Abstract
Recent advances in genome technology have enabled genome-wide searching for disease predisposition loci, using dense SNP and haplotype maps. Over the past year, such approaches have yielded positive results in human hypertension. Here we outline factors underlying the rationale for the approach, and consider reasons for false positive and negative results. While the approach has positive results, typically the trait-associated loci explain only a small fraction of the heritable fraction of trait variance. Finally, we consider alternative approaches and emerging strategies to probe the role of heredity in control of blood pressure.
Keywords: Hypertension, genomics, association
COMPLEX TRAITS: EMERGING GENOME-WIDE APPROACHES
Hypertension
Essential/idiopathic hypertension (HT), the most common cardiovascular disease, with a prevalence of >30% of adults1, is a major risk factor for stroke, heart and renal disease. Family history data of affected individuals coupled with disease concordance rate in twins has established that both genetic and environmental factors determine susceptibility to HT. The heritability (h2) of HT (and blood pressure [BP]) is typically estimated from twin and family studies in the range of up to ~50%, 2 likely with multiple contributory genes and even gene-by-gene interactions. Human population and animal studies have implicated several important physiological pathways contributing to the clinical presentation of essential HT that enable functional candidate gene association studies in addition to more comprehensive genome wide linkage or association studies.
Genetic linkage
In families or sibling pairs, linkage mapping has been used to identify chromosomal regions (genetic loci) that co-segregate during meiosis with a phenotype. This approach is indirect and in most genome-wide studies has been used to identify only relatively large genomic regions that potentially harbor genes of interest; further efforts, including fine mapping and sequencing, are necessary to identify causal genes. Further shortcomings or challenges include the susceptibility of the linkage approach to locus heterogeneity (more than one causal gene), the possibility that the final phenotype is the net effect of genes that both raise and lower the BP, phenotypic heterogeneity arising both from phenocopies (multiple forms of the same disease with different etiologies), and uncertainty as to what constitutes an appropriate phenotype (blood pressure vs. hypertension). In late-onset diseases, additional ambiguity can arise when individuals with putative variant alleles develop the disease later in life or in a much milder form (incomplete penetrance). Finally, hypertension can result from gene-by-environment (GxE) interactions, and it may be more difficult to characterizing the heterogeneous environmental exposures, particularly lifetime exposures, than to assay genotypes. There is no way to easily identify or correct for such heterogeneities in complex disease such as hypertension, and therefore, linkage analysis is best suited for single-gene (Mendelian) disorders in which genes are more highly deterministic and the correlation between genotype and phenotype is robust3; indeed, linkage remains the “go-to” technique for investigation of traits that segregate in families in such as way as to suggest a major gene process with minimal environmental contribution, e.g., the monogenic hypertensions4.
Genetic association
By contrast, genetic association correlates particular alleles, or diploid genotypes, with a trait. Association may be statistically more powerful than linkage for complex traits, especially in the setting of locus heterogeneity; however, the number of markers required to probe for association on a genome-wide (i.e., hypothesis-free) basis may be quite large (see below) 5, 6. In the case of essential hypertension, while there have been numerous reports of associated genes, only a few of these findings have been replicated. Explanations for non-replicability might include: i) statistical significance criteria may have been improperly applied, e.g., no correction for multiple comparisons, with use of a nominal p<0.05 level leading to false positives; ii) differences between the populations studied arising from differences in environmental exposures and genetic background; iii) phenotypic misclassification; and/or iv) population stratification leading to false positive associations.
Genetic architecture of complex traits: Relative role of common (high frequency) versus rare (low frequency) susceptibility variants
A priori, there is substantial uncertainty about what kind of genetic variation (or “architecture”) might underlie complex human traits, including quantitative traits such as HT or BP7, 8, 9. While HT and BP have substantial heritability, ranging up to ~50% based on twin or family studies 10, whether the bulk of the contributing allelic variation to common genetically complex diseases consists of common variants with weak effects (CD/CV) or rare variants with individually stronger effects (CD/RV), is generally unknown. Emerging results from the HapMap/CD/CV approach suggest that, while common genetic variants can be identified, they may account for only a small proportion of heritable trait variance11. Testing the CD/RV hypothesis is challenging unless rare variants produce distinctive phenotypes, e.g., PHA-II (pseudohypoaldosteronism type2) 12, because even the latest technologies are probably yet insufficiently powerful for the task of uncovering novel rare allelic variants. The question of the contribution of rare variants to the population variation of BP remains open 9.
Genome-wide association (GWA) studies
The availability increasingly dense panels of variant alleles that can be typed with automated techniques in increasingly large numbers of people have led to the development of GWA approaches to complex traits. There are a number of advantages of GWA compared to linkage disequilibrium approaches, including the hypothesis-free nature of GWA (unconstrained by prior assumptions about biological pathways contributing to the trait) and the fact that the genomic regions harboring “hypertension” genes identified by GWA are smaller and more easily dissected by direct sequencing. Indeed, it will eventually be possible to perform GWA analyses of all genomic variants and directly assess the effect of each in genomically-defined subpopulations, an approach that may help to circumvent some of the problems noted above that arise from the “phenotype-based” approaches currently employed.
Current GWA approaches are exemplified by the International HapMap project13–15 <www.HapMap.org>, which takes advantage of the effect of the historical meiotic recombination events that have fragmented the genome into stretches of DNA sequence now shared between related individuals. These “blocks” can be identified by the extent of linkage disequilibrium (or “LD”), i.e., the tendency of sufficiently nearby sequences to segregate together even during repeated meiotic recombination. The HapMap project13 first quantified genome-wide LD relationships in several populations chosen for their geographic diversity, in order to facilitate the process of selecting a minimal set of markers that could (indirectly) capture the bulk of the signals from the un-typed (functional) markers during GWA. Depending on the degree of LD in a block, the genetic information contained in the genomic segment can be captured by a few (sometimes a single) genetic markers, which markedly increases the efficiency of interrogating the genome compared to genetically tagging every gene. It is the assumption of the HapMap approach that case-control studies can identify disease-predisposing alleles of sufficiently high (minor allele) frequency can be detected by a combination of LD within blocks and appropriately spaced common marker variants that span the many LD blocks within the genome. Since the typical extent of LD in unrelated humans varies by biogeographic ancestry group, ranging from ~3–10 kbp in blacks up to ~30–50 kbp in whites16, a much denser set of variants (typically 500,000 SNPs) than that commonly used for linkage disequilibrium mapping (typically with an average spacing of ~3.3 ×109/400=~107 bp (or ~10 Mbp) is necessary. Such dense SNP mapping array sets are now available in platforms developed by Affymetrix <http://www.affymetrix.com>, Illumina <http://www.illumina.com>, or Perlegen <http://www.perlegen.com>. Because of the very large number of relatively independent LD “blocks” within the genome that can now be interrogated with SNP assays, the threshold for statistical significance (in the face of multiple potential comparisons) must be adjusted downward, typically to the level of p<5×10−8, or even lower 13–15. To augment confidence in such findings, replication of the result in an independent population sample is useful, taking advantage of the joint (or multiplicative) probability.
GWA RESULTS IN HYPERTENSION: GENOME WIDE SUCCESSES AND LIMITATIONS OF THE HAPMAP-BASED COMMON DISEASE/COMMON VARIANT (CD/CV) HYPOTHESIS
Prior to 2009
Wellcome Trust Case Control Consortium (WTCCC) 17
In a large 2007 study of several common diseases, ~2000 HT cases and ~3000 unphenotyped (BP uncertain) shared population controls in the UK were subjected to GWA with Affymetrix 500K SNP chips, which yielded ~470K usable SNPs. The WTCCC study failed to yield significant (p<5×10−7) associations with HT. However, while unphenotyped population controls should not impair a case/control study for a rare disease, in a common trait such as HT, failure to phenotype the population “controls” is likely to misclassify up to ~25% of such “controls. We elaborate on the effect of such misclassifications on the statistical power below. Even so, when the 6 variants with the highest significance (p<10−5) in the WTCCC were typed in an independent sample of 11,433 phenotyped subjects from the NHLBI Family Blood Pressure Program), one variant (rs1937506, located in a ~500 kb gene “desert” on chromosome 13q21) achieved association (p as low as 0.004) in subjects of European and Hispanic ancestry18. Two of these 6 variants (rs6997709 and rs7961152) also displayed nominal associations with SBP or DBP in a Korean population sample of 7551 subjects19. Later in (2009) the WTCCC hypertension cases were included in a larger GWA study of hypertension (see below) that did identify potential causal variants20.
Framingham Heart Study
In a 2007 report from the US NHLBI Framingham Heart Study, an initial SNP genome scan of >1000 individuals with a 100K marker Affymetrix SNP chip (yielding ~70K usable autosomal markers) was negative for hypertension 21, but as noted above this average SNP density (averaging ~30 kbp inter-marker distance) is probably insufficient to “tag” the majority of LD blocks across the human genome. Once again, at higher SNP density (~500K), the Framingham subjects contributed to a successful 2009 GWA for hypertension (see below)22.
GWA studies in hypertension have yielded positive results only since January of 2009 20, 22, 23
Amish STK39
In early 2009, a hypertension GWA in an Amish population sample 23 discovered an association with STK39, a Ser/Thr protein kinase likely involved in control of ion transport in the distal nephron; replication in >7000 individuals lends credence to the result.
CHARGE and GlobalBPgen
In mid-2009, 2 reports involving >60K people of European ancestry genotyped for ~500K SNPs (on Affymetrix or Illumina platforms) appeared in Nature Genetics: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) and GlobalBPgen (Global Blood Pressure Genetics) consortia studies 20, 22. The CHARGE consortium included 29.136 subjects, while GlobalBPgen included 34,433 subjects: both studies were primarily performed in subjects of European ancestry. These two exceptionally well-powered reports documented many novel SNP loci with significant and reproducible/replicated effects on BP and hypertension (CHARGE: 13 for SBP, 20 for DBP and 10 for hypertension; GlobalBPgen: 8 for SBP or DBP) 20, 22, although the odds ratio of each such locus on trait was very modest (typically <1.1:1), and the cumulative effects of the novel loci together explained perhaps ~1% of population BP variance 20, 22, while twin and family studies (such as our own) indicate that up to ~50% of BP variance is heritable 10.
Korea KARE
In the Korea Association Resource (KARE) project, 8842 subjects were analyzed for associations at ~350K SNP loci on the Affymetrix platform, with replication in 7641 independent samples24. Of note for the CHARGE and GlobalBPgen consortia (see above), KARE found an SBP association on chromosome 12q21 in the region of the Ca2+-translocating ATPase ATP2B1 (rs17249754, p=1.3×10−7), which had also been identified as a putative candidate in the CHARGE study.
African Americans
In a sample of 1017 African Americans genotyped at >800K SNP loci (Affymetrix 6.0 platform), genome-wide significance for SBP (though not DBP or the binary trait) was detected in/near 4 loci: PMS1, SLC24A4, YWHA7, IPO7, and CACANA1H25. Of note for the pathophysiology of hypertension, SLC24A3 encodes a sodium/potassium/calcium exchanger, while CACANA1H encodes the alpha1 (pore-forming) subunit of the T-type voltage-gated Ca2+ channel.
Germany/Estonia/UK
This study began in southern Germany, with 1644 subjects genotyped at ~400K SNPs on the Affymetrix 500K platform, and progressed to replication in Estonia and the UK for a total of 8142 subjects. Investigators discovered an association with HT (p=5.3×10−8) for a region (tagged by rs11646213) on chromosome 16q23.3 upstream of the CDH13 (T-cadherin) gene 26.
Caveats and limitations of the HapMap GWA approach: Fraction of trait variance explained by common haplotype-tagging genetic variants (estimated by the coefficient of determination, R2)
The assumptions underlying the HapMap GWA approach have been subjected to extensive critiques27, but success over the past 2 years in a variety of complex traits attest to the undeniable power of the method. Despite such successes 14, 15, it is becoming apparent that the HapMap/CD/CV approach is able to capture or explain only a very small % of common trait variability (as estimated by the coefficient of determination, or R2) when applied to even highly heritable traits such as height and BMI. The general term of a “missing heritability” problem has been applied in this setting 28.
Height
A compelling example of this gap in our knowledge arises from the trait of height, for which twin and family studies have long established heritability (or the % of trait variability attributed to genetic variance) at up to ~90% 29. However, while dense (~500K) SNP mapping identified contributions of up to 20 novel loci to height, the cumulative effect of these 20 loci explains only ~3% of population height variability11.
BMI
A recent meta-analysis of genetic effects on BMI in the GIANT (Genetic Investigation of ANthropometric Traits) consortium30, involving ~32,000 individuals genotyped at ~500K SNP loci, with replication in ~59,000 subjects, revealed association of 8 loci with BMI: 2 had been previously reported (FTO and MC4R) and 6 were novel (TMEM18, KCTD15, SH2B1, MTCH2, GNPDA2 and NEGR1). However, the cumulative effect of all 8 loci explained only ~0.84% of trait (BMI) variance, despite the fact that twin and family studies consistently reveal BMI to be a highly heritable trait, with heritability estimated at >80%31.
BP/Hypertension
Given the poor performance of GWA studies in highly heritable traits, it is perhaps not surprising that even for GWA studies that have detected variants associated with SBP, DBP or hypertension, R2 is low. As noted above, the CHARGE22 and GlobalBPgen20 consortia established and replicated the effects of up to 8 novel loci on BP. However, the cumulative effect of these 8 loci explained only ~1% of trait (BP) variance22.
The relatively small aggregate R2 values for genetic variants contributing to such traits (typically in the range of only <1–3%) are difficult to understand, given the substantially greater heritabilities (h2) of such traits, where h2 is the proportion of phenotypic variance accounted for by additive genetic variance, as estimated by family studies32. The lack of explanatory power for common genetic variants on such traits might suggest that initial CD/CV genetic discoveries might not be primarily useful for “personalized medicine”, or predictions within an individual, but instead for discovery of new pathways for physiological or pharmacological exploration. For example, initial GWA results for hypertension20, 22 suggest components of several previously unsuspected pathways to be operative in BP elevation. Thus additional approaches will be required to identify much if not most of the source of complex trait genetic determination (read below).
Why do GWAs “underperform” in hypertension and other “complex traits”?
Trait misclassification
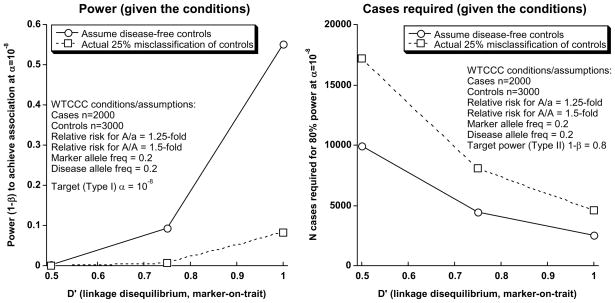
McCarthy et al14 have systematically considered the effect of trait misclassification genetic association studies. The study of such a common trait as HT may render case/control approaches especially susceptible to diluting effects of misclassification, thereby basing the approach towards the null. Misclassification is a potentially critical problem in the WTCCC where 3000 un-phenotyped (no BP measurement) controls were drawn from a population with a high (>25%) prevalence of hypertension. While the use of unselected/un-phenotyped population controls is certainly a valid procedure for many diseases (especially those that are relatively rare), for a very common malady such as hypertension, such an uncharacterized control group may result in a substantial reduction in statistical power. In Figure 1, we re-estimated power for the WTCCC hypertension case/control study (Figure 1), using variance components tools33 <http://pngu.mgh.harvard.edu/~purcell/gpc/>, and applied a spectrum of reasonable assumptions (disease allele relative risk; marker and disease allele frequencies; D’ [index of linkage disequilibrium]) to the unselected population controls. Our analysis suggests that the WTCCC GWA study does not have adequate power to discover hypertension predisposition loci: even at an overly optimistic D’=1.0 (i.e., the marker allele IS the disease allele), statistical power (1-beta) declines to only <10%.
Figure 1.
Re-estimation of statistical power for hypertension in the Wellcome Trust Case Control Consortium (WTCCC). Computation of power used variance components tools, implemented at <http://pngu.mgh.harvard.edu/~purcell/gpc>, considering a spectrum of reasonable assumptions (disease allele relative risk; marker and disease allele frequencies; D’ [linkage disequilibrium]), as well as the effects of the likely ~25% trait misclassification in the “control” group.
Common (relatively high minor allele frequency) variants as contributors to hypertension: Caveats
Biallelic SNP variants for HapMap GWA arrays are deliberately selected for relatively high minor allele frequencies, optimally >10–15%. Secondly, required sample size to detect the effect of a rare allele on a trait is necessarily greater than that for a common allele; the corollary is that any given sample size will demonstrate decreased power for detection of the rarer allele effect; for example, given an odds ratio (effect size) of 2.0 for a genetic variant, the statistical power to detect such a variant with a 30% allele frequency with 90% confidence would require a sample size (cases and controls) of ~1000, while a variant of similar effect but a 1% frequency would require a much larger sample size of ~20,000 (34, Fig 2). As we discuss above for common variants, even when such approaches are successful in identifying trait-susceptibility loci, the fraction of trait variance explained is typically far less than the heritability. Assuming that the marker SNPs selected perform as expected, i.e., capture most of the important common variants contributing to hypertension, a likely conclusion is that less common (rare, at <1–5% minor allele frequency) alleles dominate complex trait determination. The previously cited examples of height11, in which, despite trait heritability approaching ~90% that only ~3% of trait variation can be explained by CD/CV approach, as well as the corresponding examples of BMI (heritability of >80%, yet <1% of trait variance explained)30 and BP (heritability approaching ~50% yet only ~1% of trait variance explained)20, 22 strongly suggest that additional, perhaps less common genetic variation shapes trait determination. The role of rare variants has recently been confirmed for several common diseases (see below).
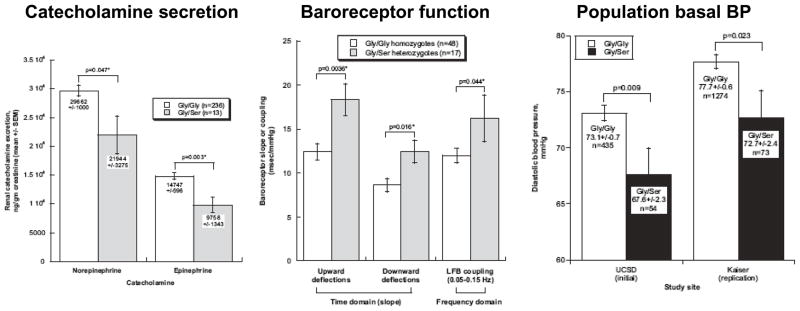
Figure 2. Common Disease/Rare Variant (CD/RV) hypothesis. Human CHGA/Catestatin Gly364Ser (~3% minor allele frequency).
Profound effects on autonomic function and BP in vivo. Left: Effects of Gly/Ser heterozygosity on catecholamine secretion. Center: Effects of Gly/Ser heterozygosity on autonomic function (baroreceptor sensitivity as measured in either the time or frequency domains). Right: Effect of Gly/Ser heterozygosity on resting DBP in the population, with replication in an independent sample. CHGA Gly364Ser is rs9658667. Reproduced from F Rao et al46.
Local SNP-by-SNP: Haplotypes
Haplotypes are ordered sequences of genetic variants along a chromosome, such as SNPs that may capture information beyond that of each single SNP. A recent re-analysis of HT in the WTCCC revealed that the C-A-A haplotype at rs11632637–rs7182413–rs11037474 (on chromosome 15q26.2) is associated with hypertension at genome-wide significance (p=2.8×10−8) 35. This compares to an association signal of lower significance (at p=5.7–7.9×10−6) that was previously detected in this chromosomal region in the WTCCC, using a single-SNP-at-a-time approach.
Genetic interactions (epistasis)
For complex and to date poorly understood traits such as HT, the genetic architecture of disease predisposition may not necessarily be monotonously limited to unifactorial marker-on-trait influences, but instead depend on gene-by-gene (or epistatic) interactions to affect phenotypic traits such as blood pressure. While such interactions are likely to exist, the introduction of one extra degree of freedom during marker-on-trait analyses may reduce statistical power to detect them.
One example of how to detect epistasis is the pathway analysis approach to the likely polygenic basis of common disease in the WTCCC GWA results, which identified several candidate pathways that were not apparent using standard single-SNP GWA statistical approaches36. For HT, even without genome-wide significant associations in the original WTCCC GWA, a network of pathways, many centering on dopaminergic transmission, was detected by analysis of the most strongly HT-associated SNPs. Apparent interconnection among these pathways suggests that multiple, but related, genetic mechanisms may underlie HT susceptibility. Understanding what pathways are etiologically important in hypertension may suggest intermediate phenotypes that can improve the utility of genotyping for identifying individuals at risk as well as approaches for tailoring antihypertensive therapy.
Gene-by-environment interactions
Ecological comparisons between Westernized and more traditional populations point to a necessary permissive role for environmental factors in the development of hypertension. Except for sodium, environmental factors promoting hypertension are poorly characterized in individuals studied to date in GWA studies. Our inability to account for the impact of environment on final phenotype introduces an element of variation which compounds that introduced by trait variation. In addition, the effects of the history of an individual’s environmental exposure may affect the final phenotype even if the environmental factor is not present at the time the individual is studied. The impact of in utero nutritional state on adult blood pressure is one such example of the chronic effect of an early environmental factor37.
PERSPECTIVES AND FUTURE DIRECTIONS
Some have declared that efforts to discover genetic variation underlying BP variation should now cease38, and instead focus upon the existing GWA results of the CHARGE22 and GlobalBPgen20 consortia; however, other prominent voices argue strongly that, for complex traits, GWA has typically discovered only a small % of the genetic variance underlying the trait, in what has been termed the “missing heritability” (h2), a problem extending to such diverse traits as height, BMI, dyslipidemia, type 2 diabetes, and early onset myocardial infarction28. Hence, new strategies may be required to move forward discovery in this setting.
Rare (relatively low minor allele frequency) genetic variants
As noted above, recent WGA studies have not identified common genetic variants with major phenotypic effects20, 22. Leading to the question of how much multiple rare alleles might contribute to variation in common traits such as BP. Even though ”uncommon” variants are individually unusual, the relative risk (as estimated by the odds ratio) conferred by each such variant may be substantially higher than that for more common variants 9. In an illustration of this strategy for hypertension, we re-sequenced a locus critical for catecholamine storage, chromogranin A (CHGA), and discovered a low (~3%) frequency functional variant, Gly364Ser (Figure 2), that had a profound effect on autonomic activity as well as risk for HT 46. Similarly, Lifton et al 47 re-sequenced three genes subserving renal tubular salt reabsorption – SLC12A3 (NCCT), SLC12A1 (NKCC2) and KCNJ1 (ROMK) – in subjects from the most extreme BPs in the population, and thereby discovered a series of rare, non-synonymous loss-of-function variants associated with lower BP 47. Resequencing at candidate genes or pathways in individuals with extreme phenotypes has also revealed evidence for rare alleles that contribute to risk for obesity48 and dyslipidemia48–52.
This approach can only be currently implemented by re-sequencing across candidate loci in phenotyped individuals 9, or ultimately by “exome”-wide39 resequencing enabled by the development of “exon capture” microarrays54, although restriction to exons may miss numerous variants conferring quantitative changes in the amount of a gene product (rather than qualitative changes in sequence). For example, we have described roles for transcriptional-regulatory variants in several genes contributing to the human sympathochromaffin phenotype and ultimately BP, including CHGA40, CHGB41, SCG242, TH43, DBH44, and NPY1R45. Eventually, genome-wide resequencing may be enabled by higher-throughput “next generation” sequencing platforms53, and pilot projects utilizing these technologies are now being undertaken by the “1000 Genomes Project” international consortium <http://www.1000genomes.org>.
Phenotypes: Trait extreme values
Better phenotyped cases and controls should improve the yield of future GWA studies of HT. One strategy to maximize the efficiency of quantitative trait studies is to derive the sample(s) from the extremes of the trait distribution55. For example, we have conducted candidate genotyping in a community-based sample from the upper and lower 5th percentiles of BP among >50,000 people in a health maintenance program. This approach has >90% statistical power to detect genes contributing as little as 3% to trait (blood pressure)variation56. The trait-extreme approach to BP genetics has already yielded progress on the CD/RV front, providing evidence that multiple rare variants in renal salt transporters influence BP in the population47. Analysis of the trait as a continuous rather than dichotomous variable also enhances statistical power, and may also enable multivariate approaches to correlated traits57.
Phenotypes: Intermediate traits
GWA analysis of “intermediate” (or “risk”) phenotypes for HT, in addition to BP itself, should assist in defining the genetic roots of HT, for several reasons. Ideal properties of such traits include pathogenic potential, substantial heritability (h2; the % of trait variance accounted for by genetic variance), and expression very early in the course of development of a complex disease trait such as HT 58. Advantages of such traits may include: enhanced statistical power for marker-on-trait associations as a result of earlier trait penetrance with greater h2; empirical partitioning of the heterogeneity within a complex trait into “endophenotypes”; and understanding the longitudinal course of disease trait development. Such approaches can best be implemented in twin or family studies, wherein relative pair correlations yield h2 estimates58.
Gene-by-environment (GxE) interactions
Studies in both humans 59 and rodents 60 indicate that organisms at genetic risk of developing hypertension display exaggerated cardiovascular responses to environmental stressors, thereby documenting the importance of GxE interaction in the development of hypertension. Indeed, GxE effects are likely to contribute to many (if not most) complex traits 61. GWA approaches to hypertension thus far have not accounted for or incorporated such interactions, which would necessitate more extensive information on such environmental triggers as diet and stress. With introduction of the necessary one additional degree of freedom in analyzing GxE effects, statistical power to detect associations will fall, unless sample size is increased substantially62. Nonetheless, understanding such GxE effects may allow more rational introduction of environmental interventions in an attempt to prevent future occurrence of disease in at-risk individuals63. For example, in studies on twin pairs we have recently documented that the BP response to environmental stress is influenced by genetic variation at several points within the adrenergic pathway, including catecholamine biosynthesis (at tyrosine hydroxylase [TH]43 and DBH44), storage (at chromogranin A [CHGA]40, 64 or B [CHGB]41, and SCG242, or post-receptor signal transduction (at rho kinase [ROCK2] 65. In addition, widespread genetic variation within the adrenergic pathway seems to influence the vascular response to environmentally-triggered increments in catecholamine release66.
Additional approaches
Manolio et al28 have summarized several additional potential areas for genomic exploration in complex traits, including exploiting the value of family studies, structural variants such as copy number variation (copy number polymorphism) and inversions, “epigenetics” or change in phenotype as a consequence of events other than DNA sequence variation (such as DNA CpG methylation, or chromatin remodeling), extension of GWAs to probes for causative variants with lower allele frequency, and systematic polymorphism discovery at loci already associated with the complex trait by GWAs. Finally, twin pairs67 (including MZ twins) may present unique opportunities for uncovering GxE interactions68.
Acknowledgments
Support: National Institutes of Health, Department of Veterans Affairs.
ABBREVIATIONS
- BP
Blood Pressure
- CD/CV
Common Disease/Common Variant
- CD/RV
Common Disease/Rare Variant
- GWAs
Genome Wide Association study
- h2
Heritability (% of trait variance accounted for by genetic variance)
- HapMap
Haplotype Map Consortium
- HT
Hypertension
- LD
Linkage Disequilibrium
- SNP
Single Nucleotide Polymorphism
- WTCCC
Wellcome Trust Case Control Consortium
Footnotes
Conflicts of interest to declare: None.
References
- 1.Rosamond W, Flegal K, Friday G, Furie K, Go A, Greenlund K, Haase N, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O’Donnell CJ, Roger V, Rumsfeld J, Sorlie P, Steinberger J, Thom T, Wasserthiel-Smoller S, Hong Y. Heart disease and stroke statistics--2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2007;115(5):e69–171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 2.Kupper N, Ge D, Treiber FA, Snieder H. Emergence of novel genetic effects on blood pressure and hemodynamics in adolescence: the Georgia Cardiovascular Twin Study. Hypertension. 2006;47(5):948–954. doi: 10.1161/01.HYP.0000217521.79447.9a. [DOI] [PubMed] [Google Scholar]
- 3.Mayeux R. Mapping the new frontier: complex genetic disorders. J Clin Invest. 2005;115(6):1404–1407. doi: 10.1172/JCI25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104(4):545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 6.Jones HB. The relative power of linkage and association studies for the detection of genes involved in hypertension. Kidney Int. 1998;53(6):1446–1448. doi: 10.1046/j.1523-1755.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 7.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant... or not? Hum Mol Genet. 2002;11(20):2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 9.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih PA, O’Connor DT. Hereditary determinants of human hypertension: strategies in the setting of genetic complexity. Hypertension. 2008;51(6):1456–1464. doi: 10.1161/HYPERTENSIONAHA.107.090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science. 2001;293(5532):1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 13.A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 15.Musunuru KKS. HapMap and Mapping Genes for Cardiovascular Disease. Circ Cardiovasc Genet. 2008;1:66–71. doi: 10.1161/CIRCGENETICS.108.813675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 17.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehret GB, Morrison AC, O’Connor AA, Grove ML, Baird L, Schwander K, Weder A, Cooper RS, Rao DC, Hunt SC, Boerwinkle E, Chakravarti A. Replication of the Wellcome Trust genome-wide association study of essential hypertension: the Family Blood Pressure Program. Eur J Hum Genet. 2008;16(12):1507–1511. doi: 10.1038/ejhg.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong KW, Jin HS, Cho YS, Lee JY, Lee JE, Cho NH, Shin C, Lee SH, Park HK, Oh B. Replication of the Wellcome Trust genome-wide association study on essential hypertension in a Korean population. Hypertens Res. 2009;32(7):570–574. doi: 10.1038/hr.2009.68. [DOI] [PubMed] [Google Scholar]
- 20.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 (Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, O’Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, Kim JD, Subramanya AR, Steinle NI, Parsa A, Ober CC, Welling PA, Chakravarti A, Weder AB, Cooper RS, Mitchell BD, Shuldiner AR, Chang YP. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106(1):226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 25.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5(7):e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, Veldre G, Doring A, Viigimaa M, Sober S, Tomberg K, Eckstein G, Kelgo P, Rebane T, Shaw-Hawkins S, Howard P, Onipinla A, Dobson RJ, Newhouse SJ, Brown M, Dominiczak A, Connell J, Samani N, Farrall M, Caulfield MJ, Munroe PB, Illig T, Wichmann HE, Meitinger T, Laan M. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18(12):2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger JD, Hiekkalinna T. An utter refutation of the “Fundamental Theorem of the HapMap”. Eur J Hum Genet. 2006;14(4):426–437. doi: 10.1038/sj.ejhg.5201583. [DOI] [PubMed] [Google Scholar]
- 28.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preece MA. The genetic contribution to stature. Horm Res. 1996;45 (Suppl 2):56–58. doi: 10.1159/000184849. [DOI] [PubMed] [Google Scholar]
- 30.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wessel J, Moratorio G, Rao F, Mahata M, Zhang L, Greene W, Rana BK, Kennedy BP, Khandrika S, Huang P, Lillie EO, Shih PA, Smith DW, Wen G, Hamilton BA, Ziegler MG, Witztum JL, Schork NJ, Schmid-Schonbein GW, O’Connor DT. C-reactive protein, an ‘intermediate phenotype’ for inflammation: human twin studies reveal heritability, association with blood pressure and the metabolic syndrome, and the influence of common polymorphism at catecholaminergic/beta-adrenergic pathway loci. J Hypertens. 2007;25(2):329–343. doi: 10.1097/HJH.0b013e328011753e. [DOI] [PubMed] [Google Scholar]
- 32.Douglas S, Falconer TFCM. Introduction to Quantitative Genetics. 4. Benjamin Cummings; 1996. [Google Scholar]
- 33.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 34.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browning BL, Browning SR. Haplotypic analysis of Wellcome Trust Case Control Consortium data. Hum Genet. 2008;123(3):273–280. doi: 10.1007/s00439-008-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92(5):265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Maternal and social origins of hypertension. Hypertension. 2007;50(3):565–571. doi: 10.1161/HYPERTENSIONAHA.107.091512. [DOI] [PubMed] [Google Scholar]
- 38.Harrap SB. Blood pressure genetics: time to focus. J Amer Soc Hypertension. 2009;3(4):231–237. doi: 10.1016/j.jash.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Rao F, Rodriguez-Flores JL, Mahapatra NR, Mahata M, Wen G, Salem RM, Shih PA, Das M, Schork NJ, Ziegler MG, Hamilton BA, Mahata SK, O’Connor DT. Common genetic variants in the chromogranin A promoter alter autonomic activity and blood pressure. Kidney Int. 2008;74(1):115–125. doi: 10.1038/ki.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K, Rao F, Rana BK, Gayen JR, Calegari F, King A, Rosa P, Huttner WB, Stridsberg M, Mahata M, Vaingankar S, Mahboubi V, Salem RM, Rodriguez-Flores JL, Fung MM, Smith DW, Schork NJ, Ziegler MG, Taupenot L, Mahata SK, DTOC Autonomic function in hypertension: Role of genetic variation at the catecholamine storage vesicle protein chromogranin B (CHGB) Circ Cardiovasc Genet. 2009;2:46–56. doi: 10.1161/CIRCGENETICS.108.785659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen G, Wessel J, Zhou W, Ehret GB, Rao F, Stridsberg M, Mahata SK, Gent PM, Das M, Cooper RS, Chakravarti A, Zhou H, Schork NJ, O’Connor DT, Hamilton BA. An ancestral variant of Secretogranin II confers regulation by PHOX2 transcription factors and association with hypertension. Hum Mol Genet. 2007;16(14):1752–1764. doi: 10.1093/hmg/ddm123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao F, Zhang L, Wessel J, Zhang K, Wen G, Kennedy BP, Rana BK, Das M, Rodriguez-Flores JL, Smith DW, Cadman PE, Salem RM, Mahata SK, Schork NJ, Taupenot L, Ziegler MG, O’Connor DT. Tyrosine hydroxylase, the rate-limiting enzyme in catecholamine biosynthesis: discovery of common human genetic variants governing transcription, autonomic activity, and blood pressure in vivo. Circulation. 2007;116(9):993–1006. doi: 10.1161/CIRCULATIONAHA.106.682302. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Wen G, Rao F, Zhang K, Wang L, Rodriguez-Flores JL, Sanchez AP, Mahata M, Taupenot L, Sun P, Mahata SK, Tayo B, Schork NJ, Ziegler MG, Hamilton BA, O’Connor DT. Human dopamine beta-hydroxylase (DBH) regulatory polymorphism that influences enzymatic activity, autonomic function, and blood pressure. J Hypertension. 2009 doi: 10.1097/HJH.0b013e328332bc87. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Rao F, Zhang K, Mahata M, Rodriguez-Flores JL, Fung MM, Waalen J, Cockburn MG, Hamilton BA, Mahata SK, O’Connor DT. Neuropeptide Y(1) Receptor NPY1R discovery of naturally occurring human genetic variants governing gene expression in cella as well as pleiotropic effects on autonomic activity and blood pressure in vivo. J Am Coll Cardiol. 2009;54(10):944–954. doi: 10.1016/j.jacc.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao F, Wen G, Gayen JR, Das M, Vaingankar SM, Rana BK, Mahata M, Kennedy BP, Salem RM, Stridsberg M, Abel K, Smith DW, Eskin E, Schork NJ, Hamilton BA, Ziegler MG, Mahata SK, O’Connor DT. Catecholamine release-inhibitory peptide catestatin (chromogranin A(352–372)): naturally occurring amino acid variant Gly364Ser causes profound changes in human autonomic activity and alters risk for hypertension. Circulation. 2007;115(17):2271–2281. doi: 10.1161/CIRCULATIONAHA.106.628859. [DOI] [PubMed] [Google Scholar]
- 47.Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40(5):592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt S, Yosef N, Ruppin E, Sharan R, Vaisse C, Sunyaev S, Dent R, Cohen J, McPherson R, Pennacchio LA. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80(4):779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 51.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 52.Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119(1):70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 54.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 55.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000;67(5):1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rana BK, Insel PA, Payne SH, Abel K, Beutler E, Ziegler MG, Schork NJ, O’Connor DT. Population-based sample reveals gene-gender interactions in blood pressure in White Americans. Hypertension. 2007;49(1):96–106. doi: 10.1161/01.HYP.0000252029.35106.67. [DOI] [PubMed] [Google Scholar]
- 57.Tenesa A, Visscher PM, Carothers AD, Knott SA. Mapping quantitative trait loci using linkage disequilibrium: marker- versus trait-based methods. Behav Genet. 2005;35(2):219–228. doi: 10.1007/s10519-004-0811-5. [DOI] [PubMed] [Google Scholar]
- 58.O’Connor DT, Insel PA, Ziegler MG, Hook VY, Smith DW, Hamilton BA, Taylor PW, Parmer RJ. Heredity and the autonomic nervous system in human hypertension. Curr Hypertens Rep. 2000;2(1):16–22. doi: 10.1007/s11906-000-0053-8. [DOI] [PubMed] [Google Scholar]
- 59.Schneider GM, Jacobs DW, Gevirtz RN, O’Connor DT. Cardiovascular haemodynamic response to repeated mental stress in normotensive subjects at genetic risk of hypertension: evidence of enhanced reactivity, blunted adaptation, and delayed recovery. J Hum Hypertens. 2003;17(12):829–840. doi: 10.1038/sj.jhh.1001624. [DOI] [PubMed] [Google Scholar]
- 60.Folkow B. Early structural changes in hypertension: pathophysiology and clinical consequences. J Cardiovasc Pharmacol. 1993;22 (Suppl 1):S1–6. [PubMed] [Google Scholar]
- 61.Dempfle A, Scherag A, Hein R, Beckmann L, Chang-Claude J, Schafer H. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16(10):1164–1172. doi: 10.1038/ejhg.2008.106. [DOI] [PubMed] [Google Scholar]
- 62.Hein R, Beckmann L, Chang-Claude J. Sample size requirements for indirect association studies of gene-environment interactions (G × E) Genet Epidemiol. 2008;32(3):235–245. doi: 10.1002/gepi.20298. [DOI] [PubMed] [Google Scholar]
- 63.Khoury MJ, Davis R, Gwinn M, Lindegren ML, Yoon P. Do we need genomic research for the prevention of common diseases with environmental causes? Am J Epidemiol. 2005;161(9):799–805. doi: 10.1093/aje/kwi113. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Rao F, Rodriguez-Flores JL, Mahata M, Fung MM, Stridsberg M, Vaingankar SM, Wen G, Salem RM, Das M, Cockburn MG, Schork NJ, Ziegler MG, Hamilton BA, Mahata SK, Taupenot L, O’Connor DT. Naturally occurring human genetic variation in the 3′-untranslated region of the secretory protein chromogranin A is associated with autonomic blood pressure regulation and hypertension in a sex-dependent fashion. J Am Coll Cardiol. 2008;52(18):1468–1481. doi: 10.1016/j.jacc.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seasholtz TM, Wessel J, Rao F, Rana BK, Khandrika S, Kennedy BP, Lillie EO, Ziegler MG, Smith DW, Schork NJ, Brown JH, O’Connor DT. Rho kinase polymorphism influences blood pressure and systemic vascular resistance in human twins: role of heredity. Hypertension. 2006;47(5):937–947. doi: 10.1161/01.HYP.0000217364.45622.f0. [DOI] [PubMed] [Google Scholar]
- 66.Fung MM, Nguyen C, Mehtani P, Salem RM, Perez B, Thomas B, Das M, Schork NJ, Mahata SK, Ziegler MG, O’Connor DT. Genetic variation within adrenergic pathways determines in vivo effects of presynaptic stimulation in humans. Circulation. 2008;117(4):517–525. doi: 10.1161/CIRCULATIONAHA.107.706317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3(11):872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 68.Wray NR, Coventry WL, James MR, Montgomery GW, Eaves LJ, Martin NG. Use of monozygotic twins to investigate the relationship between 5HTTLPR genotype, depression and stressful life events: an application of Item Response Theory. Novartis Found Symp. 2008;293:48–59. doi: 10.1002/9780470696781.ch4. discussion 59–70. [DOI] [PubMed] [Google Scholar]