Abstract
Background
Several lines of evidence have suggested that estrogen receptor α (ERα)–negative breast tumors, which are highly aggressive and nonresponsive to hormonal therapy, arise from ERα-positive precursors through different molecular pathways. Because microRNAs (miRNAs) modulate gene expression, we hypothesized that they may have a role in ER-negative tumor formation.
Methods
Gene expression profiles were used to highlight the global changes induced by miRNA modulation of ERα protein. miRNA transfection and luciferase assays enabled us to identify new targets of miRNA 206 (miR-206) and miRNA cluster 221-222 (miR-221-222). Northern blot, luciferase assays, estradiol treatment, and chromatin immunoprecipitation were performed to identify the miR-221-222 transcription unit and the mechanism implicated in its regulation.
Results
Different global changes in gene expression were induced by overexpression of miR-221-222 and miR-206 in ER-positive cells. miR-221 and -222 increased proliferation of ERα-positive cells, whereas miR-206 had an inhibitory effect (mean absorbance units [AU]: miR-206: 500 AU, 95% confidence interval [CI]) = 480 to 520; miR-221: 850 AU, 95% CI = 810 to 873; miR-222: 879 AU, 95% CI = 850 to 893; P < .05). We identified hepatocyte growth factor receptor and forkhead box O3 as new targets of miR-206 and miR-221-222, respectively. We demonstrated that ERα negatively modulates miR-221 and -222 through the recruitment of transcriptional corepressor partners: nuclear receptor corepressor and silencing mediator of retinoic acid and thyroid hormone receptor.
Conclusions
These findings suggest that the negative regulatory loop involving miR-221-222 and ERα may confer proliferative advantage and migratory activity to breast cancer cells and promote the transition from ER-positive to ER-negative tumors.
CONTEXTS AND CAVEATS
Prior knowledge
Expression of microRNA 206 (miR-206) is increased in estrogen receptor (ER)–negative tumors, and miR-206 has been shown to target ERα. The miRNA cluster 221-222 (miR-221-222) also targets ERα and is highly expressed in tamoxifen-resistant breast cancer cells. These miRNAs may play a role in the development of ERα-negative tumors from ERα-positive precursors.
Study design
The mutual interactions of ERα, miRNAs 206, 221, and 222, and various transcriptional cofactors were examined by microarray analyses, RNA and protein gel blots, chromatin immunoprecipitation, and luciferase assays in human breast cancer and control cell lines.
Contribution
miR-206 strongly reduced ERα expression and inhibited the proliferation of ERα-positive cells, whereas miR-221 and -222 increased proliferation of ERα-positive cells. In turn, ERα negatively modulated miR-221-222 by recruiting corepressors to the miR-221-222 transcription site. miR-221-222 expression also reduced the expression of various tumor-suppressor proteins.
Implications
The miRNAs 221, 222, and 206 may participate in a regulatory loop with ERα, which binds to the miR-221-222 transcription start site and recruits cofactors that suppress their transcription. In turn, ERα expression is repressed by miR-206. Overexpression of miR-221-222 may confer a proliferation advantage to cancer cells and induce resistance to therapeutic agents through targeting of tumor suppressors.
Limitations
The ERα and miRNAs 221, 222, and 206 molecular pathway was not confirmed in vivo. In addition, direct interactions of all of the components of this pathway remain to be verified.
From the Editors
Approximately 75% of diagnosed breast tumors express estrogen receptor α (ERα), and this ERα-positive status is associated with a better prognosis because of response to hormonal treatment (1). Several studies suggest that a fraction of ER-negative tumors arise from ER-positive precursors (reviewed in 2). Different molecular events have been reported to suppress ERα expression, such as estrogen withdrawal (3), hypoxia (4), overexpression of epidermal growth factor receptor or v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2), which results in hyperactivation of mitogen-activated protein kinase (5), and DNA methylation at the ERα promoter (6).
MicroRNAs (miRNAs) are small noncoding RNAs that suppress gene expression posttranscriptionally by base pairing to the 3′ untranslated region (3′UTR) of the target mRNAs (7). There is a large body of evidence that dysregulation of miRNAs is a hallmark of cancer (8). We have previously determined miRNA expression profiles of breast cancer tissues (9) and demonstrated that miRNAs are aberrantly expressed in breast cancer and that their expression pattern could discriminate between breast tumors with different biopathological phenotypes, such as ERα status. We previously showed that the expression of miRNA 206 (miR-206) is increased in ER-negative tumors (9), and it has also been shown to target ERα (10). Zhao et al. (11) reported that miR-221 and -222 also target ERα and strongly reduce tamoxifen sensitivity in ER-positive cells. Moreover, Miller et al. (12) reported that miR-221 and -222 are highly expressed in tamoxifen-resistant MCF7 breast cancer cells and that their expression is related to ERBB2 overexpression in primary breast tumors generally resistant to tamoxifen therapy (12). In this study, we examined the role of ERα in modulation of miR-221-222 and miR-206.
Methods
Cell Culture
Human breast cancer cell lines T47D, MCF7, MDA-MB-231, and MDA-MB-436 were purchased from the American Type Culture Collection and grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine and 100 U/mL penicillin–streptomycin. ERα and ERBB2 status were confirmed for all cell lines by western blot analyses, and all experiments were performed at the third passage after thawing. All transfections were carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
MCF7 cells expressing ERBB2 (MCF7-ERBB2) and MCF7 empty control (MOCK) stable clones were obtained from Dr Elda Tagliabue. These cells were grown in DMEM supplemented with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 U/mL penicillin–streptomycin, and 0.5 mg/mL geneticin. For the treatment with the ERBB2 ligand Heregulin (heregulin β1 from Neomarker, Freemont, CA), MCF7-ERBB2 and MOCK cells were grown in serum-free medium for 48 hours and then stimulated for the following 3 days. Cells were collected for RNA and protein extraction at the reported time points. For estradiol (E2) treatments, MCF7, T47D, and MDA-MB-231 cells were grown to 70% confluency in phenol red–free DMEM supplemented with 5% charcoal–dextran-stripped FBS for at least 5 days before treatment.
The megakaryoblastic cell line (Meg01) was purchased from the American Type Culture Collection and grown in Roswell Park Memorial Institute 1640 medium containing 10% heat-inactivated FBS, 2 mM L-glutamine, and 100 U/mL penicillin–streptomycin.
Proliferation Assays
MCF7 cells (3000 per well) were plated in 96-well plates and grown for 96 hours after transfection (final miRNA concentration of 100 nM) in normal culture conditions. Cell proliferation was documented every 24 hours for 4 days using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay kit (Promega, Madison, WI), and absorbance at 490 nm was evaluated by a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA).
Cell Cycle Analyses
MCF7 cells were fixed with 70% ethanol at 72 hours after transfection and stained with 25 μg/mL of propidium iodide (Roche Molecular Biochemicals, Indianapolis, IN) in fluorescence-activated cell sorting buffer (phosphate-buffered saline containing 0.1% bovine serum albumin, 0.05% of Triton X-100, and 50 μg/mL of RNase A). Cells were analyzed using FACSCalibur and Cell Quest Pro Software (BD Biosciences, San Jose, CA).
Plasmid Construction
To generate hepatocyte growth factor receptor (MET) and forkhead box O3 (FOXO3) luciferase reporter constructs, the 3′UTRs were amplified by polymerase chain reaction (PCR) and cloned downstream of the luciferase-coding sequence in the pGL3-control vector at the XbaI restriction site (Promega). Mutations were introduced into the miRNA-binding sites by using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA). To map the miR-221-222 and cyclin-dependent kinase inhibitor 1B (CDKN1B, formerly known as p27/Kip1) promoters, the upstream genomic sequence of the miR-222 hairpin and the first CDKN1B exon, respectively, were amplified by PCR and cloned at the NheI and XhoI sites of the pGL3-basic vector (Promega). For the pCruz-HA-ERα construct, full-length ERα cDNA was synthesized from MCF7 mRNA by RT-PCR and cloned at the KpnI and EcoRV sites of pCRUZ-HA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). All constructs were sequenced to verify integrity.
Microarray Analysis to Identify miRNA Targets
All microarray data have been deposited in the Array Express database with the accession number E-TABM-601. The hybridized Human Genome U133A 2.0 Array was scanned and analyzed with the Affymetrix Microarray Analysis Suite version 5.0. The average density of hybridization signals from three independent samples was used for data analysis, and genes with signal density less than 300 pixels were omitted from the analysis. P values were calculated with two-sided t tests with unequal variance assumptions. To correct for multiple hypothesis testing, the false discovery rate was calculated according to Benjamini and Hochberg (13). Differentially expressed genes were selected using both a false discovery rate of less than 0.1 and a fold-change greater than 1.5 or less than −1.5. A dimensionality reduction approach, the singular value decomposition analysis, was used to visualize gene expression data (14). A tree cluster was generated by hierarchical cluster analysis to classify the miR-transfected cells; for this analysis, we used average linkage metrics and centered Pearson correlation (Cluster 3.0). Java Treeview 1.1 (http://sourceforge.net/projects/jtreeview/) was used for tree visualization. The associations between gene modulation by two miRNAs were examined using a two-sided Fisher exact test. The association between modulation by any two miRNAs was statistically significant if P was less than .001. The online program Pathway-Express (Onto-Tools; Wayne State University, Detroit, MI) (http://vortex.cs.wayne.edu/Projects.html) was used to explore the most biologically relevant pathways affected by a list of input genes. Specific biological pathways were defined by the Kyoto Encyclopedia of Genes and Genomes database (Kanehisa Laboratories, Kyoto, Japan) (http://www.genome.jp/kegg/pathway.html). Given a list of genes, Pathway-Express selects pathways based on impact analysis that considers not only conventional statistical analysis but also other biological factors, such as expression levels (ie, fold change) of input genes, type and position in a given pathway, and protein–protein interactions, among other variables. Pathways were considered statistically significant if the corrected gamma P was less than .01. Pathways were then ranked according to impact factor (15). To identify common pathways shared by miRNAs 221, 222, and 206, as well as potential regulatory effects of these three miRNAs, the overlapping and nonoverlapping pathways were examined and selected for further analysis. Investigation of the enrichment of gene sets as predicted miRNA targets was conducted using the L2L microarray analysis tool 2007.1 (http://depts.washington.edu/l2l/about.html). The targets list used for the L2L analyses was extracted by TargetScan 5.1 (http://www.targetscan.org/) regardless of the conservation site.
Quantitative Real-Time PCR for miRNA and mRNA Quantification
Quantitative real-time PCR (qRT-PCR) was performed with the TaqMan PCR Kit (Applied Biosystems, Foster City, CA), followed by detection with the Applied Biosystems 7900HT Sequence Detection System (P/N: 4329002, Applied Biosystems). PCR was carried out in 10 μL of reaction buffer containing 0.67 μL RT product, 1 μL TaqMan Universal PCR Master Mix (P/N: 4324018, Applied Biosystems), 0.2 mM TaqMan probe, 1.5 mM forward primer, and 0.7 mM reverse primer. The reaction mixture was incubated in a 96-well plate at 95°C for 10 minutes, followed by 40 cycles of denaturation (95°C for 15 seconds) and extension (60°C for 1 minute). All reactions were performed in triplicate. Simultaneous quantification of small endogenous nucleolar RNA U44/U48 was used as a reference for TaqMan assay data normalization. For quantification of ERα, progesterone receptor, caveolin (CAV) 1, CAV2, DNA polymerase alpha 1 (POLA1), phosphatase and tensin homolog (PTEN), tuberous sclerosis 1, bone morphogenetic protein (BMP) 4, BMP7, FOXO3, MET, BCL2-like 11 (BIM), CDKN1B, trefoil factor 1 (TFF1/pS2), and glyceraldehyde-3-phosphate dehydrogenase mRNAs, the appropriate TaqMan probes were purchased from Applied Biosystems. The comparative cycle threshold (Ct) method for relative quantization of gene expression (User Bulletin #2; Applied Biosystems) was used to determine miRNA or mRNA expression level. For all the qRT-PCR experiments, values on the y-axis equal to 2−(ΔCt),where ΔCt is the difference between gene Ct and normalizer gene Ct. Ct represents the threshold cycle at which fluorescence rises statistically significantly above the baseline.
Luciferase Assays for Target and Promoter Identification
To confirm that MET and FOXO3 harbor responsive seed regions (complementary sequences) so that miR-206 and miR-221 and -222, respectively, can bind to their 3′UTRs, 250 ng of pGL3 reporter vector carrying the miR-221 and -222 binding site (see plasmid construct, Figure 3, C and E), 25 ng of the phRL-SV40 control vector (Promega), and 100 nM miRNA precursors or scrambled sequence miRNA control (Ambion, Inc, Austin, TX) were cotransfected into human megakaryoblastoma cell line (Meg01) cells in 24-well plates. To ascertain CDKN1B promoter responsiveness to miR-221, -222, and -206, 250 ng of pGL3 reporter vector carrying the CDKN1B promoter (see plasmid construct, Figure 2, C), 25 ng of the phRL-SV40 control vector, and 100 nM miR precursors or scrambled sequence miRNA control (Ambion, Inc) were cotransfected into ER-positive cells in 24-well plates. To map the miR-221-222 promoter, 250 ng of pGL3 reporter vector carrying the upstream genomic sequence of the miR-222 hairpin (see plasmid construct, Figure 5, D) and 25 ng of the phRL-SV40 control vector were cotransfected into Meg01 cells in 24-well plates. Firefly luciferase activity was measured with a Dual Luciferase Assay Kit (Promega) 24 hours after transfection and normalized with a Renilla luciferase reference plasmid. Reporter assays were carried out in quadruplicate. Statistical significance was analyzed by the unpaired Student t test.
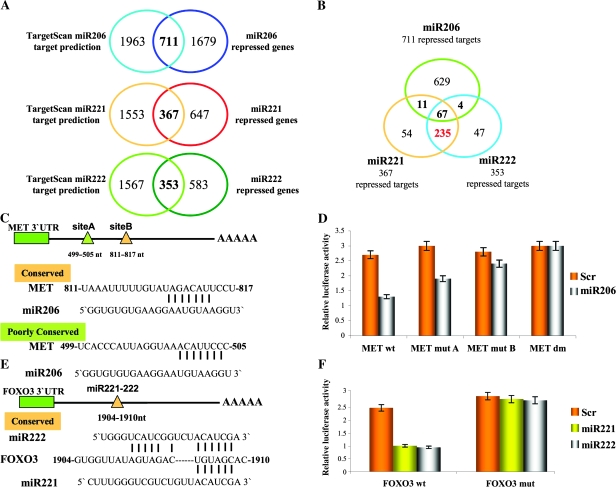
Figure 3.
Identification of new miR-221, -222, and -206 targets. A) Intersection of predicted miR-221, -222, and -206 targets (from TargetScan) with the 2390, 1014, and 936 genes statistically significantly repressed by miR-206, -221, and -222, respectively. B) Intersection of repressed targets of miR-221, -222, and -206. C) Schematic diagram depicts two potential binding sites for miR-206 in the hepatocyte growth factor receptor (MET) 3′ untranslated region (3′UTR), predicted by the TargetScan database; “Poorly conserved” (green) and “conserved” (orange) seed regions were mapped at 499–505 and 811–817 nucleotides, respectively, from the translation stop codon. D) Luciferase activities for MET wild type (wt) and MET mutant (mut) plasmids were determined after transfection of Meg01 cells; all luciferase experiments were performed in quadruplicate, and the luciferase activity was read in triplicate. Data are presented as means (bars) of relative luciferase activity and 95% confidence intervals (error bars). E) Schematic diagram depicts the potential binding for miR-221 and -222, predicted by TargetScan database, in the forkhead box O3 (FOXO3) 3′UTR. F) Luciferase assays for FOXO3 were performed and represented as previously described in (D). Scr = scrambled sequence microRNA control.
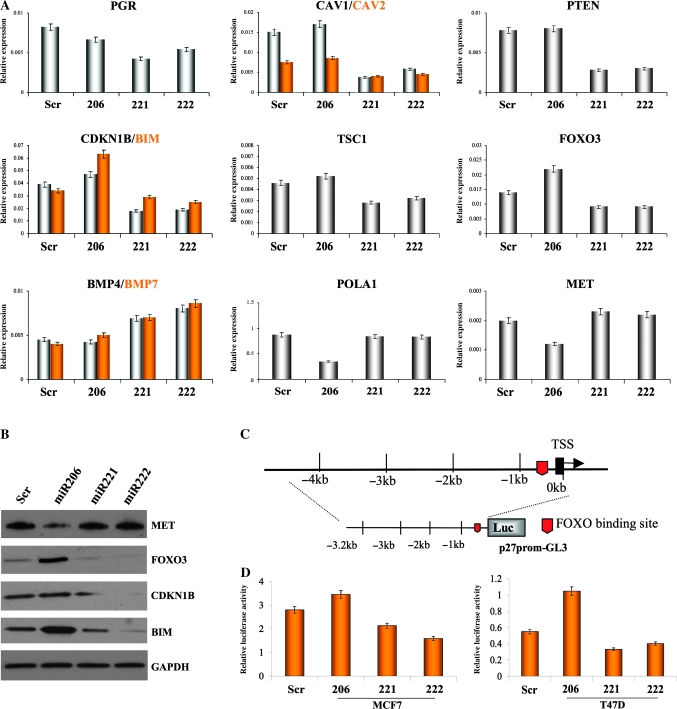
Figure 2.
Effects of miR-206, -221, and -222 on expression of tumor-suppressor and metastasis genes. A) Quantitative real-time polymerase chain reaction to confirm the expression levels of multiple genes statistically significantly modulated in the microarray analyses: progesterone receptor (PGR), caveolin (CAV) 1, CAV2, DNA polymerase alpha 1 (POLA1), tuberous sclerosis 1 (TSC1), phosphatase and tensin homolog (PTEN), bone morphogenetic protein 4 (BMP) 4, BMP7, forkhead box O3 (FOXO3), hepatocyte growth factor receptor (MET), cyclin-dependent kinase inhibitor 1B (CDKN1B), and BCL2-like 11 apoptosis facilitator (BIM). The results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. Experiments were repeated twice in triplicate with similar results. Means (bars) and 95% confidence intervals (error bars) are shown. B) Western blot analyses for MET, FOXO3, CDKN1B, and BIM expression in MCF7 cells after miR-206, -221 and -222 overexpression. GAPDH was used as a loading control. The following antibodies were used: rabbit polyclonal anti-MET (Santa Cruz, Inc., sc-10; dilution 1:500), rabbit polyclonal anti-FOXO3 (Millipore [Billerica, MA], 07-702; dilution 1:1000), rabbit polyclonal anti-CDKN1B (Santa Cruz, Inc, sc-528; dilution 1:500), rabbit polyclonal anti-BIM (Santa Cruz, Inc, sc-11425; dilution 1:1500), and mouse monoclonal anti-GAPDH (Calbiochem CB1001; dilution 1:10000). C) Schematic representation of the CDKN1B promoter and the reported FOXO3-binding site. D) Luciferase assay of CDKN1B promoter in two estrogen receptor–positive cell lines, MCF7 and T47D, after miR-221, -222, and -206 overexpression; luciferase experiments were performed in quadruplicate, and luciferase activity was read in triplicate. Data are presented as means of relative luciferase activity (bars) and 95% confidence intervals (error bars). Scr = scrambled sequence microRNA control; TSS = transcription start site.
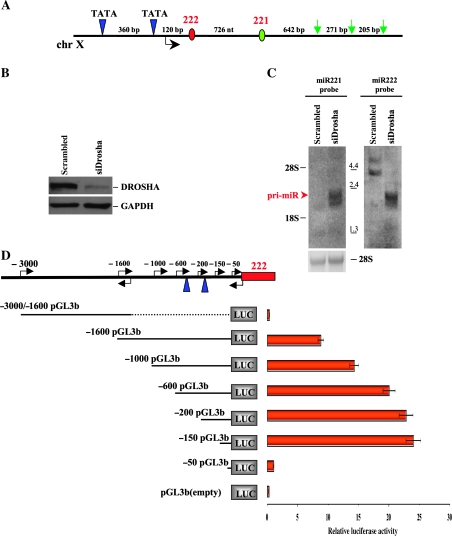
Figure 5.
Identification of miR-221-222 transcriptional unit. A) Eleven-kilobase genomic region spanning miR-221 and -222 was analyzed using the Promoter 2.0 and human polyA signals databases to search for promoter and polyA signal sequences. The schematic diagram represents two canonical TATA boxes (blue triangles) located 550 and 190 base pair (bp) upstream of pre-miR-222 (red circle) and three polyA signals (green arrows) located downstream of pre-miR-221 (green circle). (B and C) MDA-MB-231 cells overexpressing miR-221 and -222 were transfected with small interfering RNA (siRNA) against DROSHA or control siRNA against green fluorescent protein (100 nM). B) Seventy-two hours after transfection, siRNA-treated cells were subjected to western blot analysis for the detection of DROSHA protein, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were used as a loading control. The following antibodies were used: rabbit polyclonal anti-Drosha (Santa Cruz, Inc, sc-33778; dilution 1:500), mouse monoclonal anti-GAPDH (Calbiochem CB1001; dilution 1:10000). C) Seventy-two hours after transfection, siRNA-treated cells were also subjected to northern blot analysis for the detection of miR-221 (left panel) and 222 (right panel) primary transcript. Positions of the RNA marker (Invitrogen) are shown between the panels. Ethidium bromide staining of 28S ribosomal RNA is shown as a loading control. The experiments were repeated twice with similar results. D) Luciferase assays were carried out to identify the miR-221-222 promoter. Genomic fragments located upstream of pre-miR-222 and cloned into the pGL3-basic vector are shown on left. Twenty-four hours after transfection, luciferase activity assays of Meg01 cells transfected with respective reporter constructs were performed. All luciferase experiments were performed four times in duplicate, and data are represented with 95% of confidence intervals. ChrX = chromosome X; pri-miR = primary transcript of microRNAs.
Acrylamide and Agarose Northern Blot Analyses for miRNA Detection
Acrylamide northern blotting was performed as previously described (9). Briefly, 10 μg aliquots of total RNA were resolved on a 15% denaturing polyacrylamide gel (Bio-Rad, Hercules, CA) and were electrophoretically transferred to BrightStar blotting membrane (Ambion, Inc). The oligonucleotide encoding the complementary sequence of the mature miRNA annotated in the miRNA Registry (release 14: September 2009) was end-labeled with [γ32 P]-ATP by T4 polynucleotide kinase (USB, Cleveland, OH). RNA-blotted membrane was prehybridized in Ultrahyb Oligo solution (Ambion, Inc) and subsequently hybridized in the same solution containing probe at a concentration of 106 cpm/mL at 37°C overnight. The membrane was washed at high stringency in the solution containing 2× standard saline citrate and 1% sodium dodecyl sulphate at 37°C. To detect the primary transcript of miR-221 and -222, total RNA (10 μg per sample) was size-fractionated through 1.2% agarose–formaldehyde gels and transferred to nylon filters (GE Healthcare Biosciences, Piscataway, NJ). The northern blot was hybridized with random-primed and α-32P-labeled genomic DNA fragments spanning either the miR-221 or the miR-222 hairpin as a probe (Roche Molecular Biochemicals). Northern hybridization signals were captured and converted to digital images with the Typhoon Scanner (GE Healthcare Biosciences).
5′ and 3′ Rapid Amplification of cDNA Ends to Identify miR-221-222 Primary Precursor
To map the 5′ and 3′ termini of the primary precursor of miR-221 and -222, 5′ and 3′ rapid amplification of cDNA ends (RACE) analyses were performed using a FirstChoice RLM-RACE Kit (Ambion, Inc). RACE experiments were carried out by using total RNA prepared from Drosha knockdown MDA-MB-231 cells according to the manufacturer's instructions.
Transcriptional Elements Analyses
For miR-221 and -222 promoter prediction, an 11 000 base pair (bp) DNA genomic region spanning miR-221-222 was used as input for the online software Promoter 2.0 (http://www.cbs.dtu.dk/services/promoter/), and only highly likely predictions (positions scoring above 1.0) were considered. For polyA signal identification, a DNA sequence 5000 bp downstream of miR-221 was used as input for the online software Poly-ADQ (http://rulai.cshl.org/tools/polyadq/polyadq_form.html). Default cutoffs were used, and only positive predictions (64% sensitivity, 83% specificity, and a correlation coefficient of .512) were considered. For estrogen response element (ERE) identification, the 5000 bp DNA genomic region spanning the miR-221-222 region was used as input for the Transcriptional Element Search Software database (http://www.cbil.upenn.edu/cgi-bin/tess/tess).
Chromatin Immunoprecipitation Assay to Evaluate ERα-Binding Sites
Chromatin immunoprecipitation (ChIP) assays were performed with the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) with minor modifications. Briefly, MCF7 and MDA-MB-231 cells were grown to 70% confluency in DMEM supplemented with 10% FBS. The cross-linking was performed with 1% formaldehyde at 37°C for 10 minutes. Cells were then rinsed with ice-cold PBS and resuspended in 0.4 mL of lysis buffer containing 1% sodium dodecyl sulphate, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1, 1× protease inhibitor cocktail (Roche Molecular Biochemicals), and sonicated. A 30 μL aliquot of the preparation was treated to reverse the cross-linking, deproteinized with proteinase K, extracted with phenol–chloroform, and the DNA concentration determined by Nanodrop 2000c (Thermo Scientific, Wilmington, DE) measurements. An aliquot of chromatin preparation containing 25 μg DNA was used per ChIP. DNase-free RNase (Roche Molecular Biochemicals) was added at a concentration of 200 μg/mL during reverse cross-linking. After deproteinization with proteinase K, DNA was purified in 50 μL of Tris–EDTA with a PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. A 2-μL aliquot was used for PCR. Primer sequences are listed in Supplementary Materials and Methods (available online). The primary antibodies used for immunoprecipitation were rabbit polyclonal ERα (Bethyl Laboratories [Montgomery, TX] A300-498A), rabbit polyclonal silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) (Santa Cruz, Inc, sc-20778), rabbit polyclonal nuclear receptor corepressor (NCoR) (Santa Cruz, Inc, sc-8994), rabbit IgG control (Zymed, Carlsbad, CA), rabbit polyclonal acetyl-H3 and acetyl-H4 (Upstate Biotechnology). In some experiments, ChIP-enriched DNA was also subjected to SYBR green qPCR (Applied Biosystems). In this case, results were expressed as relative enrichment according to the following formula: 2−[(ctChIP − ctinput) − (ctIgG − ctinput)], where ctChIP, ctIgG, and ctinput indicate the cycle threshold for the specific antibody, IgG control, and input (5% of the total amount of immunoprecipitated material), respectively.
Western Blot Analyses
All cell lysates were prepared by using RadioImmuno Precipitation Assay Buffer (Pierce, Rockford, IL). Fifty micrograms of cell lysates was separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and then electroblotted onto a polyvinylidene fluoride membrane (Hybond P; Amersham Biosciences, Piscataway, NJ). All primary antibodies used for western blot analyses are reported in Supplemental Materials and Methods (available online). Detection was performed with horseradish peroxidase–conjugated secondary antibodies (specific to rabbit and mouse) and enhanced chemiluminescence (Pierce).
Statistical Analysis
Data are represented as means with 95% confidence intervals (CIs). Statistical significance was determined with unpaired Student t tests, except as noted for analyses of microarray data, which were examined with Fisher exact tests. P values less than .05 were considered statistically significant. All statistical tests were two-sided.
Results
Differential Effects of miR-221 and -222 and miR-206 in Breast Cancer
To determine the differential effect of miR-221 and -222 and miR-206 on ERα production and their impact on MCF7 breast cancer cells, which lack or have very low miR-221, -222, and -206 expression (Supplementary Figure 1, A–C, available online), MCF7 cells were transfected with all three miRNAs and a scrambled sequence miRNA control. qRT-PCR showed efficient accumulation of all miRNAs (mean expression levels, relative units: miR-206: 1400, 95% CI = 1257 to 1609, Scr: 0.0029; miR-221: 1656, 95% CI = 1436 to 1942, Scr: 0.023; miR-222: 1500, 95% CI = 1348 to 1768, Scr: 0.032. All Ps of difference between the samples and the control <.05) (Figure 1, A). This was followed, as expected, by a suppression of ERα expression (Figure 1, B). qRT-PCR demonstrated that only miR-206 substantially reduced (approximately 52%) expression of ERα mRNA (mean expression levels, relative units: Scr: 0.042; miR-206: 0.02; miR-221: 0.038; miR-222: 0.037) (Figure 1, C).
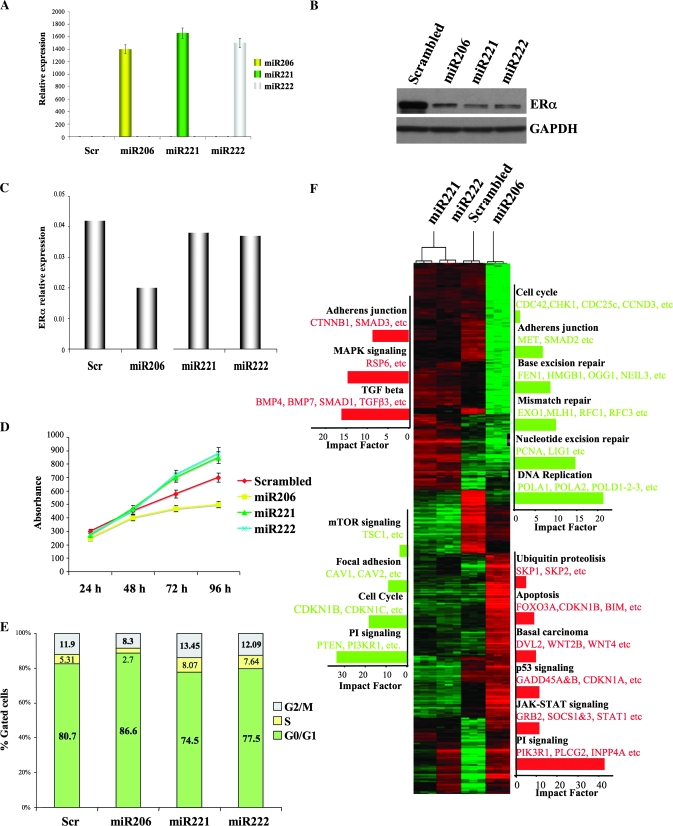
Figure 1.
Effect of miR-221, -222, and -206 on gene expression. MCF7 cells were transfected with scrambled sequence mRNA control and miR-206, -221, and -222 (100 nM) and collected 72 hours after transfection. A) miR-206, 221, and 222 levels were assayed by quantitative real-time polymerase chain reaction (qRT-PCR) analyses. The experiment was repeated twice, each with three replicates. Means (bars) and 95% confidence intervals (error bars) are shown. B) Estrogen receptor α (ERα) expression levels were analyzed by western blot, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were used as a loading control. The following antibodies were used: mouse monoclonal anti-ERα (Santa Cruz, Inc, sc-8002; dilution 1:1000) and mouse monoclonal anti-GAPDH (Calbiochem [Gibbstown, NJ], CB1001; dilution 1:10000). C) ERα mRNA expression levels were analyzed by qRT-PCR. GAPDH levels were used as a normalizer. The experiments were performed twice with similar results. D) Cell growth was measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide–based colorimetric cell proliferation assay. The experiment was repeated four times, each with eight replicates, and the data are expressed as mean absorbance units with 95% confidence intervals (error bars). E) miR-transfected MCF7 cells were also subjected to fluorescence-activated cell sorting analysis, and the relative G1, S, and G2/M compartments calculated. Percentages of cells in each compartment are means of three independent experiments performed in triplicate. F) Unsupervised clustering of genes differentially expressed between scrambled sequence mRNA control, miR-206, -221, and -222; representative miR-activated genes (red) and miR-repressed genes (green) are listed under each molecular pathway. Impact factor strength of miR-activated (red bars) and repressed (green bars) genes is shown. Scr = scrambled sequence microRNA control.
It was recently shown that miR-206 inhibits cell growth of ER-positive cells, whereas inhibition of miR-221 and -222 in ER-negative cells induces apoptosis (16,17). MCF7 proliferation assays to assess the effects of miR-206, -221, and -222 on ER-positive cells revealed that miR-221 and -222 increased cell proliferation, whereas miR-206 had an inhibitory effect (mean absorbance units [AU] at 96 hours: miR-206: 500 AU, 95% CI = 480 to 520; miR-221: 850 AU, 95% CI = 810 to 873; miR-222: 879 AU, 95% CI = 850 to 893. All Ps of difference between the samples and the control <.05) (Figure 1, D).
Cell cycle analyses indicated that only miR-221 and miR-222 induced a statistically significant increase in the transition from G1 to S phase (mean percentage of cells in S phase: miR-221: 8.07%, 95% CI = 7.96% to 8.3%; miR-222: 7.64%, 95% CI = 7.5% to 7.78%. All Ps of difference between the samples and the control <.05) (Figure 1, E). Indeed, miR-206 induced a statistically significant block in G1 (mean percent cells in G1: miR-206: 86.6%, 95% CI = 86.3% to 89%. All Ps of difference between the samples and the control <.05) (Figure 1, E).
Finally, we profiled the global changes in gene expression after miRNA expression in MCF7 cells. Singular value decomposition and unsupervised clustering analyses showed that genes with increased and decreased expression in MCF7 cells overexpressing miR-221 and -222 cluster together and are separate from those expressed in cells overexpressing miR-206 or the scrambled sequence control (Figure 1, F, and see Supplementary Figure 2, available online).
Statistical analyses identified 1966, 1091, and 1079 genes with statistically significantly increased expression and 2390, 1014, and 936 genes with statistically significantly decreased expression levels after overexpression of miR-206, -221, and -222, respectively (see Supplementary Table 1, available online). We determined that almost 75% of the genes modulated by miR-221 and -222 are overlapping, but only 30% are modulated in common with miR-206 (Supplementary Figure 3, available online). Moreover, functional profiling of these genes revealed that the processes modulated by miR-221 and -222 are statistically significantly overlapping (P < .001) and different from those of miR-206 (P206 vs 221 = .628, P 206 vs 222 = .215) (Supplementary Figure 4, A, available online). Notably, miR-221 and -222 comodulated processes include apoptosis, mitogen-activated protein kinase and transforming growth factor-beta signaling pathways, focal adhesion, and colorectal cancer (Supplementary Figure 4, B, available online).
We used qRT-PCR to confirm the expression of different genes identified in microarray analyses of MCF7 cells (Figure 2, A). With respect to reductions in ERα expression, first we searched for the modulation of estrogen-regulated genes; as expected, expression of progesterone receptor, a direct target of ERα, was reduced by all three miRNAs (mean expression levels, relative units: miR-206: 0.0067, 95% CI = 0.0064 to 0.0069; miR-221: 0.0042, 95% CI = 0.0038 to 0.0048; miR-222: 0.0054, 95% CI = 0.0050 to 0.0056. All Ps of difference between the samples and the control <.05).
Next, the expression of many genes known to have important functions in suppressing tumor growth and metastasis was found to be repressed in miR-221– and -222–expressing cells. These genes include focal adhesion proteins CAV1 and CAV2, which are strong tumor suppressors in mammary glands (18), as well as suppressors of cytokine signaling; PTEN, which blocks cell proliferation, motility, and migration (19); CDKN1B, which blocks cells in G1 phase by preventing CDK-dependent phosphorylation of retinoblastoma tumor-suppressor protein (pRb) (20); BIM, the expression of which is sufficient to trigger apoptosis in a variety of cell types (21); tuberous sclerosis 1, which inhibits the mechanistic target of rapamycin pathway that reduces angiogenesis and tumor growth (22); and FOXO3, which increases expression of many genes involved in tumor suppression, such as CDKN1B and BIM, two well-known targets of miR-221 and 222 (17,23,24) (mean expression levels, relative units: CAV1: miR-221: 0.0038, 95% CI = 0.0034 to 0.0040; miR-222: 0.0058, 95% CI = 0.0057 to 0.0060. CAV2: miR-221: 0.004, 95% CI = 0.0039 to 0.0042; miR-222: 0.0045, 95% CI = 0.0044 to 0.0048. PTEN: miR-221: 0.0028, 95% CI = 0.0025 to 0.0032; miR-222: 0.003, 95% CI = 0.0027 to 0.0035. CDKN1B: miR-221: 0.018, 95% CI = 0.0125 to 0.023; miR-222: 0.019, 95% CI = 0.017 to 0.02. BIM: miR-221: 0.029, 95% CI = 0.025 to 0.03; miR-222: 0.025, 95% CI = 0.017 to 0.027. Tuberous sclerosis 1: miR-221: 0.0028, 95% CI = 0.0025 to 0.003; miR-222: 0.0032, 95% CI = 0.003 to 0.0033. FOXO3: miR-221: 0.009, 95% CI = 0.005 to 0.010; miR-222: 0.009, 95% CI = 0.007 to 0.011. All Ps of difference between the samples and the control <.05) (Figure 2, A).
Many genes involved in promoting metastasis were found to have increased expression in miR-221– and -222–expressing cells, including BMP4 and BMP7 (Figure 2, A), transforming growth factor, beta 3 (TGFB3), and SMAD family member 1 (SMAD1) (data not shown), which are important for bone metastasis and invasiveness (25,26) (mean expression levels, relative units: BMP4: miR-221: 0.0069, 95% CI = 0.0067 to 0.0073; miR-222: 0.008, 95% CI = 0.0076 to 0.0083. BMP7: miR-221: 0.007, 95% CI = 0.0068 to 0.0077; miR-222: 0.0086, 95% CI = 0.0081 to 0.0088. All Ps of difference between the samples and the control <.05).
In contrast, miR-206 overexpression blocked the activity of genes involved in DNA synthesis and repair, including PCNA and LIG1 (data not shown) and the large subunit of DNA polymerase alpha (POLA1), reported to be a target of miR-206 (27) (mean expression levels, relative units: miR-206: 0.35, 95% CI = 0.26 to 0.38, P206 vs Scr = .035). The activity of the MET oncogene, which is known to stimulate invasive growth of epithelial cells and is associated with decreased survival of breast cancer patients (28), was also blocked by miR-206 overexpression (mean expression levels, relative units: miR-206: 0.012, 95% CI = 0.009 to 0.013, P206 vs Scr = .003).
The expression of tumor-suppressor FOXO3 and its direct targets CDKN1B and BIM (Figure 2, A) was increased in miR-206–expressing MCF7 cells and decreased in those expressing miR-221 and -222 compared with scrambled sequence miRNA control (mean expression levels, relative units: FOXO3: miR-206: 0.022, 95% CI = 0.018 to 0.025; CDKN1B: miR-206: 0.047, 95% CI = 0.045 to 0.054; BIM: miR-206: 0.063, 95% CI = 0.058 to 0.070. All Ps of difference between the samples and the control <.05). qRT-PCR for the same genes in T47D cells gave similar results (see Supplementary Figure 5, A and B, available online). Western blot analyses confirmed that miR-206–expressing cells repressed the expression of MET but activated that of FOXO3, CDKN1B, and BIM. Conversely, miR-221– and -222–expressing cells repressed the expression of FOXO3, CDKN1B, and BIM but not that of MET (Figure 2, B). Finally, because FOXO3 was reported to directly activate the transcription of CDKN1B (29), the promoter of CDKN1B, containing the previously identified FOXO3-binding site (30), was cloned in the promoterless luciferase vector (Figure 2, C). Overexpression of miR-206 in two different ER-positive cells statistically significantly increased CDKN1B promoter activity relative to control, whereas miR-221– and -222–expressing cells showed an inhibitory effect (Figure 2, D) (mean activity, relative luciferase units: MCF7: miR-206: 3.45, 95% CI = 3.33 to 3.6, P206 vs Scr = .003; miR-221: 2.1, 95% CI = 2 to 2.2, P221 vs Scr = .005; miR-222: 1.58, 95% CI = 1.45 to 1.60, P222 vs Scr = .003. T47D: miR-206: 1.04, 95% CI = 0.95 to 1.2, P206 vs Scr = .002; miR-221: 0.33, 95% CI = 0.24 to 0.35, P221 vs Scr = .0048; miR-222: 0.4, 95% CI = 0.37 to 0.41, P222 vs Scr = .004). Disruption of the reported FOXO3-binding site completely abolished the effect of the miRNAs on the CDKN1B promoter luciferase activity (Supplementary Figure 6, A and B, available online).
MET and FOXO3 as Targets of miR-206 and miR-221-222
Underexpressed genes containing miRNA putative targets were analyzed to identify new direct targets of miR-221-222 and miR-206, which might explain the opposite biological effects of these three miRNAs on ER-positive cells. We used the L2L analysis tool to investigate the enrichment of target genes based on the target list of the TargetScan database. A substantial enrichment of miRNA targets was found for all three miRNAs, which validated our microarray approach as a valuable tool for miRNA target identification (Figure 3, A). Because miR-221 and 222 share an identical seed sequence, 75% of their underexpressed targets were in common. Less than 10% of the underrepressed targets were found in common between miR-206 and miR-221-222 (Figure 3, B). MET and FOXO3, previously found to be repressed at the mRNA and protein levels by miR-206 and miR-221-222, respectively (Figure 2, A and B), were also found in our list of underexpressed targets.
MiR-206 was predicted by TargetScan software to bind the MET 3′UTR at two different sites, A and B (Figure 3, C). Site B is highly conserved across several species, whereas site A is poorly conserved (Figure 3, C, and Supplementary Figure 7, A, available online). To verify whether miR-206 targets the MET 3′UTR, a luciferase reporter in the pGL3-control vector containing the full-length MET 3′UTR was constructed. Overexpression of miR-206 statistically significantly inhibited MET 3′UTR luciferase activity relative to the scrambled sequence control, whereas the MET 3′UTR containing mutations at site A or B or at both lost this inhibition (mean activity, relative luciferase units: MET wt: Scr: 2.7, 95% CI = 2.5 to 3; miR-206: 1.3, 95% CI = 1.24 to 1.45; P206 vs Scr < .001. MET mutA: Scr: 3, 95% CI = 2.6 to 3.5; miR-206: 1.9, 95% CI = 1.75 to 2.1; P206 vs Scr = .016. MET mutB: Scr: 2.8, 95% CI = 2.6 to 3.1; miR-206: 2.4, 95% CI = 2.3 to 2.6; P206 vs Scr = .078. MET DM: Scr: 3, 95% CI = 2.8 to 3.2; miR-206: 3, 95% CI = 2.79 to 3.2; P206 vs Scr = .8) (Figure 3, D, and Supplementary Figure 7, B, available online). To verify that miR-221 and -222 directly target the FOXO3 3′UTR, an 800 bp region containing the conserved miR-221-222 predicted binding site was cloned after the luciferase reporter gene (mean activity, relative luciferase units: FOXO3 wt: Scr: 2.2, 95% CI = 2.1 to 2.3; miR-221: 0.9, 95% CI = 0.75 to 1.01; miR-222: 0.85, 95% CI = 0.8 to 0.94; P221 vs Scr < .001, P222 vs Scr < .001. FOXO3 mut: Scr: 2.6, 95% CI = 2.4 to 2.8; miR-221: 2.5, 95% CI = 2.3 to 2.65; miR-222: 2.46, 95% CI = 2.3 to 2.94; P221 vs Scr = .49, P222 vs Scr = .45) (Figure 3, E, and Supplementary Figure 7, C, available online). Overexpression of miR-221-222 reduced the luciferase activity of the FOXO3 reporter to approximately 60% of that of the scrambled sequence control (Figure 3, F, and Supplementary Figure 7, D, available online). Importantly, disruption of the predicted binding site completely restored luciferase activity (Figure 3, F). We concluded that miR-206 targets MET and that miR-221 and -222 target FOXO3 by binding sites within the 3′UTRs.
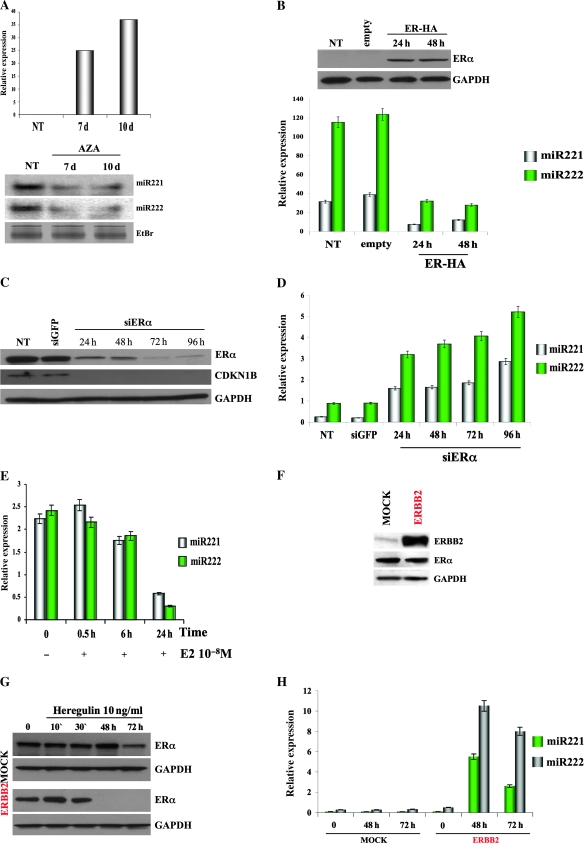
ERα Regulation of miR-221-222 Expression
Loss of miR-221-222 expression in ER-positive cells (11) raised the possibility that ERα negatively regulates transcription of these miRNAs. To test this hypothesis, ER-negative cells MDA-MB-231, carrying the hypermethylated ERα promoter (6), were treated for 10 days with demethylating agent 5-AZA-2′-deoxycytidine (5′AZA). Analysis by qRT-PCR showed a time-dependent induction of ERα mRNA (Figure 4, A, upper panel) (mean expression levels, relative units: not treated: 0.75; 7 days: 25; 10 days: 37), followed by a reduction in miR-221 and -222 expression (Figure 4, A, lower panel, and Supplementary Figure 8, A, available online). Moreover, restoration of ERα protein in ER-negative MDA-MB-231 cells (Figure 4, B, upper panel) also induced a strong reduction of miR-221 and -222 expression (Figure 4, B, lower panel) (mean expression levels, relative units: miR-221 [48 hours]: 12.05, 95% CI = 10.5 to 13.6; miR-222 [48 hours]: 27.85, 95% CI = 21.5 to 29.6. All Ps of difference between the samples and the control <.05), indicating that ERα inhibits miR-221 and -222. To corroborate these findings, silencing of ERα by using small interfering RNA was carried out in MCF7 cells. Strong suppression of ERα protein was accompanied by decreased expression of CDKN1B (Figure 4, C); analysis by qRT-PCR demonstrated that reduction in ERα protein, in turn, increased the expression of miR-221 and -222 (Figure 4, D) (mean expression levels, relative units: miR-221 (96 hours): 2.9, 95% CI = 2.68 to 3.2; miR-222 (96 hours): 5.2, 95% CI = 4.9 to 5.6. All Ps of difference between the samples and the control <.05). Silencing of ERα was also performed in T47D cells with similar results (Supplementary Figure 8, B, available online). In summary, the enforced expression of ERα in ER-negative cells and the elimination of ERα from ER-positive cells caused suppression and induction, respectively, of miR-221 and -222, suggesting a novel function of ERα as a negative regulator of the expression of these miRNAs. Because ERα activity is regulated by the hormone E2 (1), we determined the effect of E2-activated ERα on miR-221-222 transcription. Hormone-starved MCF7 cells were treated with 10 nM E2. Analysis by qRT-PCR determined a time-dependent induction of TFF1 transcript, a known target of ERα activity (see Supplementary Figure 8, C, available online), followed by a marked inhibition of miR-221 and -222 expression at 24 hours after treatment (Figure 4, E) (mean expression levels, relative units: miR-221 [24 hours]: 0.58, 95% CI = 0.43 to 0.6; miR-222 [24 hours]: 0.29, 95% CI = 0.25 to 0.33. All Ps of difference between the samples and the control <.05). The same ligand-dependent repression of miR-221 and -222 was also found in T47D cells (Supplementary Figure 8, D and E, available online). Similar repression of miR-221 and -222 expression was not detected after E2 treatment in ER-negative cells (Supplementary Figure 8, F, available online).
Figure 4.
Negative regulation of miR-221 and -222 expression by estrogen receptor α (ERα). A) MDA-MB-231 cells were treated with 5-aza-2′-deoxycytidine (5′AZA) at a concentration of 10 μM for 10 days. 5′AZA-treated cells were subjected to quantitative real-time polymerase chain reaction (qRT-PCR) analysis for the detection of ERα transcript, and the data are expressed relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression (top). miR-221 and -222 expression levels were detected by northern blot analysis. Ethidium bromide (EtBr) staining is shown as a loading control (bottom). The experiments were repeated twice with similar results. B) MDA-MB-231 cells were cultured for 5 days in hormone-deprived media and were transfected with the pCruz-HA-ERα construct and an empty control. Twenty-four hours after transfection, cells were stimulated by adding estradiol (E2) at a concentration of 10 nM and were kept in culture for an additional 48 hours. Transfected cells were subjected to western blotting analysis for the detection of HA-ERα (top) and to qRT-PCR analysis (bottom) for the quantitative detection of mature miR-221 and 222. (C and D) ERα-positive MCF7 cells were transfected with small interfering RNA (siRNA) against ERα or with a control siRNA-targeting green fluorescent protein. Twenty-four, 48, 72, and 96 hours after transfection, transfected cells were subjected to western blotting analysis (C) for the detection of ERα and cyclin-dependent kinase inhibitor 1B (CDKN1B) protein. GAPDH was used as a loading control for the experiments. qRT-PCR analyses (D) were also used for the quantitative detection of mature miR-221-222. E) MCF7 cells were hormone starved for 5 days and were then stimulated by adding E2 at a concentration of 10 nM. Total RNAs from E2-treated cells were subjected to qRT-PCR for detection of mature miR-221-222. (F, G, H) MCF7-mock and MCF7-HER2 cell lines were analyzed by western blotting for the detection of HER2 and ERα (F); MCF7-mock and MCF7-HER2 cell lines were starved for 48 hours and then activated with heregulin β1 at a concentration of 10 ng/mL. Cells were collected and analyzed for ERα (G) and miR-221 and 222 expression levels (H). Except where noted, all data are expressed as means of three experiments (bars) and 95% confidence intervals (error bars). For the western blot analyses, the following antibodies were used: mouse monoclonal anti-HA-HRP conjugates (Roche Molecular Biochemicals; dilution 1:500), mouse monoclonal anti-ERα (Santa Cruz, Inc, sc-8002; dilution 1:1000), rabbit polyclonal anti-CDKN1B (Santa Cruz, Inc, sc-528; dilution 1:500), rabbit polyclonal anti–v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2) (Santa Cruz, Inc, sc-134481; dilution 1:500), mouse monoclonal anti-GAPDH (Calbiochem CB1001; dilution 1:10000). NT = not treated; Scr = scrambled sequence microRNA control.
Continuous activation of receptor tyrosine kinases, including epidermal growth factor receptor and ERBB2, induces loss of ERα expression in MCF7 cells (5,31). Because overexpression of these receptor tyrosine kinases is frequently observed in ER-negative tumors (32) and positively associated with miR-221 and -222 expression (12), we sought to determine whether activation of the receptor tyrosine kinases induced miR-221 and -222 expression. To this end, the previously described (33) MCF7-ERBB2 and MOCK cells were analyzed for miR-221-222 and ERα expression. MCF7-ERBB2 cells exhibited increased expression of ERBB2, whereas neither ERα nor miR-221-222 expression was suppressed (Figure 4, F, and data not shown). Activation of ERBB2 signaling with growth factor heregulin β1 strongly reduced ERα expression but not in MCF7-MOCK cells (Figure 4, G), followed by a substantial induction of miR-221 and -222 expression in these ERα-repressed MCF7-ERBB2 cells compared with MOCK MCF7 cells (Figure 4, H) (mean expression levels, relative units: miR-221 [ERBB2-72 hours]: 2.6, 95% CI = 2.3 to 3; miR-222 [ERBB2-72 hours]: 8.15, 95% CI = 7.5 to 8.68. All Ps of difference between the samples and the control <.05).
Transcriptional Unit of miR-221 and -222
MiR-221 and 222 are closely located on human chromosome X (Figure 5, A), and because the genomic region lacks any protein-coding gene, both miRNAs appear to represent a single transcription unit. To produce global accumulation of pri-miRNAs (34), knockdown against the RNAse Drosha was performed in MDA-MB-231 cells expressing high levels of miR-221 and -222 (Figure 5, B). Northern blot analysis, probed with pre-miR-221, detected a transcript of approximately 2.1 kb that is hardly detectable in control cells (Figure 5, C). Detection of the 2.1 kb transcript by reprobing the blot with pre-miR-222 (Figure 5, C) indicated that miR-221 and -222 are transcribed into a single species of pri-miRNA. To specifically define the 5′ and 3′ termini of the transcriptional unit of pri-miR-221 and -222, we carried out 5′ and 3′ RACE analyses and found a 5′ extension of about 230 nucleotides from the 3′ end of pre-miR-222 and a 3′ extension of about 1000 nucleotides from the 5′ end of pre-miR-221, respectively (data not shown). Analysis of miR-221-222 with the Promoter.2 prediction server and the polyA signals databases predicted two canonical TATA boxes located approximately 550 and 190 nucleotides upstream from the 5′ end of pre-miR-222 and multiple polyadenylation sites close to the 3′ end of the primary transcript (Figure 5, A). To determine whether this upstream region could be the miR-221-222 transcriptional promoter, we constructed reporter plasmids by inserting the fragments spanning minus 1600 bp to approximately minus 3000 bp and plus 3 to approximately minus 1600 bp (the plus 1 position corresponds to the 5′ terminus of pre-miR-222) into the promoterless vector pGL3basic (Figure 5, D). The subsequent luciferase assay showed that the promoter became functional starting at minus 1600 bp, with the minus 1600 pGL3b construct giving an approximately ninefold induction of luciferase activity compared with the empty vector (Figure 5, D). A series of 5′ end deletion mutants from minus 1600 to minus 50 bp enabled us to map the minimal promoter of miR-221-222 at minus 150 bp to approximately minus 50 bp, where the proximal TATA box is excluded (Figure 5, D). Taken together, these results suggest that both miR-221 and -222 are transcribed into a single species of 2.1 kb RNA, and their expression is related to ERα levels.
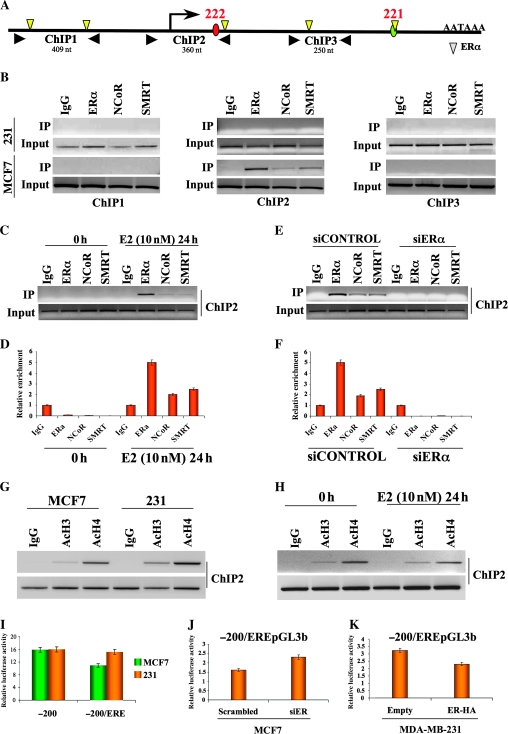
Recruitment of ERα, NCoR, and SMRT at the miR-221-222 Locus
E2 activation of ERα confers two important functions, localization of ERα at the target genes by binding specific DNA sequences, EREs, followed by recruitment of additional cofactors that have either activator or repressor functions on target genes (35). To investigate whether ERα binds directly to miR-221-222, we carried out a ChIP analysis. A search of the Transcriptional Element Search Software database found five potential EREs within the 5-kb genomic region spanning miR-221 and -222 (Figure 6, A). Three chromatin regions were analyzed for EREs (Figure 6, A). A ChIP assay of ER-positive MCF7 cells identified ERα binding at ChIP region 2, which spans the promoter of miR-221-222 (Figure 6, B). No ERα binding was observed in ER-negative MDA-MB-231 cells. Next, ChIP assays against NCoR and SMRT revealed colocalization of these corepressors with ERα in region 2 (Figure 6, B). Because no apparent binding of NCoR and SMRT was detected in region 1 or 3 of MCF7 or in any ChIP-analyzed regions of MDA-MB-231, we suggest that ERα is responsible for the colocalization of the corepressors.
Figure 6.
Binding of estrogen receptor α (ERα) to miR-221-222 and recruitment of corepressor proteins nuclear receptor corepressor (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT). A) Schematic diagram of miR-221-222 transcription unit with transcription start site (red circle: miR-222, green circle: miR-221), polyA signal, and canonical ERα-binding sites (yellow triangles). Chromatin immunoprecipitation (ChIP)–analyzed regions are indicated with three pairs of black arrowheads. B) Cross-linked chromatin was prepared from MDA-MB-231 (ER-negative) and from MCF7 (ER-positive) cells and subjected to ChIP assays. The polymerase chain reaction (PCR) products were analyzed on 2% agarose gels. Input indicates a 5% portion of the ChIP input. All the experiments were performed in triplicate with similar results. C) MCF7 cells were hormone starved for 5 days and then stimulated by adding estradiol (E2) at a concentration of 10 nM. Twenty-four hours after stimulation, cells were cross-linked and subjected to ChIP assays. D) The amount of ChIP-enriched DNA was quantified by quantitative real-time PCR (qRT-PCR), and the results were shown as relative enrichment. The data are presented as means of three experiments. E) MCF7 were transfected with small interfering RNA (siRNA) against ERα or control siRNA. Seventy-two hours after transfection, siRNA-treated cells were cross-linked and were subjected to ChIP assay. F) The amount of ChIP-enriched DNA was quantified by qRT-PCR, and the results were shown as relative enrichment. The data represent mean of three experiments. G) MCF7 and MDA-MB-231 cells were grown to 70% confluence and subjected to ChIP analyses to analyze histone H3 and H4 acetylation status. The experiment was performed in duplicate with the same results. H) Estradiol-stimulated MCF7 cells were subjected to ChIP assays to check the acetylation status of miR-221-222 promoter. The experiment was performed in duplicate with the same results. I) The minus 200/ERE-pGL3b and minus 200 pGL3b constructs were transfected into MCF7 and MDA-MD-231 cells; 24 hours after transfection, luciferase assays were performed and results represented as relative luciferase activity. J) Luciferase activity of minus 200/ERE in ER-positive cells, MCF7, after ERα knockdown. K) Luciferase activity of minus 200/ERE in ER-negative cells, MDA-MB-231, after ERα restoration. All luciferase experiments were performed in triplicate, and the luciferase activity was read in triplicate. Error bars = 95% confidence intervals. AcH3 = acetyl histone H3; AcH4 = acetyl histone H4; ERE = estrogen response element; IgG = immunoglobulin control; nt = nucleotides.
Next, we showed that in hormone-starved MCF7 cells, neither ERα nor corepressors are recruited to miR-221-222 (Figure 6, C), consistent with the increase of miR-221 and -222 expression after hormone starvation (Supplementary Figure 9, A, available online). In contrast, following E2 stimulation, all three proteins localized to the miR-221-222 promoter. Analyses by qRT-PCR showed a strong enrichment of ERα, NCoR, and SMRT (mean of enrichment level, relative unit: ERα0h: 0.1, 95% CI = 0.08 to 0.015; NCoR0h: 0.02, 95% CI = 0.015 to 0.025; SMRT0h: 0.0091, 95% CI = 0.008 to 0.015; ERα24h: 5, 95% CI = 4.56 to 5.8; NCoR24h: 2.0, 95% CI = 1.87 to 2.5; SMRT24h: 2.5, 95% CI = 2.37 to 2.8. All Ps of difference between the samples and the control are <.05) (Figure 6, D), whereas miR-221 and -222 expression was suppressed (Figure 4, E). Silencing of ERα in MCF7 cells resulted in loss of the corepressors from the miR-221-222 promoter (Figure 6, E), confirmed by qRT-PCR analysis (mean of enrichment level, relative unit: ERαsiControl: 4.89, 95% CI = 4.58 to 5.1; NCoRsiControl: 1.9, 95% CI = 1.5 to 2.5; SMRTsiControl: 0.0091, 95% CI = 0.008 to 0.015; ERαsiER: 0.002, 95% CI = 0.0015 to 0.0024; NCoRsiER: 0.02, 95% CI = 0.0187 to 0.025; SMRTsiER: 0.01, 95% CI = 0.0089 to 0.015. All Ps of difference between the samples and the control are <.05) (Figure 6, F). Furthermore, compared with ERα silencing, knockdown of SMRT and NCoR induced a partial activation of miR-221 and -222 (Supplementary Figure 9, B, available online). Because both SMRT and NCoR complexes contain histone deacetylases (36), ChIP analyses showed a strong difference in the acetylation status of histones in the promoter region of miR-221-222 between ER-positive and ER-negative cells (Figure 6, G). Furthermore, E2-mediated recruitment of Erα, NCoR, and SMRT to the miR-221-222 promoter substantially reduced its acetylation status (Figure 6, H).
Finally, because the previously analyzed miR-221-222 promoter vectors (Figure 5, D) did not show any sensitivity to E2 (data not shown), the ERα-enriched ChIP region 2, containing a predicted ERE, was cloned into the promoterless pGL3basic vector, minus 200/ERE-pGL3b.
Both vectors, minus 200/ERE-pGL3b and minus 200 pGL3b, were transfected into ER-positive and ER-negative cells, but a statistically significant increase in luciferase activity was observed only for minus 200/ERE–GL3b in ER-negative cells (Figure 6, I) (mean activity, relative luciferase units: for MCF7: minus 200: 15.8, 95% CI = 15 to 16.5; minus 200/ERE: 10.9, 95% CI = 10.3 to 11; for MDA-MB-231: minus 200: 15.96, 95% CI = 15.4 to 16; minus 200/ERE: 15.2, 95% CI = 15 to 15.7. All Ps of difference between the samples and the control <.05). Moreover, knockdown of ERα in ER-positive cells increased luciferase activity of minus 200/ERE-pGL3b (mean activity, relative luciferase units: minus 200/ERE-pGL3b in ER-positive cells, MCF7, after knockdown of ERα; Scr: 1.6, 95% CI = 1.4 to 1.73; siER: 2.3, 95% CI = 2.23 to 2.67. P = .008) (Figure 6, J), whereas the restoration of ERα in ER-negative cells induced a statistically significant reduction in activity (mean activity, relative luciferase units: minus 200/ERE-pGL3b in ER-negative cells, MDA-MB-231, after restoration of ERα: empty: 3.2, 95% CI = 3.1 to 3.6; ER-HA: 2.3, 95% CI = 2.2 to 2.4. P = .004) (Figure 6, K). These results lead us to conclude that ERα recruits NCoR and SMRT to suppress miR-221-222 expression.
Discussion
We showed that overexpression of miR-221 and -222 in ER-positive cells induces a global change in gene expression that differs from the miR-206 signature and may account for the generation of a more invasive and deadly tumor phenotype. Furthermore, we demonstrated that ERα directly represses miR-221 and -222 by recruiting the corepressors NCoR and SMRT. Whether or not miR-206 blocks proliferation and suppresses oncogenic signaling in ER-positive cells, miR-221 and -222 may trigger a malignant transformation that confers ability to survive apoptotic pressure under anchorage-independent conditions (decrease in CAV1 and CAV2 expression) and induce an increase in cell cycle progression (decrease in PTEN, CDKN1B, and BIM expression) and tumor invasion and metastasis (increase in BMP4, BMP7, and TGFbeta3 expression). Gene expression profiles enabled us to identify the tumor-suppressor FOXO3 and the oncogenic MET protein as new players involved in the effects of miR-221-222 and miR-206, respectively.
The FOXO3 transcription factor, one of the members of the large forkhead family (37), activates multiple target genes involved in tumor suppression, such as BIM (21) and CDKN1B (20), which are targets of miR-221 and -222 (17,24). We demonstrated that miR-221 and -222 induce not only posttranscriptional repression of BIM and CDKN1B but also transcriptional repression of their activator FOXO3. These findings provide evidence that a single miRNA, through its ability to modulate different genes involved in the same pathway, may act as a strong inhibitor of the entire cellular pathway, suggesting a possible greater therapeutic potential for miRNAs than for single gene–directed drugs.
Conversely, we showed that miR-206 causes an overall decrease in cell proliferation in ER-positive cells. Two different aspects of this tumor-suppressor activity were explored in our study. First, we showed that miR-206 suppresses the oncogenic MET receptor, which is highly overexpressed in many solid tumors (38) and associated with basal and ERBB2-positive subtypes of breast cancer (28). Moreover, we found that FOXO3 and its direct transcriptional targets, BIM and CDKN1B, are strongly stimulated, albeit indirectly, by miR-206.
Lack of in vivo confirmation of our molecular pathway in breast cancer is a limitation of this study. However, the roles of miR-221 and -222 in lung (39) and liver (39,40) cancers in vivo have been recently highlighted. More studies will be required to elucidate all of the aspects of the contrasting effects of the miR-221-222 cluster and miR-206 in ER-positive cells.
This work demonstrates that ERα reduces expression of miR-221 and -222. We found that ERα is bound to miR-221-222 and is responsible for the recruitment of the corepressor proteins NCoR and SMRT. Other examples of corepressor recruitment have also been recently reported, for example, transcriptional repression of the cyclin G2 (CCNG2) gene (41) and the vascular endothelial growth factor receptor 2 (VEGFR2) gene (42). It is intriguing that both studies showed an involvement by the SP1 transcription factor in the recruitment of corepressor proteins at CCNG2 and VEGFR2. A GC-rich stretch, a favorable site for specificity protein binding, was also found in the region of the miR-221-222 promoter. Further studies will be necessary to determine whether these proteins are also involved in the transcriptional repression of miR-221-222 mediated by ERα.
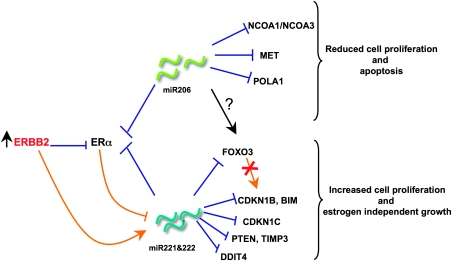
Overall, our results suggest a negative transcriptional regulatory loop in which miR-221 and -222 target ERα, which, in turn, represses miR-221 and -222 expression (Figure 7). Overexpression of miR-221-222 suppresses the expression of ERα at the posttranscriptional level, conferring estrogen-independent growth. It also suppresses the expression of different tumor suppressors, such as CDKN1B, BIM, CDKN1C, PTEN, TIMP3, DNA damage–inducible transcript 4, and FOXO3, promoting high proliferation. Repression of FOXO3 by miR-221-222, in turn, blocks transcriptional activation of CDKN1B and BIM, creating a double suppressive effect on these two genes. Like miR-221-222, miR-206 suppresses the expression of ERα and two of its coactivators, nuclear receptor coactivator (NCOA) 1 (also known as SRC1) and NCOA3 (also known as SRC3) (43), indicating that miR-206 strongly reduces estrogenic response through targeting of multiple proteins. Interestingly, inhibition of the oncogenic tyrosine-kinase receptor MET and activation of the FOXO3 transcriptional network could partially explain the inhibitory effect on proliferation of miR-206 in ER-positive cells. Finally, aberrant activation of the ERBB receptor signaling network, by inducing a strong repression of ERα, activates miR-221-222, which, in turn, represses ERα signaling and the network of tumor suppressors mentioned above (Figure 7). Further studies will be necessary to identify the mediator of the miR-206 effect on FOXO3.
Figure 7.
Effects of miR-206, -221, and -222 on oncogenic activity in breast cancer cells. Schematic representation of the differential effects of miR-221-222 and miR-206 on estrogen receptor (ER)-positive cells. The red cross indicates double suppressive effect of forkhead box O3 (FOXO3) on cyclin-dependent kinase inhibitor 1B (CDKN1B) and BIM (bcl2 interacting mediator of cell death). Orange lines indicate transcriptional regulation; blue lines indicate posttranscriptional regulation. DDIT4 = DNA damage–inducible transcript 4.
We showed that continuous activation of ERBB2, frequently observed in ER-negative tumors (32) and also associated with miR-221 and -222 overexpression (12), perturbs the miR-221-222/ERα regulatory loop by inducing miR-221 and -222 expression. This increased expression of miR-221-222 may confer a proliferation advantage to cancer cells and subsequent resistance to therapeutic agents by repressing the expression of not only ERα but also that of proteins like CDKN1B, CDKN1C (44), BIM, FOXO3, CAV1, CAV2, PTEN, and progesterone receptor (11,12,45). The molecular circuitry composed of miR-221-222 and ERα may provide a basis for understanding how activation of miRNA expression can induce ER-positive breast tumors to become ER negative, a frequent observation with profound impact on clinical outcomes in recurrent breast cancer.
Funding
National Institutes of Health (R01 CA115965 and R01 CA124541 to C.M.C.).
Supplementary Material
Footnotes
G. Di Leva and P. Gasparini contributed equally to this work.
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
The authors want to acknowledge Dr Elda Tagliabue for providing the previously characterized MCF-ERBB2 and MCF7-Mock cell lines (33).
References
- 1.Elledge RM, Allred DC. Clinical aspect of estrogen and progesterone receptors. In: Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Disease of the Breast. Philadelphia, PA: Lippincott Williams and Wilkins; 2004. pp. 601–617. [Google Scholar]
- 2.Allred DC, Brown P, Medina D. The origin of estrogen receptor alpha-positive and estrogen receptor alpha-negative human breast cancer. Breast Cancer Res. 2004;6(6):240–245. doi: 10.1186/bcr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santen RJ, Song RX, McPherson R, et al. The role of mitogen-activated protein (MAP) kinase in breast cancer. J Steroid Biochem Mol Biol. 2002;80(2):239–256. doi: 10.1016/s0960-0760(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 4.Stoner M, Saville B, Wormke M, et al. Hypoxia induces proteasome-dependent degradation of estrogen receptor alpha in ZR-75 breast cancer cells. Mol Endocrinol. 2002;16(10):2231–2242. doi: 10.1210/me.2001-0347. [DOI] [PubMed] [Google Scholar]
- 5.Oh AS, Lorant LA, Holloway JN, et al. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol. 2001;15(8):1344–1359. doi: 10.1210/mend.15.8.0678. [DOI] [PubMed] [Google Scholar]
- 6.Yan L, Yang X, Davidson NE. Role of DNA methylation and histone acetylation in steroid receptor expression in breast cancer. J Mammary Gland Biol Neoplasia. 2001;6(2):183–192. doi: 10.1023/a:1011308707512. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Veronese A, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 11.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple to multiple testing. J R Stat Soc Ser B. 1995;57(2):289–300. [Google Scholar]
- 14.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97(18):10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo N, Toyama T, Sugiura H, et al. miR-206 Expression is downregulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68(13):5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 17.Le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SW, Reimer CL, Oh P, et al. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene. 1998;16(11):1391–1397. doi: 10.1038/sj.onc.1201661. [DOI] [PubMed] [Google Scholar]
- 19.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6(3):184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 20.Musgrove EA, Davidson EA, Ormandy CJ. Role of the CDK inhibitor p27 (Kip1) in mammary development and carcinogenesis: insights from knockout mice. J Mammary Gland Biol Neoplasia. 2004;9(1):55–66. doi: 10.1023/B:JOMG.0000023588.55733.84. [DOI] [PubMed] [Google Scholar]
- 21.Kang MH, Reynolds CP. Bcl2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15(4):1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mak BC, Yeung RS. The tuberous sclerosis complex genes in tumor development. Cancer Invest. 2004;22(4):588–603. doi: 10.1081/cnv-200027144. [DOI] [PubMed] [Google Scholar]
- 23.Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15(3):752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terasaka K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J. 2009;276(12):3269–3276. doi: 10.1111/j.1742-4658.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 25.Alarmo EL, Kuukasjarvi T, Karhu R, Kallioniemi A. A comprehensive expression survey of bone morphogenetic proteins in breast cancer highlights the importance of BMP4 and BMP7. Breast Cancer Res Treat. 2007;103(2):239–246. doi: 10.1007/s10549-006-9362-1. [DOI] [PubMed] [Google Scholar]
- 26.Katsuno Y, Hanyu A, Kanda H, et al. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through SMAD pathway. Oncogene. 2008;27(49):6322–6333. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 27.Kim HK, Lee YS, Sivaprasad U, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174(5):677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graveel CR, DeGroot JD, Su Y, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12909–12914. doi: 10.1073/pnas.0810403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl M, Dijkers PF, Kops GJ, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168(10):5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 30.Chandramohan V, Mineva ND, Burke B, et al. c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem. 2008;104(6):2091–2106. doi: 10.1002/jcb.21765. [DOI] [PubMed] [Google Scholar]
- 31.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66(7):3903–3911. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 32.Sainsbury JR, Farndon JR, Sherbet GV, Harris AL. Epidermal-growth-factor receptors and oestrogen receptors in human breast cancer. Lancet. 1985;1(8425):364–366. doi: 10.1016/s0140-6736(85)91385-6. [DOI] [PubMed] [Google Scholar]
- 33.Giani C, Casalini P, Pupa SM, et al. Increased expression of c-erbB-2 in hormone-dependent breast cancer cells inhibits cell growth and induces differentiation. Oncogene. 1998;17(4):425–432. doi: 10.1038/sj.onc.1201954. [DOI] [PubMed] [Google Scholar]
- 34.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 35.Lonard DM, O’malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27(5):691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Minucci S, Nervi C. Lo Coco F, Pelicci PG. Histone deacetylase: a common molecular target for differentiation treatment of acute myeloid leukemias? Oncogene. 2001;20(24):3110–3115. doi: 10.1038/sj.onc.1204336. [DOI] [PubMed] [Google Scholar]
- 37.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 38.To CT, Tsao MS. The roles of hepatocyte growth factor/scatter factor and met receptor in human cancer. Oncol Rep. 1998;5(5):1013–1024. doi: 10.3892/or.5.5.1013. [DOI] [PubMed] [Google Scholar]
- 39.Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stossi F, Likhite VS, Katzenellenbogen JA, et al. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem. 2006;281(24):16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- 42.Higgins KJ, Liu S, Abdelrahim M, et al. Vascular endothelial growth factor receptor-2 expression is downregulated by 17beta-estradiol in MCF-7 breast cancer cells by estrogen receptor alpha/Sp proteins. Mol Endocrinol. 2008;22(2):388–402. doi: 10.1210/me.2007-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF7 breast cancer cells. Mol Endocrinol. 2009;23(8):1215–1230. doi: 10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 45.Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.