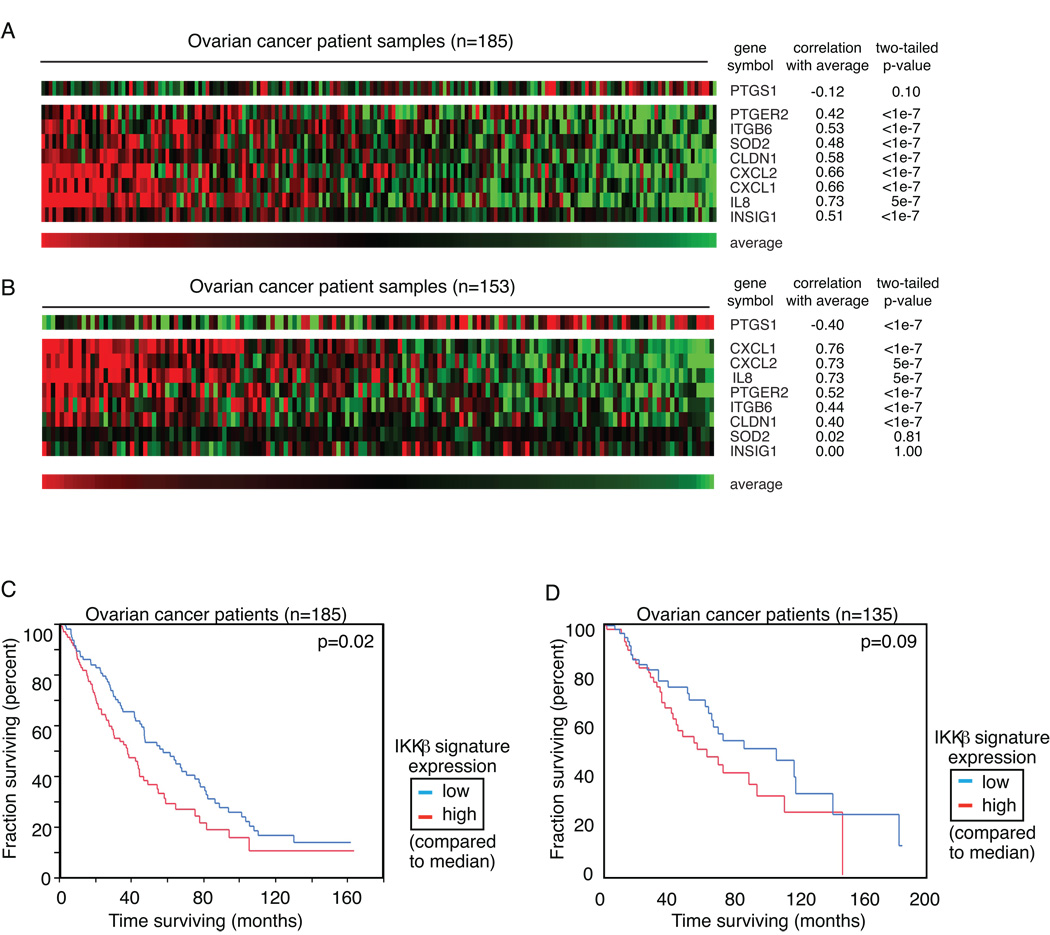
The gene set was then evaluated in two independently collected cohorts of ovarian tumor specimens profiled at the time of initial surgical resection (4, 12). Both of these datasets are publicly available at http://www.ncbi.nlm.nih.gov/geo/. The 9 genes in the ovarian cancer-specific IKKβ signature were highly correlated with each other, suggesting that they are co-regulated in ovarian cancer (Figure 3A and B). The expression of IKKβ gene itself was highly correlated with the group of target genes, (R=0.20, p=0.006, data not shown) suggesting a positive relationship between expression of IKKβ and its activity as represented by downstream target gene regulation. Elevated IKKβ target gene expression was statistically related to worse outcome in the datasets of women with high grade, stage III or IV ovarian cancer at diagnosis (p=0.02; Figure 3C). Patient samples were separated into the high or low cohort based on the median expression of the ovarian cancer IKKβ signature within the group. Patients whose tumors showed activation of IKKβ, based on expression of the signature above the median, had a lower median survival than those who showed signature expression below the median. A second previously published dataset, independently collected and comprised of tumor samples from women with ovarian cancer of any stage, was used to validate these findings. A similar trend was observed in the second dataset, without reaching statistical significance (p=0.09; Figure 3D). The more modest effect in the independent analysis of the second dataset may be due in part to the different characteristics of the patients. The first dataset includes a more homogeneous set of only high grade (III), late stage (IIIC or IV) cancers, while the second set includes all stages, and all grades.
Figure 3. IKKβ target genes are co-regulated in primary ovarian cancers.
A - Affymetrix U133plus2.0 gene expression profiling data from 185 purified cancer cell populations harvested from untreated patients with ovarian cancer (4). Samples are ranked according to the average expression of the 9 IKKβ target genes. Expression was centered based on the median value. Correlation of each gene with the signature average is noted. B - Expression of the 9-gene IKKβ signature was evaluated in an independently collected dataset of ovarian cancers profiled on Affymetrix U133A gene expression arrays (12). Correlation was strongest with 7 of the 9 genes. C – Patients from dataset 1 were separated into 2 groups based on the median expression of the 9-gene IKKβ signature. Overall survival is plotted. D – Patients from dataset 2 were separated into two groups based on the median expression of the genes in the IKKβ signature, and overall survival is again plotted.
IKKβ mediates properties of aggressiveness in ovarian cancer
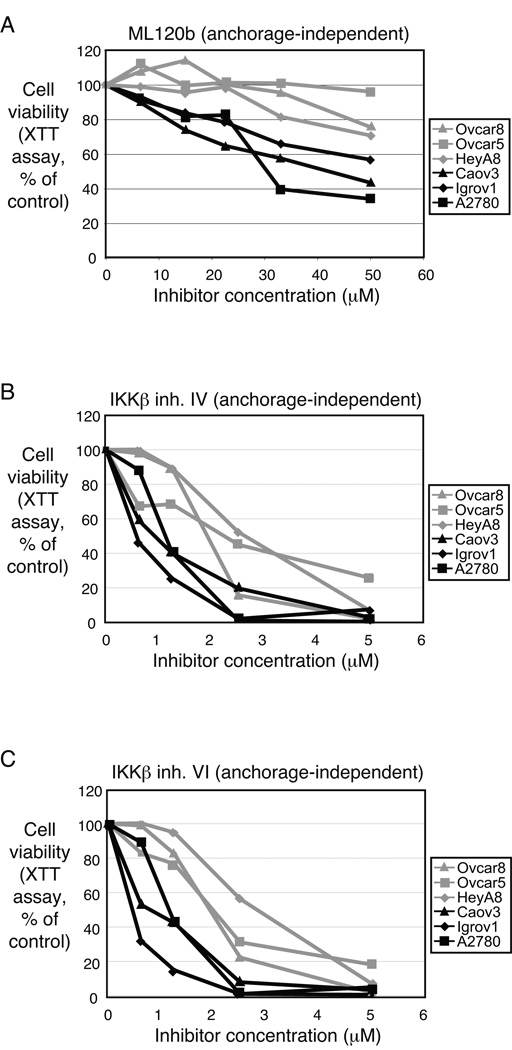
Inhibition of IKKβ differentially suppressed growth of ovarian cancer cell lines. A panel of 6 different ovarian cancer cell lines was treated with three different small molecule inhibitors of IKKβ (Figure 4). Each compound is reportedly highly specific to IKKb kinase inhibition (10, 11). All three inhibitors resulted in a similar pattern of anchorage-independent growth inhibition. The cell lines CAOV3, Igrov1 and A2780 showed at least a 40–60% decrease in viability with IKKβ inhibition, while the cell lines OVCAR5, OVCAR8 and HeyA8 were less susceptible.
Figure 4. IKKβ regulates anchorage-independent growth in ovarian cancer cell lines.
Six ovarian cancer cell lines were treated with small molecule IKKβ inhibitors and assayed for their ability to grow in soft agar. The same profile of sensitivity was apparent in cells treated with (A) ML120b, (B) IKKβ inhibitor IV, or (C) IKKβ inhibitor VI.
Blockade of IKKβ by either small molecule inhibition (Figure 5A, left panel) or targeted RNA interference (Figure 5A, right panel) decreased invasiveness of most ovarian cancer cell lines. Baseline invasion varied modestly between the cell lines and generally correlated with IKKβ protein expression level (Figure S2). A decrease in invasive capacity was seen in OVCAR8 and OVCAR5, cell lines that were insensitive to the growth inhibitory effects of the pathway inhibition, indicating that the pleotropic effects triggered by IKKβ may be regulated differently across ovarian cancer subtypes. This is consistent with the expression patterns demonstrated in the primary tumors (Figure 3A), where the genes were highly correlated across the entire dataset, but individual patients did not have exactly the same pattern in every case.
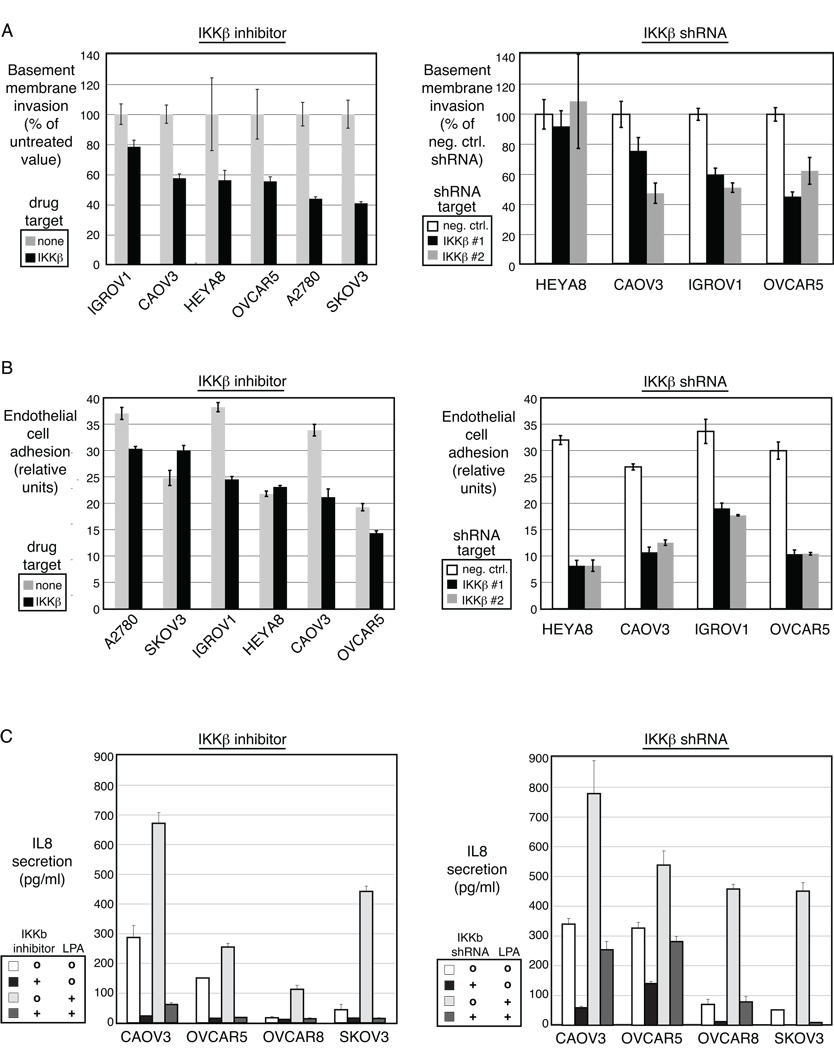
Figure 5. IKKβ mediates properties of aggressiveness in ovarian cancer.
A – Invasion through basement membrane was measured after interruption of IKKβ signaling with the small molecule inhibitor (left panel) or the shRNA directed to IKKβ (right panel). B – Adhesion of ovarian cancer cells to endothelial cells was measured after interruption of IKKβ signaling with the small molecule inhibitor (left panel) or the shRNA directed to IKKβ (right panel). C – IL8 secretion was measured after interruption of IKKβ signaling with the small molecule inhibitor (left) or the shRNA directed to IKKβ (right).
The attachment of ovarian cancer cells to human vascular endothelial cells represents an early function required for initiation of tumor dissemination. Such heterotypic cell adhesion was assessed in the presence of small molecule IKKβ inhibition, or with IKKβ knockdown by shRNA. Heterotypic adhesion of tumor cells to endothelial cells was decreased in the absence of IKKβ signaling by either small molecule inhibition (Figure 5B, left panel) or shRNA depletion of IKKβ (Figure 5B, right panel).
Similarly, secretion of IL-8 was decreased after IKKβ inhibition by either method (Figures 5C). IL-8 gene expression was regulated by IKKβ (Figure 2) and contributed to the ovarian cancer IKKβ signature (Figure 3). Two of the cell lines (A2780 and IGROV1) did not secrete IL-8 at baseline, consistent with prior studies, and therefore this property was not affected by IKKβ blockade (data not shown). LPA is a serum cytokine that is upregulated in ovarian cancer. LPA is known to stimulate IL-8 secretion (17), and the LPA-induced rise in IL-8 secretion was attenuated by IKKβ inhibition by either small molecule or shRNA (Figure 5C). Again, the range of biological effect downstream of IKKβ-signaling in the cell lines mirrors the individual patient samples, which demonstrated varying levels of the 9 IKKβ target genes.
DISCUSSION
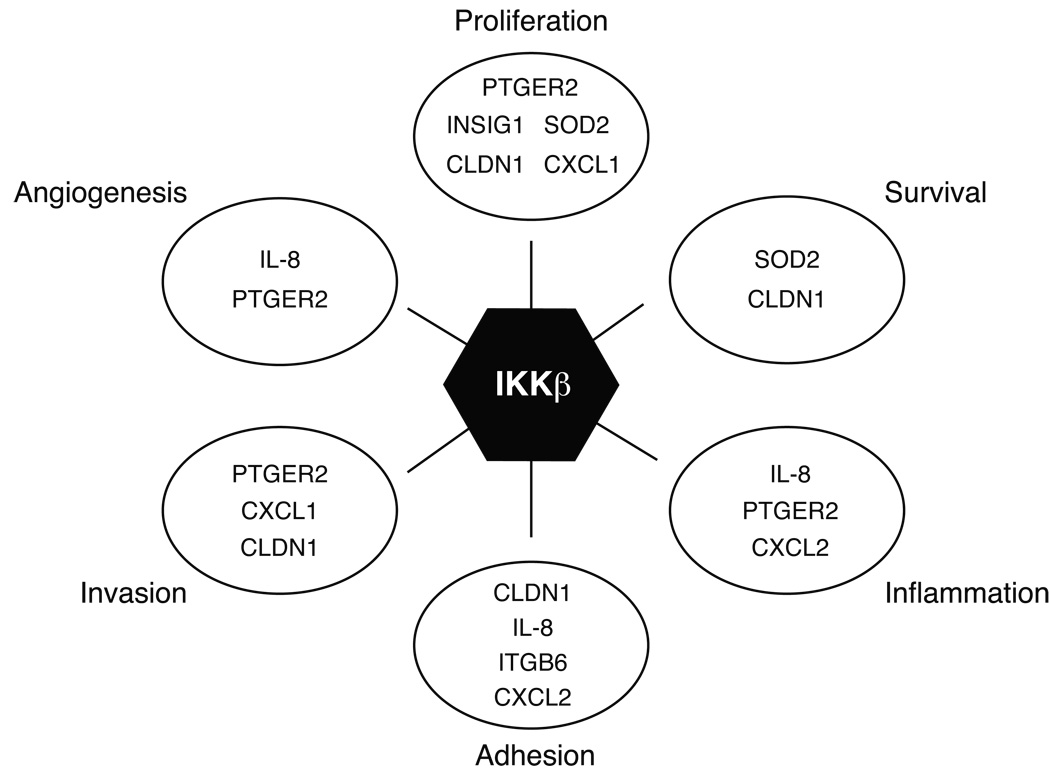
We have shown here that the expression of IKKβ protein itself and the ovarian cancer-specific signature of IKKβ-regulated genes defined a subset of ovarian cancer susceptible to therapeutic targeting of this pathway. The gene signature regulated by IKKβ in cell lines was related to poor outcome in independently collected sets of primary ovarian cancers, suggesting that IKKβ activity contributes to an aggressive phenotype of this disease. The genes regulated by IKKβ signaling in ovarian cancer are known to be involved in a wide range of cellular effects that increase the aggressive nature of the cells, including proliferation, invasion, adhesion, and IL-8 secretion (Figure 6). Overactivation of these cellular activities is consistent with the poor prognostic association in patients. We functionally validated the effect of IKKβ signaling on stimulating proliferation, invasion and adhesion in ovarian cancer cell lines. IKKβ activity promoted all of these cellular functions in ovarian cancer cell lines, reflecting its modulation of the target genes identified.
Figure 6. Pleotropic effects of IKKβ target genes on ovarian cancer aggressiveness.
IKKβ promotes multiple cellular functions in ovarian cancer cells. The genes regulated by IKKβ signaling in ovarian cancer are known to be involved in a wide range of cellular effects that increase the aggressive nature of the cells. IKKβ target genes are indicated in groups based on known functions of proliferation, survival, inflammation, adhesion, invasion and angiogenesis.
This ovarian cancer-specific IKKβ gene signature was developed using an un-biased approach of global gene expression profiling after pharmacologic or genetic manipulation of IKKβ in ovarian cancer cell lines. Therefore, it reflects downstream effects of IKKβ activity, and represents the functions that are regulated by IKKb in these ovarian cancer cells. An overlapping but not identical gene list was recently reported to be IKKβ-dependent in breast cancer cell lines when the E3-ligase KEAP1 was silenced, allowing IKKβ activity to ensue (18). The breast cancer KEAP1-IKKβ gene list was selected based on genes previously known to be NF-κB targets, and lends further confidence to the independently generated signature that we have discovered in ovarian cancer.
The significance of IKKβ activity in ovarian cancer has been characterized by demonstration of its role in multiple biological events necessary for the composite activity of ovarian cancer proliferation and dissemination. Although each cellular process interrogated was blocked by 50% or less in most of the cell lines, such modest effects on individual cellular properties, when taken as a whole, have the potential to result in even more profound suppression of tumor growth in vivo. Interestingly, the pattern of functions mediated by IKKβ was not uniform across the cell lines tested, indicating a differential predominance of each effect among distinct ovarian cancers. For example, the IGROV1 and A2780 cell lines express very little of the IKKβ target gene IL-8, yet they showed sensitivity to IKKβ inhibition in the phenotypes of proliferation and invasion, likely affected by changes in the IKKβ targets CLDN1 and CXCL1 (also known as Growth-related oncogene, GRO1) (19, 20). In other cell lines such as OVCAR5 and OVCAR8, IL-8 was expressed downstream of IKKβ, yet the proliferative rate was less influenced by IKKβ. These findings underscore the heterogeneous nature of ovarian cancer, supporting the hypothesis that there are multiple molecularly defined subtypes of this disease.
It is also interesting to note that the small molecule inhibitor of IKKβ kinase activity showed a slightly different pattern of effect on the panel of ovarian cancer cell lines compared to the shRNA depleting IKKβ protein. One potential reason for this difference could lie in a differential effect caused by inhibiting the kinase activity of the IKKβ protein, versus taking away the entire protein from the cell. This might suggest that the protein can have some function independent of its kinase activity, perhaps as a scaffold for other signaling cascades in the cell.
Diverse mechanisms can trigger NF-κB signaling, and extensive crosstalk exists between major signaling pathways within a cell that can augment the NF-κB signaling cascade (21). Activation of NF-κB in ovarian cancer could be provided by signals in the tumor microenvironment. Inflammatory cytokines and growth factors have been associated with NF-κB activation. One potential cause of IKKβ activation may derive from microenvironmental or paracrine influences such as LPA (22). LPA is found at high levels in the serum of ovarian cancer patients, and is known to activate several cell signaling cascades promoting cancer cell survival and proliferation (23). Our results confirmed the role of IKKβ in relaying LPA-induced signaling in ovarian cancer.
Downstream of cell surface receptors and immediately upstream of IKKβ, many different signaling nodes influence the kinase activity of the IKK complex. Interaction with the stroma, via ICAMs or integrins, may be amplified or perpetuated by mutations in NF-κB activators such as IKKε, or by loss of NF-κB attenuators such as CYLD (24). We previously identified diverse genetic abnormalities that activated IKKβ in multiple myeloma (9). These mechanisms included inactivation of negative regulators CYLD, BIRC2/3 or TRAF3, or overactivation of positive regulators NIK or CD40. Additionally, RAS signaling may activate IKKb and NF-kB through TBK1 (25, 26). Potential activators of IKKβ do not appear in the signature that we present in the current work, due to the manner in which the signature was generated. Activators of IKKβ are of great interest in the context of ovarian cancer, and disease-specific dysregulation of such pathways are the subject of ongoing investigations.
The diversity of functions controlled by IKKβ in ovarian cancer has implications for treatment of this malignancy. Small molecule inhibitors of IKKβ are under preclinical development, yet, like many kinase inhibitors tried thus far in ovarian cancer, they may not exhibit a strong anti-cancer effect as a single agent in most forms of ovarian cancer. Therefore, IKKβ inhibitor therapy, alone or in combination with chemotherapy, should be focused to patients whose tumors express a molecular profile suggestive of dependence on IKKβ activity. This profile could then be monitored in the tumor during therapy for validation of drug effect or mechanisms of resistance.
Supplementary Material
Acknowledgments
Financial support: This work was supported by the Intramural Program of the Center for Cancer Research, NCI (CMA, ECK), the Marsha Rivkin Foundation for Ovarian Cancer Research (CMA) and by grants from the Norwegian Cancer Society and the Health Region of South-Eastern Norway (BD).
Footnotes
Conflicts of interest: none
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Annunziata CM, Azad N, Dhamoon AS, Whiteley G, Kohn EC. Ovarian cancer in the proteomics era. Int J Gynecol Cancer. 2008;18 Suppl 1:1–6. doi: 10.1111/j.1525-1438.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 3.Martin LP, Schilder RJ. Management of recurrent ovarian carcinoma: current status and future directions. Semin Oncol. 2009;36:112–125. doi: 10.1053/j.seminoncol.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bonome T, Levine DA, Shih J, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo JL, Tan W, Ricono JM, et al. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 6.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehm JS, Zhao JJ, Yao J, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Alvero AB, Silasi DA, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27:4712–4723. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagashima K, Sasseville VG, Wen D, et al. Rapid TNFR1-dependent lymphocyte depletion in vivo with a selective chemical inhibitor of IKKbeta. Blood. 2006;107:4266–4273. doi: 10.1182/blood-2005-09-3852. [DOI] [PubMed] [Google Scholar]
- 11.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 12.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 13.Lorenzi PL, Reinhold WC, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8:713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scudiero DA, Shoemaker RH, Paull KD, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- 15.Dent MF, Hubbold L, Radford H, Wilson AP. Use of XTT for quantitating clonogenic growth in soft agar. Cytotechnology. 1996;18:219–225. doi: 10.1007/BF00767769. [DOI] [PubMed] [Google Scholar]
- 16.Ngo VN, Davis RE, Lamy L, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 17.So J, Navari J, Wang FQ, Fishman DA. Lysophosphatidic acid enhances epithelial ovarian carcinoma invasion through the increased expression of interleukin-8. Gynecol Oncol. 2004;95:314–322. doi: 10.1016/j.ygyno.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Lee DF, Kuo HP, Liu M, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dos Reis PP, Bharadwaj RR, Machado J, et al. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer. 2008;113:3169–3180. doi: 10.1002/cncr.23934. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Hendricks DT, Wamunyokoli F, Parker MI. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res. 2006;66:3071–3077. doi: 10.1158/0008-5472.CAN-05-2871. [DOI] [PubMed] [Google Scholar]
- 21.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Lin X. Beta-arrestin 2 is required for lysophosphatidic acid-induced NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:17085–17090. doi: 10.1073/pnas.0802701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang X, Gaudette D, Furui T, et al. Lysophospholipid growth factors in the initiation, progression, metastases, and management of ovarian cancer. Ann N Y Acad Sci. 2000;905:188–208. doi: 10.1111/j.1749-6632.2000.tb06550.x. [DOI] [PubMed] [Google Scholar]
- 24.Hutti JE, Shen RR, Abbott DW, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.