Abstract
While bio-labeled immunoglobulin (IgG) or antibodies are extensively used in imaging studies and therapeutic modality, there have been no reports describing atomic force microscopy (AFM) micrographs of these labeled IgG molecules. Here, AFM studies of phycoerythrin (PE)-conjugated IgG were undertaken to examine whether PE conjugation induced conformational changes or molecular interaction, and subsequently resulted in an alteration in nano-structures of PE-conjugated IgG complex. Once immobilized on mica, single PE-conjugated IgG molecule exhibited globular shape with approximately 60 microns in diameter and 5 microns in height. PE-conjugated IgG were able to form monomers, spindle-like trimers, and hexamers that developed through an end–end connection of two trimers, after they were continuously immobilized on mica. Interestingly, these multimers could aggregate in different directions to form circular monolayers with a highly dense core of PE-conjugated IgG polymers. The formation of these well-organized polymers and aggregates of PE-conjugated IgG may be attributed to the PE conjugation as well as the air–liquid tension on subtract. These findings may help to understand the nano-structures of bio-labeled IgG or antibodies, and facilitate the potential use of PE-conjugated antibodies as markers or immunosensers for AFM bio-analytics of biomolecules in cells and membranes.
Keywords: Mouse phycoerythrin conjugated immunoglobulin G (PE-IgG), Polymer, Aggregation, Atomic force microscopy (AFM)
1. Introduction
Atomic force microscopy (AFM) is emerging as extremely powerful nanobiotechnology for imaging studies of biomolecules (Frederix et al., 2003). Over the past decade, AFM has been broadly used in biology to reveal subnanometer details and surface dynamics of biomolecules such as DNA, RNA, proteins, and saccharides. It has also been applied in chemistry to studies of Langmuir–Blodgett (LB) films and self-assembled structures of organic amphiphiles. Continuous progress in sample preparation and instrumentation will make it possible to image and dissect single biomolecules in conjunction with their biological function in living cells or tissues.
AFM has recently been employed to study immunoglobulin G (IgG). In humans, IgG constitutes most antibodies that are essential for protective immunity against pathogen infections. In biotechnology, IgG are usually high-affinity antibodies that are used as reagents for bioassays or detection and for therapeutic intervention. It is important to note that the majority of those IgG antibodies for bioassay or therapy are labeled either with radioactive isotopes, toxins or biomarkers including fluorescent materials. To date, AFM studies have been done only for simple non-labeled IgG molecules, but not for bio-labeled IgG. Those studies have addressed four aspects of non-labeled IgG: observation of single IgG molecules (Hansma, 1999; Leatherbarrow et al., 1991), adsorption of IgG molecules with different substrates (Diederich and Losche, 1997; Ortega-Vinuesa et al., 1998; You and Lowe, 1996), interaction of IgG with its antigen or anti-IgG (Delamarche et al., 1997; Ouerghi et al., 2002; Perrin et al., 1998; Zhang et al., 1997), and immobilization of IgG on a self-assembled monolayer (Boncheva and Vogel, 1997; Wadu-Mesthrige et al., 2000). Nevertheless, it is not known whether bio-labeled IgG molecules are different in nano-structures from those of non-labeled IgG. Since bio-labeled IgGs are frequently used for studies in the fields of immunology, cell biology, biomedicine, and antibody-based therapy, it is important to gain some information regarding the nanometer structures of bio-labeled IgG molecules. To this end, we sought to examine the nano-structures and surface dynamics of PE-conjugated IgG molecules using AFM nanotechnology. The rationale of these studies is that PE-conjugated IgG antibodies are routinely used in flow cytometry or immunohistochemistry, and potentially employed as markers or immunosensers in AFM studies of biomolecules in cells and cell membranes. We proposed that the PE conjugation of IgG molecules could induce conformational changes or molecular interaction, and such changes would result in an alteration in nano-structures of PE-conjugated IgG. To test this possibility, PE-conjugated IgGs were immobilized for AFM studies using Langmuir–Blodgett (LB) technology(Barraud et al., 1993). LB technique has proved to be useful chemical immobilization method for AFM-based application. To our knowledge, there have been no reports describing the nanostructures of PE-conjugated IgG immobilized on LB films.
2. Materials and methods
100 mg/L R-PE-conjugated IgG (Becton Dinckison Biosciences Pharmingen, San Diego, CA) was diluted into 0.1 mg/L with distilled water. Five micro liters (μl) of diluted PE-conjugated IgG were introduced to freshly cleaved mica surfaces and then air-fixed rapidly through vigorous waving of the slide. The film was assessed for quality under an optical microscope. Atomic force microscopy was performed in air and in tapping mode using a commercial AFM (AutoProbe CP, Thermomicroscopes, USA). The mica carrying PE-conjugated IgG molecules was mounted onto the XY stage of the AFM, and an integral video camera was used to locate the regions of interest. Microfabricated silicon cantilevers (Park Scientific Instruments) with a force constant of approximately 25 N/m were used. Repeated scanning of the samples confirmed that no physical damage occurred in samples during imaging. AFM images were obtained at a line scan rate of 1 Hz (humidity: 75%, temperature: 25 °C). The images were stable and reproducible during repeated scanning. The AFM images were planar-leveled using the software provided with the instrument (Thermomicroscopes Proscan Image Processing Software Version 2.1). The quantification of images was carried out as routine. The average width and height (i.e. thickness) of fluorescence-conjugated IgG molecules and polymers were determined using the line-analytical function of the software. The sizes and unique end–end connection, but not the simple shapes, of detectable molecules were used as morphological criteria for defining a single monomer and trimer and multimer (hexamer) of PE-labeled IgG.
3. Results and discussion
3.1. Single PE-conjugated IgG molecule exhibited a globular shape and a size of 60 nm × 5nm
AFM images of individual PE-conjugated IgG at a nanometer level were readily observable when these molecules were initially immobilized on mica (Fig. 1a). The individual PE-conjugated IgG molecules were globular with an approximate diameter of 60 nm and a height of around 5 nm. These single PE-conjugated IgG molecules were different in size and shape from the unlabeled IgG molecules. X-ray diffraction analyses showed that single unlabeled IgG proteins were about 14.2 nm in diameter, whereas AFM revealed a diameter of 25–40 nm and a height of around 2 nm for those unlabeled IgG (Zhang et al., 1997). Our AFM studies of non-labeled IgG showed that a single non-labeled IgG molecule is 20–40 nm in diameter and about 2 nm in height (Fig. 1b). While some non-labeled IgG molecules could be globular due to their intrinsic complexity, they also exhibited a characteristic Y shape (Fig. 1b).
Fig. 1.
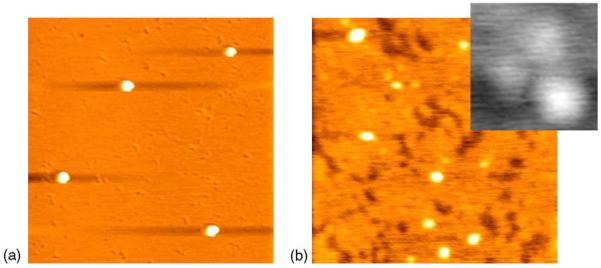
AFM image in (a) shows that single PE-conjugated IgG molecule is globular, with a diameter of about 60 nm and a height of about 5 nm (scanning size: 1.5 μm). (b) shows that single non-labeled IgG molecule is 20–40 nm in diameter and about 2 nm in height (scanning size: 0.5 μm). The insect image in the upper right corner reveals the characteristic Y shape of an IgG molecule with a diameter of about 40 nm (scanning size for inset, 50 nm).
The scanning tunneling microscopy (STM) showed that the IgG molecule exhibited a trilobed shape when unlabeled IgG were deposited from buffer onto HOPG (Leatherbarrow et al., 1991). Similarly, a Y-type structure of IgG molecules under aqueous fluid was revealed by AFM (Hansma, 1999).
The difference in size and shape between unlabeled IgG and PE-conjugated IgG is attributed to at least two factors. First, PE conjugation of IgG protein results in an increase in the size of IgG molecules, since PE is a protein with a molecular weight of about 240 kDa (Galland-Irmouli et al., 2000). The difference in molecular weights between PE-conjugated IgG and non-conjugated IgG can certainly explain why the size of PE-conjugated IgG molecules seen in the present study is about twice as large as that of non-conjugated IgG proteins (Zhang et al., 1997). Second, a different size or shape of PE-conjugated IgG may be due to the effect of air–liquid interface under AFM. When AFM is performed at air–liquid interface, a layer of water can cover the molecular surface (Drake et al., 1989). If several monolayers of condensed water are lying on the surface of a rigid sample, AFM will give rise to an image reflecting the surface of the liquid layer in non-contact or intermittent-contact mode (Fig. 2). Further studies are needed to clarify such speculation.
Fig. 2.
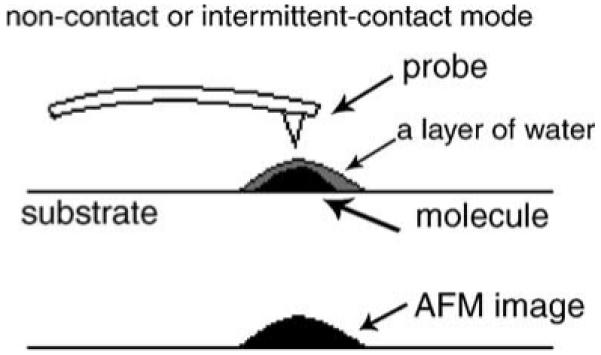
A model for explaining a change in AFM images on a surface with a droplet of water in non-contact or intermittent-contact mode.
3.2. PE-conjugated IgG molecules were able to form polymers through an end–end interaction or connection
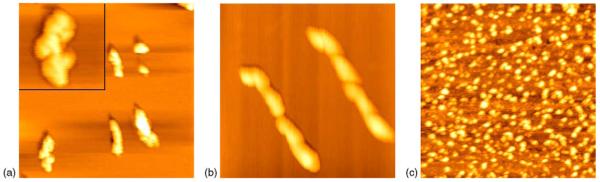
Polymers formed by PE-conjugated IgG were observed in AFM micrograph following a continuous immobilization of PE-conjugated IgG on mica at air–liquid interface (Fig. 3). Monomers, trimers, and hexamers of PE-conjugated IgG molecules were apparent under AFM analyses. The trimer was approximately 200 nm in length, 75 nm in width, and 5 nm in height. Interestingly, while a spindle-like trimer was composed of three PE-conjugated IgG molecules, two trimers could be connected through an end–end interaction to form a hexamer. Such a hexamer was about 400 nm in length, 50 nm in width, and 14 nm in height (Fig. 3b and Fig. 4). It is important to point out that non-conjugated IgG molecules cannot form end–end connected multimers of IgG complex even at high concentrations [(Zhang et al., 1997) and Fig. 3c]. This is also contrasted to non-labeled IgM or IgA, which form multimers through self-assembling. Therefore, PE-conjugated IgG molecules appeared to acquire the capacity to form polymers through an end—end interaction. Based on biological characteristics of PE and IgG molecules, we propose several models for the formation of polymers of PE-conjugated IgG (Fig. 4). It is likely that the formation of PE-conjugated IgG polymers on mica occurs as a result of the conjugation of PE fluorescent polypeptide to IgG. The PE protein, which is bio-labeled on the Fab fragments in the PE-conjugated IgG used in the present study, is comprised of 2α, 2β and 1γ chains with a MW of about 240 kDa. The PE molecule has 36 chromophores, a quantum efficiency of about 0.85, and a pI of 3.5 (Chang et al., 1996; Kang and Yeung, 2002). Because α and β subunits of PE are not stable in neutral medium (in our experiment, the medium is distilled water with pH of 7.0), one may expect to see a change in nano-structures that results from conformational change and molecular interaction of PE-conjugated IgG molecules during their immobilization on subtract.
Fig. 3.
PE-conjugated IgG molecules were able to form a monomer, trimer or polymer with different sizes and shapes. (a) The monomer and trimer are indicated by long and short arrows, respectively. An enlarged image of trimer with a scanning size of 250 nm was shown on the top left corner; three circles underline three PE-conjugated IgG molecules making up the trimer. (b) Shown are images of two hexamers that were formed by an end–end connection of two trimers; three circles indicate three molecules making up a trimer of a hexamer. (c) The image indicates that non-labeled IgG molecules are unable to form end–end connected polymers and circular monolayers even at high concentrations. These molecules distribute dispersedly and irregularly, with no evidence of non-labeled IgG polymers. Scanning size: (a) 1 μm; (b) 0.6 μm; (c) 1 μm. Single monomer and trimer and multimer (hexamer) of PE-labeled IgG were judged based on the size and unique end–end connection of detectable molecules. The monomer indicated by the large arrow in (a) has a diameter of approximately 50–60 nm, which is consistent with the single molecule of PE-labeled IgG. The multimer indicated by the small arrow has a length of approximately 200 nm, which was three-fold larger than that of the monomer of a PE-labeled IgG molecule (about 60 nm). Such a multimer was interpreted as trimer, since its length or size was equivalent to an elongation of three PE-labeled IgG monomers. Similarly, the elongated multimers in (b) was interpreted as a hexamer, since it had an end–end connected length of approximately 400 nm, six-fold longer than the length of a single PE-labeled IgG monomer.
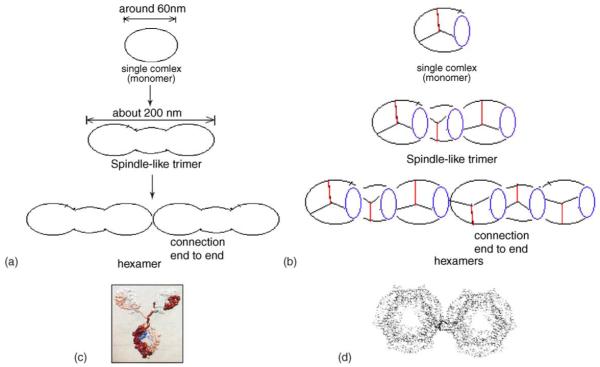
Fig. 4.
Proposed models for formation of PE-conjugated IgG polymers. (a) A simple model of polymerization and (b) a model showing possible connections among molecules. The purple circles mean one PE molecule or two adjacent PE molecules; the red lines show the Fc fragments of IgG molecules; and the other two black lines represent the two Fab fragments of IgG molecules. (c) A model of a non-conjugated IgG antibody based on structures determined by X-ray crystallography and (d) a model of two adjacent PE hexamers.
Multimers of IgM and IgA usually develop in association with an additional polypeptide chain, the J chain. Here in our studies, PE is likely to function as the J chain for the development of polymers of PE-conjugated IgG. It is possible that the end–end connection of PE-conjugated IgG molecules during the formation of polymers occurs as a result of the interaction between α and β subunits of PE proteins labeled on Fab fragments of PE-conjugated IgG. It is also possible that such end–end connection for polymer development is attributed to the interaction between PE molecules and the Fab fragments of adjacent IgG molecules.
3.3. PE-conjugated IgG molecules were able to form circular aggregates comprised of well-assembled polymers
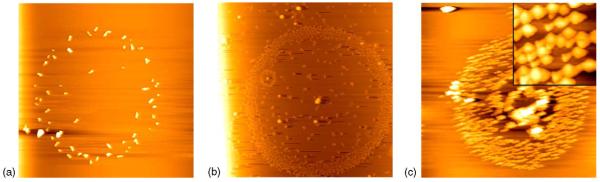
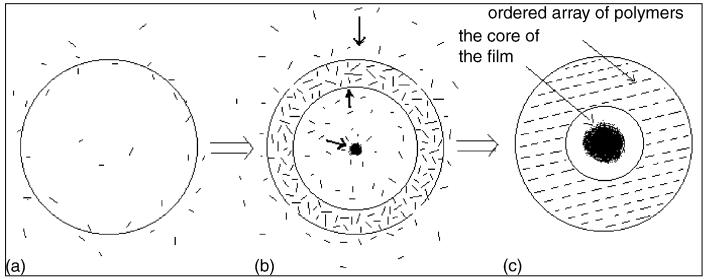
With PE-conjugated IgG immobilized longer on mica, a variety of circular aggregates was seen in AFM micrographs (Fig. 5). While PE-conjugated IgGs were able to aggregate in different directions and to form a ring of polymers, most of formed aggregates had a core of polymer mass (Fig. 5a,b). The asymmetrical distribution of these aggregated polymers was likely due to the uneven concentration of PE-conjugated Ig molecules immobilized on mica. The differing concentrations may also contribute to the different density and extent of aggregations. When the concentration of molecules is high enough, a circular film comprised of dense polymers was seen on the deposited mica (Fig. 5c). The formation of well-aggregated polymers represented a unique feature for PE-labeled IgG, since non-labeled IgG was not able to form circular film even at high concentrations (Fig. 3c). It was worth to mention that the film remained a thickness of about 5 nm as a monolayer, which resembled the same 5 nm height of a single PE-conjugated IgG molecule. The formation of circular film appeared to occur as a result of the volatilization of condensed water-monolayer overlaying on the surface of PE-conjugated IgG polymers. During volatilization, polymers moved in different directions under the air–liquid tension. Such movement of polymers resulted in the formation of the ring of circular film as well as the assembling of a polymer core in the center of the film (Fig. 6b). The combined effect of volatilization and high-concentration might result in the highly ordered arrays of polymerization or substantial aggregation (Fig. 6c). Therefore, the circular monolayer of PE-conjugated IgG polymers (Fig. 5c) may represent dissipative and self-assembled phenomenon of bio-labeled IgG molecules.
Fig. 5.
PE-conjugated IgG molecules are able to form circular aggregates comprised of well-assembled polymers. (a) AFM images of circular aggregates identified in PE-conjugated IgG polymers immobilized on mica. Note a ring of polymers formed. (b) A core of polymer mass, as indicated by an arrow, was developed in the circular polymers. (c) A monolayer of circular polymers with a thickness of around 5 nm. Shown on the top right corner is an enlarged image with a scanning size of 500 nm, in which hexamers are assembled or arrayed orderly or synchronously in end-to-end and side-by-side fashions. Scanning size: (a) 7 μm; (b) 25 μm; (c)7 μm.
Fig. 6.
A proposed model of aggregation of PE-conjugated IgG polymers. A short rod means a PE-conjugated IgG polymer, and the circle means the dense region of PE-conjugated IgG polymers. The arrows in (b) mean the directions in which PE-conjugated IgG polymers move.
3.4. Biological significance of PE-conjugated IgG nanostructures
Elucidation of nano-structures of PE-conjugated IgG is important for understanding and applying molecular characteristics of bio-labeled IgG molecules. It appears that PE-conjugated IgG is unique for the formation of trimers, polymers and well-assembled aggregates in biomonolayers, since non-labeled IgG were unable to form such multimers and aggregates [(Zhang et al., 1997) and Fig. 3c]. PE-conjugated IgG as primary or secondary antibodies have been extensively used in routine image studies including flow cytometry- and histology-based analyses. Importantly, fluorescence-conjugated IgG/antibodies are being used as markers of immunosensers to image biomolecules in cells and cell membranes at a nano-structure level or even in a real-time fashion using advanced optical instruments including AFM. Thus, the information regarding the polymerization and aggregation of PE-conjugated IgG will be useful for designing, prediction and interpretation of the atomic force bio-analytics of important biomolecules in cells.
4. Conclusions
AFM micrographs showed that PE-conjugated IgG molecules were able to undergo conformational changes or molecular interaction in biomonolayers, which resulted in the formation of trimers, polymers and well-assembled aggregates. PE polypeptides that were labeled on the Fab fragments of IgG appeared to be the driving force for the induction of well-organized polymerization and aggregation. The air–liquid tension upon the PE-conjugated IgG on mica might also play a role in the formation of polymers and aggregates. The structural information elucidates the nanometer architecture of the well-organized polymers and circular aggregates of PE-conjugated IgG molecules. The findings provide useful information regarding the potential use of bio-labeled antibodies as markers or immunosensors for AFM bio-analytics of biomolecules in cells and cell membranes.
Acknowledgements
This research project is supported by the National 973 Program of China (2001CB510101), and the National Natural Science Foundation of China (60278014 and 30230350). ZWC is supported by NIH R01 grant HL64560.
References
- Barraud A, Perrot H, Billard V, Martelet C, Therasse J. Study of immunoglobulin G thin layers obtained by the Langmuir–Blodgett method: application to immunosensors. Biosens. Bioelectr. 1993;8:39–48. doi: 10.1016/0956-5663(93)80042-n. [DOI] [PubMed] [Google Scholar]
- Boncheva M, Vogel H. Formation of stable polypeptide monolayers at interfaces: controlling molecular conformation and orientation. Biophys. J. 1997;73:1056–1072. doi: 10.1016/S0006-3495(97)78138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WR, Jiang T, Wan ZL, Zhang JP, Yang ZX, Liang DC. Crystal structure of R-phycoerythrin from Polysiphonia urceolata at 2.8 A resolution. J. Mol. Biol. 1996;262:721–731. doi: 10.1006/jmbi.1996.0547. [DOI] [PubMed] [Google Scholar]
- Delamarche E, Bernard A, Schmid H, Michel B, Biebuyck H. Patterned delivery of immunoglobulins to surfaces using microfluidic networks. Science. 1997;276:779–781. doi: 10.1126/science.276.5313.779. [DOI] [PubMed] [Google Scholar]
- Diederich A, Losche M. Novel biosensoric devices based on molecular protein hetero-multilayer films. Adv. Biophys. 1997;34:205–230. doi: 10.1016/s0065-227x(97)89641-4. [DOI] [PubMed] [Google Scholar]
- Drake B, Prater CB, Weisenhorn AL, Gould SA, Albrecht TR, Quate CF, Cannell DS, Hansma HG, Hansma PK. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science. 1989;243:1586–1589. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- Frederix P, Akiyama T, Staufer U, Gerber C, Fotiadis D, Muller D, Engel A. Atomic force bio-analytics. Curr. Opin. Chem. Biol. 2003;7:641–647. doi: 10.1016/j.cbpa.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Galland-Irmouli AV, Pons L, Lucon M, Villaume C, Mrabet NT, Gueant JL, Fleurence J. One-step purification of R-phycoerythrin from the red macroalga Palmaria palmata using preparative polyacrylamide gel electrophoresis. J. Chromatogr. B Biomed. Sci. Appl. 2000;739:117–123. doi: 10.1016/s0378-4347(99)00433-8. [DOI] [PubMed] [Google Scholar]
- Hansma HG. Varieties of imaging with scanning probe microscopes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14678–14680. doi: 10.1073/pnas.96.26.14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Yeung ES. Dynamics of single-protein molecules at a liquid/solid interface: implications in capillary electrophoresis and chromatography. Anal. Chem. 2002;74:6334–6339. doi: 10.1021/ac0261202. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow RJ, Stedman M, Wells TN. Structure of immunoglobulin G by scanning tunnelling microscopy. J. Mol. Biol. 1991;221:361–365. doi: 10.1016/0022-2836(91)80056-z. [DOI] [PubMed] [Google Scholar]
- Ortega-Vinuesa JL, Tengvall P, Lundstrom II. Aggregation of HSA, IgG, and fibrinogen on methylated silicon surfaces. J. Colloid Interf. Sci. 1998;207:228–239. doi: 10.1006/jcis.1998.5624. [DOI] [PubMed] [Google Scholar]
- Ouerghi O, Touhami A, Othmane A, Ouada HB, Martelet C, Fretigny C, Jaffrezic-Renault N. Investigating specific antigen/antibody binding with the atomic force microscope. Biomol. Eng. 2002;19:183–188. doi: 10.1016/s1389-0344(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Perrin A, Elaissari A, Theretz A, Chapot A. Atomic force microscopy as a quantitative technique: correlation between network model approach and experimental study. Colloid Surf. B. 1998;11:103–112. [Google Scholar]
- Wadu-Mesthrige K, Amro NA, Liu GY. Immobilization of proteins on self-assembled monolayers. Scanning. 2000;22:380–388. doi: 10.1002/sca.4950220607. [DOI] [PubMed] [Google Scholar]
- You HX, Lowe CR. Progress in the application of scanning probe microscopy to biology. Curr. Opin. Biotechnol. 1996;7:78–84. doi: 10.1016/s0958-1669(96)80099-1. [DOI] [PubMed] [Google Scholar]
- Zhang PC, Bai C, Ho PK, Dai Y, Wu YS. Observing interactions between the IgG antigen and anti-IgG antibody with AFM. IEEE Eng. Med. Biol. 1997;16:42–46. doi: 10.1109/51.582175. [DOI] [PubMed] [Google Scholar]