Abstract
Volume and change in volume of the hippocampus are both important markers of Alzheimer's disease (AD). Delineation of the structure on MRI is time-consuming and therefore reliable automated methods are required. We describe an improvement (multiple-atlas propagation and segmentation (MAPS)) to our template library-based segmentation technique. The improved technique uses non-linear registration of the best-matched templates from our manually-segmented library to generate multiple segmentations and combines them using the simultaneous truth and performance level estimation (STAPLE) algorithm. Change in volume over 12 months (MAPS-HBSI) was measured by applying the boundary shift integral using MAPS regions. Methods were developed and validated against manual measures using subsets from Alzheimer's Disease Neuroimaging Initiative (ADNI). The best method was applied to 682 ADNI subjects, at baseline and 12-month follow-up, enabling assessment of volumes and atrophy rates in control, mild cognitive impairment (MCI) and AD groups, and within MCI subgroups classified by subsequent clinical outcome. We compared our measures with those generated by SNT (Surgical Navigation Technologies) available from ADNI. The accuracy of our volumes was one of the highest reported (mean(SD) Jaccard Index 0.80(0.04) (N=30)). Both MAPS baseline volume and MAPS-HBSI atrophy rate distinguished between control, MCI and AD groups. Comparing MCI subgroups (reverters, stable and converters): volumes were lower and rates higher in converters compared with stable and reverter groups (p≤0.03). MAPS-HBSI required the lowest sample sizes (68 subjects) for a hypothetical trial. In conclusion, the MAPS and MAPS-HBSI methods give accurate and reliable volumes and atrophy rates across the clinical spectrum from healthy aging to AD.
Introduction
A diagnosis of Alzheimer's disease (AD), the most common cause of dementia, can only be confirmed pathologically by the presence of intracellular neurofibrillary tangles made of tau protein and extracellular amyloid plaques. The hippocampus is affected early in the disease (Braak and Braak, 1991) and hippocampal atrophy using magnetic resonance imaging (MRI) has been shown to be a marker of AD pathology (Likeman et al., 2005). Hippocampal atrophy is also predictive of clinical decline at a mild cognitive impairment (MCI) stage (Henneman et al., 2009; Jack et al., 1999) and even presymptomatically in familial AD (Fox et al., 1996; Ridha et al., 2006). As a result, reduced hippocampal volume using MRI has recently been proposed as part of new criteria to allow a diagnosis of AD to be made earlier than would be possible on purely clinical grounds (Dubois et al., 2007).
Not only is there interest in single time-point assessment of hippocampal integrity using structural imaging, but there is also interest in measuring volume change over time. Significantly increased hippocampal atrophy rates have been shown by many studies in subjects with AD (Barnes et al., 2008b; Henneman et al., 2009; Jack et al., 2000; Jack et al., 2004; Thompson et al., 2004) and MCI (Henneman et al., 2009; Jack, Jr. et al., 2005; Schuff et al., 2009) compared with control subjects of a similar age. Atrophy rates have been shown to increase gradually early in the course of both familial (Ridha et al., 2006) and sporadic AD (Jack, Jr. et al., 2008b) and to be predictive of future decline from MCI to AD (Henneman et al., 2009). Hippocampal rates of atrophy have been used to assess putative disease-modifying treatments for AD (Fox et al., 2005; Hashimoto et al., 2005; Jack et al., 2003).
However, the hippocampus is a complex anatomical structure and manual segmentation, even with some degree of computer assistance, requires around 45 minutes per hippocampus by trained raters in order to achieve reasonable reproducibility (e.g. less than 5% of difference in volume both within and between raters) (Fox et al., 1996). Consequently, many attempts have been made to automate or reduce manual involvement in the segmentation process. These techniques include using deformable models (Ashton et al., 1997; Chupin et al., 2009b; Duchesne et al., 2002; Kelemen et al., 1999; Patenaude et al., 2007; Pitiot et al., 2004; Shen et al., 2002) or voxel classification (Fischl et al., 2002; Gosche et al., 2001). These techniques are usually combined with anatomical and probabilistic priors to aid segmentations. Most of the deformable model techniques are based on statistical shape models to constrain label generation (Duchesne et al., 2002; Kelemen et al., 1999; Patenaude et al., 2007; Shen et al., 2002) whereas others employ anatomical priors and competitive deformation of neighbouring structures to segment the structure (Ashton et al., 1997; Chupin et al., 2009b). Other techniques utilize some form of registration and region propagation with most using nonlinear (Aljabar et al., 2009; Carmichael et al., 2005; Collins et al., 1996; Schuff et al., 2009) rather than linear (Barnes et al., 2008a; Webb et al., 1999) registration. A hybrid technique combining the voxel classification and region propagation was also proposed and shown to improve the results from either method (Collins et al., 1999).
Techniques which utilise atlases or templates vary between making a probabilistic atlas from a set of images (Hammers et al., 2003; Shattuck et al., 2008) to using a single subject template (Haller et al., 1997). The main drawback with the use of single subject templates is that they cannot encompass the very wide inter-individual variability (Figure 1) which will be present within the study. This can be partially circumvented by deforming the individual template to the average shape of all images in the study (Kochunov et al., 2002), but no single template could be adequately warped to all potential anatomical variations. Average templates or atlases built from multiple subjects include the necessary variability but do not necessarily preserve the anatomical resolution required for small structures such as the hippocampus. Using an average of all labelled subjects also typically means that individual subjects in the study will be poorly matched (in terms of anatomy and/or acquisition properties) to some subjects. By selecting one or more templates from a library of labelled images (multi-atlas selection or fusion) (Aljabar et al., 2009; Barnes et al., 2008a; Klein et al., 2008) it is possible to include variability without loss of resolution or quality of matching. The disease status of subjects used in the atlas system or training dataset may affect results obtained on a different dataset; most studies have atlas systems based on normal controls (Fischl et al., 2002; Hammers et al., 2002; Hammers et al., 2007; van der Lijn et al., 2008; Webb et al., 1999) while few include both normal and specific patient groups (Barnes et al., 2008a). Furthermore, a recent publication by Wolz et al. (2010) addressed this problem by propagating the initial set of atlases of normal controls to all images in the dataset (containing normal controls, MCI and AD subjects) through a succession of multi-atlas segmentation steps – effectively breaking down the problem of registering “dissimilar” images into a problem of registering a series of relatively “similar” images (Wolz et al., 2010).
Figure 1.
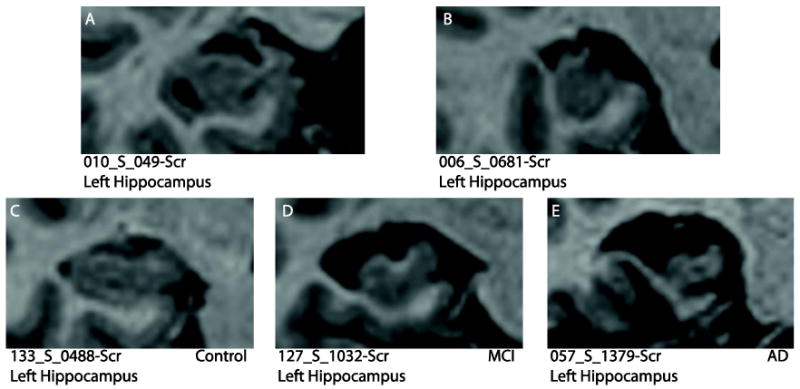
These examples illustrate the wide range of morphological variation in hippocampi from subjects in the Alzheimer's Disease Neuroimaging Initiative database (http://www.loni.ucla.edu/ADNI/). (A) A large hippocampal cyst and lack of temporal horn and (B) malrotation of the hippocampus (tall and narrow). Atrophy causing changes from (C) normal hippocampus to (D) MCI hippocampus (considerable atrophy) and (E) AD hippocampus (marked atrophy).
Very few fully-automated systems of measuring hippocampal change have been generated: most have some level of intervention from manually segmenting baseline hippocampi and using fluid registration (Crum et al., 2001) or linear registration combined with boundary shift integral (BSI) (Barnes et al., 2004) to measure change directly within the region. Other methods include application of the cross-sectional technique to baseline and repeat images separately to measure change indirectly (Schuff et al., 2009; Wang et al., 2003).
In our previously published multi-atlas single-site study we described a leave-one-out experiment where for each individual we found the best match from all other subjects in the study based on the similarity of images in the hippocampal area (Barnes et al., 2008a). This best match was then used as a single-person template together with linear registration, morphological operations and intensity thresholding. This technique was able to generate single time-point hippocampal regions of sufficient accuracy to generate relative rates of atrophy using serial images. In this study, we select top matches from our multi-atlas system to generate multiple segmentations (Aljabar et al., 2009) and combine them using label fusion methods (Heckemann et al., 2006; Rohlfing and Maurer, Jr., 2007; Warfield et al., 2004). For brevity, we refer to the technique as multiple-atlas propagation and segmentation (MAPS). We evaluate MAPS on multi-site data of over 680 subjects with serial volumetric MRI from the Alzheimer's Disease Neuroimaging Initiative (ADNI, http://www.loni.ucla.edu/ADNI/). Our aim was first to determine the ability of MAPS to distinguish between normal controls, MCI and AD subjects; and between subgroups of subjects diagnosed as MCI at baseline that were subsequently diagnosed as normal (“reverters”), MCI (“stable”) or AD (“converters”). We also wished to assess its ability to track change in the hippocampus in controls, MCI and AD subjects, and to estimate sample sizes that would be needed in a putative disease-modifying clinical trial.
Methods
Overview
We first trained the segmentation algorithm on the left hippocampi of a subset of 15 manually labelled images, to optimise the various methodological options and parameters. Segmentation accuracy was then directly measured on the left hippocampi of an independent test set of 30 further manually labelled images. We then indirectly evaluated performance on a much larger set of 682 (unlabelled) images, using metrics such as sample size for a hypothetical clinical trial. We finally compared directly estimated MAPS-HBSI atrophy rates to indirect rates from differences in volumes from applying MAPS to the two time-points.
Image data
We downloaded pre-processed baseline and 12-month repeat volumetric T1-weighted MR scans acquired using 1.5T scanners (General Electric Healthcare, Philips Medical Systems or Siemens Medical Solutions) at multiple sites from the ADNI website. Representative imaging parameters were TR = 2400ms, TI = 1000ms, TE = 3.5ms, flip angle = 8°, field of view = 240 × 240mm and 160 sagittal 1.2mm-thick-slices and a 192 × 192 matrix yielding a voxel resolution of 1.25 × 1.25 × 1.2 mm3, or 180 sagittal 1.2mm-thick-slices with a 256 × 256 matrix yielding a voxel resolution of 0.94 × 0.94 × 1.2 mm3. The details of the ADNI MR imaging protocol are described in (Jack, Jr. et al., 2008a), and listed on the ADNI website (http://www.loni.ucla.edu/ADNI/Research/Cores/). The T1-weighted volumetric scans that were designated to be the “best” after quality control were processed using the standard ADNI image processing pipeline, which included post-acquisition correction of gradient warping (Jovicich et al., 2006), B1 non-uniformity correction (Narayana et al., 1998) depending on the scanner and coil type, intensity non-uniformity correction (Sled et al., 1998) and phantom based scaling correction (Gunter et al., 2006) - the geometric phantom scan having been acquired with each patient scan.
Clinical and demographic data are shown in Tables 1-4. Table 1 shows the demographic data of the 15 randomly-selected subjects (5 control, 5 MCI and 5 AD) used for method optimisation. Table 2 shows the demographic data of the 30 randomly-selected subjects (10 control, 10 MCI and 10 AD) used for method validation (note that this subset of 30 subjects was separate from the subset of 15 subjects used for method optimisation). Table 3 shows the demographic data of 682 subjects (200 control, 335 MCI and 147 AD) in our full dataset. We also sub-divided the MCI subjects into three subgroups (8 reverters, 204 stable and 123 converters) based on their follow-up clinical diagnoses determined up to 36 months after baseline. Table 4 shows the demographic data of the MCI subgroups.
Table 1.
Subject demographics in subset of 15 randomly selected subjects used to establish optimal methods and parameters. Mean (SD) unless specified otherwise.
| Control (n=5) | MCI (n=5) | AD (n=5) | |
|---|---|---|---|
| Age, years | 82.7 (4.3) | 76.6 (7.1) | 75.6 (7.2) |
| Gender male (%) | 2 (40%) | 1 (20%) | 3 (60%) |
| Scanning interval, days | 380.8 (20.5) | 379.8 (17.4) | 385.4 (12.2) |
| MMSE, /30 | 29.8 (0.4) | 27.4 (1.7) | 23.2 (1.8) |
Table 4.
Subject demographics of the MCI subgroups. Mean (SD) unless specified otherwise.
| MCI reverters (n=8) |
MCI stable (n=204) |
MCI converters (n=123) |
|
|---|---|---|---|
| Age, years | 70.7 (8.9) | 75.3 (7.1) | 74.5 (7.2) |
| Gender male (%) | 5 (63%) | 133 (65%) | 75 (61%) |
| Scanning interval, days | 396.3 (46.0) | 396.3 (24.3) | 390.1 (22.6) |
| MMSE, /30 | 28.0 (1.3) | 27.3 (1.8) | 26.6 (1.7) |
| Mean years of clinical follow-up | 1.8 (0.5) | 1.8 (0.7) | 2.1 (0.6) |
| TIV, ml | 1584 (144) | 1567 (149) | 1554 (154) |
Table 2.
Subject demographics in subset of 30 randomly selected subjects used for method validation. Mean (SD) unless specified otherwise.
| Control (n=10) | MCI (n=10) | AD (n=10) | |
|---|---|---|---|
| Age, years | 78.6 (5.4) | 75.3 (8.8) | 77.2 (6.8) |
| Gender male (%) |
6 (60%) | 7 (70%) | 7 (70%) |
| MMSE, /30 | 29.5 (0.7) | 27.4 (1.8) | 27.0 (2.7) |
Table 3.
Subject demographics of the full dataset. Mean (SD) unless specified otherwise.
| Control (n=200) | MCI (n=335) | AD (n=147) | |
|---|---|---|---|
| Age, years | 76.0 (5.1) | 74.9 (7.2) | 75.3 (7.3) |
| Gender male (%) | 106 (53%) | 213 (64%) | 78 (53%) |
| Scanning interval, days | 396.3 (25.8) | 394.0 (24.5) | 392.6 (23.3) |
| MMSE, /30 | 29.1 (1.0) | 27.0 (1.8) | 23.4 (1.9) |
| Mean years of clinical follow-up | 2.2 (0.6) | 1.9 (0.7) | 1.8 (0.4) |
| TIV, ml | 1538 (142) | 1563 (150) | 1536 (168) |
ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a 5-year public-private partnership. The aims of ADNI included assessing the ability of imaging and other biomarkers to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD).
The Principal Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California - San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research -- approximately 200 cognitively normal older individuals, 400 people with MCI, and 200 people with early AD. For up-to-date information see www.adni-info.org.
Template library creation
We used a previously-described hippocampal template library of manually-segmented regions from 55 subjects scanned at a single site 1.5T GE scanner using a volumetric T1-weighted acquisition (Barnes et al., 2008a). The left and right hippocampal regions were manually segmented by an expert segmentor S1. A detailed description of the template library creation is included in the Appendix.
Method optimisation using a manually-segmented subset of 15 subjects
Based on the same manual segmentation protocol as in the template library creation, the expert segmentor S1 manually delineated the left hippocampus on the baseline and repeat T1-weighted MR images of 15 randomly-selected subjects (5 AD, 5 MCI, 5 controls; details in Table 1) from the ADNI dataset, in order to assess which methods and parameters provided the most accurate and reliable segmentation using the baseline left hippocampi (cross-sectional analysis). In the longitudinal analysis, we assessed any differences between automated longitudinal changes in volume calculated by the boundary shift integral (BSI) (Freeborough and Fox, 1997) using automated baseline regions and two measures of changes: i) BSI using manual baseline regions and ii) manual rates of atrophy derived from the volume difference of manual segmentations at both time-points. Furthermore, we assessed separately, in both baseline and repeat images, the intra-rater and inter-rater variability of manual hippocampal segmentation in 15 subjects.
Cross-sectional analysis
Each subject's baseline scan and its flipped mirror image (along the mid-sagittal plane) were registered to the control to which all the template library scans were registered (12 dof brain to brain followed by 6 dof hippocampus to hippocampus). This flipping effectively doubled the size of the template library by allowing, for example, the left hippocampus in the ADNI subject to be matched to the right hippocampus in the template library. Best matches for each hippocampus were ranked as to their similarity over the corresponding dilated hippocampal regions using the cross-correlation (R2) between the ADNI subject images (flipped and unflipped) and the template library. Cross correlation has been shown to provide a good criterion for template selection in the hippocampal region in multi-centre imaging data (Aljabar et al., 2009). Once a rank of best to worst matches for each hippocampus was established, a subset of the highest ranking matchers could be used to propagate the undilated hippocampal regions onto the subset of images to be segmented.
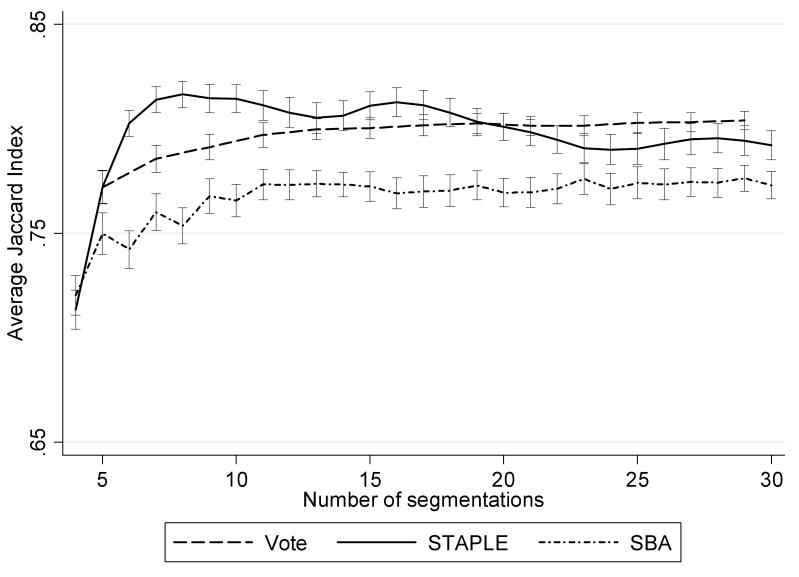
The process of optimising the method and parameters for the automated hippocampal segmentation is depicted in Figure 2. At each stage we found the method and parameters which produced the most accurate region as compared with the manual region determined by the mean Jaccard index (JI) (Jaccard, 1907), which was defined as JI(A, B) = | A ∩ B | / | A ∪ B |, where A is the set of voxels in the automated region and B is the set of voxels in the manual region. We began by assessing the registration algorithm and compared linear 12 dof registration (Woods et al., 1998a) with non-linear registration based on free form deformation (FFD) (Rueckert et al., 1999), in which multiple control point spacings (16mm→8mm→4mm) were used in order to model increasingly local deformations. Since intensity thresholds were used in the semi-automated hippocampal segmentation to exclude white matter and CSF (Barnes et al., 2008a), the regions from the better registration algorithm were identically thresholded to assess if this improved the segmentation accuracy. We then compared the combination of segmentations from the top 4 to the top 30 matches using the “vote rule” (Heckemann et al., 2006), simultaneous truth and performance level estimation (STAPLE) (Warfield et al., 2004) and shape-based average (SBA) (Rohlfing and Maurer, Jr., 2007) to determine the optimal number of matches. Furthermore, we assessed whether the use of a Markov random field (MRF) model in STAPLE to incorporate spatial smoothness would further improve the segmentation accuracy. We tested the interaction strength parameter between neighbouring voxels from 0 to 0.5 using increments of 0.1. Note that we followed Heckemann et al. (2006) and only used odd numbers of segmentations in the “vote rule”.
Figure 2.
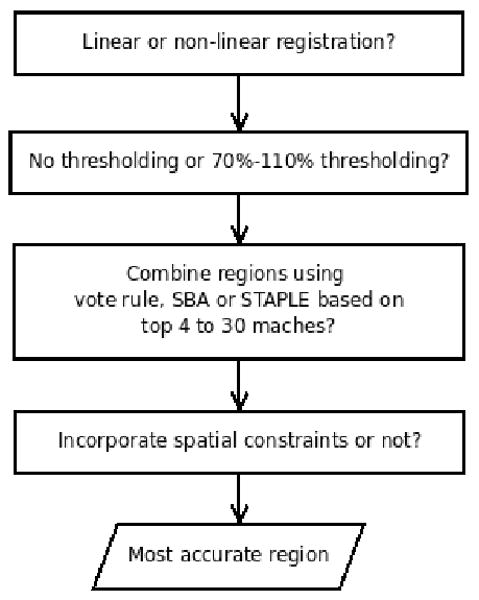
Flow chart showing how the best methods and parameters for the automated hippocampal segmentation are selected.
Longitudinal analysis
Using the most accurate baseline hippocampal region, we then assessed whether using local registration (Woods et al., 1998a) of serial hippocampi combined with BSI (Freeborough and Fox, 1997) produced reasonable change in volume measures in the subset. Hippocampal BSI (HBSI) was calculated using a double intensity window approach (Hobbs et al., 2009) (see Appendix for details), in order to capture changes across both the CSF-hippocampal border and the white matter-hippocampal border. Since the images were acquired from different scanner manufacturers and models in ADNI, we computed the intensity windows for the CSF-hippocampal border and the white matter-hippocampal border for each image pair, and used them to compute HBSI of that image pair. We referred to this method of assessing longitudinal change in hippocampal volume using MAPS and HBSI as MAPS-HBSI. We compared MAPS-HBSI against a HBSI generated using the manual baseline region (manualHBSI), and a completely manual atrophy rate (difference of manual hippocampal volumes at baseline and repeat) in the subset of 15 subjects using only the left hippocampus.
Intra-rater variability of manual hippocampal segmentation
The same baseline and repeat images from the 15 subjects were segmented manually again by the same expert segmentor S1 following an interval of more than two months to allow assessment of manual intra-rater reliability in both volumes and change in volumes. To assess reliability of the HBSI measure, a second manualHBSI was calculated using the second manually-segmented baseline region.
Inter-rater variability of manual hippocampal segmentation
Based on the manual segmentation protocol in the template library creation, another expert segmentor S2 manually delineated the left hippocampus on the same baseline and repeat images from the 15 subjects, in order to assess manual inter-rater reliability in both volumes and change in volumes.
Statistical methods
All analyses were performed using STATA (version 10). To examine whether the average magnitude and variability of rates differed between the manual and automated methods, we calculated the differences in mean and ratios of standard deviation (SD) of volumes and atrophy rates between manual and automated measurements, according to subject group. Confidence intervals were found for the mean differences assuming normality of the paired differences, and for the ratio of SDs using Pitman's method (Pitman, 1939).
To assess intra-rater reliability, intra-class correlations (ICCs) and JIs for pairs of manually-segmented volumes delineated by the expert segmentor S1 were calculated, ignoring subject group, at baseline and repeat. In addition an intra-rater ICC was calculated for the difference in volumes over time generated from the first segmentation of the 15 pairs delineated by S1 compared with the difference in volumes over time generated from the second segmentation of the 15 pairs delineated by S1. Similarly an intra-rater ICC was calculated for the two HBSIs generated from the first and second baseline mask delineated by S1. Confidence intervals were found using the iccconf command in STATA.
To assess inter-rater reliability, intra-class correlations (ICCs) and JIs for pairs of manually-segmented volumes delineated by the expert segmentors S1 and S2 were calculated, ignoring subject group, at baseline and repeat. An inter-rater ICC was calculated for the difference in volumes over time generated from the first segmentation of the 15 pairs delineated by S1 compared with the difference in volumes over time generated from the second segmentation of the 15 pairs delineated by S2. Similarly an inter-rater ICC was calculated for the two HBSIs generated from the baseline masks delineated by S1 and S2. Inter-rater ICCs and 95% confidence intervals were found using the icc23 command in STATA, assuming random rater effects.
Method validation using a manually-segmented subset of 30 subjects
For the method validation, the expert segmentor S1 manually delineated the left hippocampus in the baseline images of another subset of 30 randomly selected subjects in the ADNI database (10 AD, 10 MCI and 10 controls; details in Table 2) that differed from the subset of 15 subjects used for the method optimisation. We applied the optimised methods and parameters as determined above to generate left hippocampal regions in the baseline images for this subset, in order to assess any differences between manual and automated hippocampal regions.
Statistical methods
To examine whether the average magnitude and variability differed between the manual and automated methods, we calculated the differences in mean and ratios of standard deviation (SD) of volumes between manual and automated measurements, according to subject group. Confidence intervals were found for the mean differences assuming normality of the paired differences, and for the ratio of SDs using Pitman's method.
Analysis of the full dataset
We used the optimised methods and parameters as determined above to generate baseline and repeat hippocampal volume from MAPS and rate of change from MAPS-HBSI for each subject in our full dataset (detailed in Table 3 and 4). All baseline hippocampal regions were visually checked for large segmentation errors by the expert segmentor S1. We assessed volumes and changes in subjects by baseline diagnosis (controls, MCI and AD), and by the MCI subgroups (reverters, stable and converters).
We generated a head size measure by estimating total intracranial volume (TIV) from the summation of the volumes of grey matter (GM), white matter (WM) and CSF. Each of these volumes was computed by summing (over voxels) the values of probabilistic tissue segmentations produced using the new segmentation toolbox available in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8), multiplied by the voxel volume in ml. SPM8's new segmentation is an extension of the unified segmentation model (Ashburner and Friston, 2005); importantly, for the purpose of TIV estimation, one of the extensions is the use of tissue (prior) probability maps (TPMs) for non-brain tissue. The version used here (SPM8 rev.3164) has six TPMs: GM, WM, CSF, bone, non-brain soft-tissue, and air. The additional TPMs improve the segmentation of CSF, in contrast to earlier versions of SPM, where this has been observed to be a problem (Shuter et al., 2008).3
Hippocampal volumes at baseline and 12 months as calculated by using a non-linear warping technique from a template aided by the placement of manual landmarks (Haller et al., 1997) were downloaded from the ADNI website4. The technique (referred to as SNT) is commercially available from Medtronic Surgical Navigation Technologies (Louisville, CO) and has been validated in elderly subjects including MCI and AD patients (Hsu et al., 2002), and results using ADNI data have been reported recently (Schuff et al., 2009). Annualised hippocampal atrophy rates calculated by normalising the difference between baseline and 12-month hippocampal volumes by the baseline hippocampal volume and scan interval were referred to as indirect atrophy rates. We compared MAPS to SNT in terms of volumes, indirect atrophy rates, correlations with cognitive scores, and sample size estimates. Finally, we compared the indirect and direct methods of estimating atrophy rates using the automated regions from MAPS, namely MAPS indirect atrophy rate (calculated from the baseline and 12-month hippocampal volumes) and MAPS-HBSI (calculated from the boundary shift integral), in terms of atrophy rates, correlations with cognitive scores and sample size estimates.
Statistical methods
Again, all analyses were performed using STATA (version 10). To examine differences between groups in baseline volume, we fitted a linear regression model, using age, gender, and TIV as covariates in the regression model. We estimated between-group differences in atrophy rates using a similar linear regression model, with adjustment for age and gender. We fitted regression models using data only from MCI subjects to investigate differences across MCI subgroups according to follow-up diagnosis (reverters, stable and converters), for both volumes and atrophy rates, using the same methods to those described above for baseline diagnosis.
We compared the distributions of age, gender, baseline MMSE, and TIV between the subset of subjects for which an SNT measure was available and those subjects for which an SNT measure was not available. In the subset of subjects for which an SNT measure was available (98 controls, 143 MCI and 62 AD), and separately by subject group, we compared the mean and SD of measures using MAPS with those from the SNT method. Specifically, we calculated the mean of the paired differences between the measurements of baseline volume using MAPS compared to SNT, and calculated 95% CIs assuming normality of these paired differences. Pitman's method was used to calculate 95% confidence intervals for the ratio of SDs between the two methods.
We estimated sample sizes required for a hypothetical clinical trial in AD (or MCI) subjects, using either our methods or the SNT method to calculate atrophy rate. The sample sizes were calculated using the following formula: sample size = (u + v)2 × (2σ2)/(Δμ)2, where u = 0.841 to provide 80% power and v = 1.96 to test at the 5% significance level, Δμ is the change in the annualised percentage atrophy between the treatment groups and σ is the SD of rates of atrophy in the treatment and placebo groups (assuming SD is the same in the treatment and placebo groups) (Fox et al., 2000). In addition, sample sizes were reported with or without controlling for normal aging:
Based on AD (or MCI) atrophy rates alone: sample sizes were calculated to detect a 25% reduction in atrophy rates in AD subjects (Schuff et al., 2009). This implied a 100% effective treatment would reduce atrophy to zero.
Controlling for normal aging: this was assumed that the difference in atrophy rates between age-matched controls and AD (or MCI) subjects represents the maximum possible treatment effect. A 25% reduction in disease progression was thus considered to be equal to 25% of this difference rather than 25% of the atrophy rates in AD subjects (Fox et al., 2000).
Confidence intervals for the ratio of sample sizes using the two methods were found using bias-corrected and accelerated (BCa) bootstrap CIs (100,000 bootstrap samples), using STATA's bootstrap command (Efron and Tibshirani, 1993). This procedure created 100,000 samples by sampling subjects (and their data) from the original dataset (with replacement). Since the distribution of the ratio of estimated sample sizes was non-normal, we report whether p<0.05 on the basis of whether the BCa bootstrap CI for the ratio includes the null value of 1.
We used Pearson's correlation coefficient to estimate the correlations between baseline volume (and atrophy rates), measured either with our methods or SNT, and two cognitive test scores likely to be associated with hippocampal function: Auditory Verbal Learning Test score (AVLT) (Rey, 1964) and Logical Memory I scale of the Wechsler Memory scale (Wechsler, 1981) at baseline. For the AVLT we used the total score of trials I-V. We also estimated the correlation between hippocampal atrophy rates and the annualised change in AVLT score, using the baseline and 12-month results. Confidence intervals for the correlation estimates were found using the STATA corrci command. For differences in correlations between the methods we used bootstrapping (100,000 bootstrap samples) to estimate the standard error, and found Wald-type confidence intervals and p-values, assuming normality.
We also compared the MAPS indirect atrophy rates with the MAPS-HBSI rates, using the same statistical analyses as for the comparison with SNT rates.
Results
Method optimisation using a manually-segmented subset of 15 subjects
Table 5 and Figure 3 report the mean (SD) JI for each stage in our baseline left hippocampal region accuracy assessment. We found that the most accurate regions (with a mean JI of 0.83) were generated using non-linear FFD registration, thresholding and combining the top 8 matches using STAPLE with Markov random field smoothing of interaction strength parameter 0.2. Table 6 shows the means (SD) of the manual and automated hippocampal volumes. The mean (SD) of differences in the manual and automated hippocampal volumes by baseline diagnostic group was -56 (126) mm3 (automated > manual) for controls, 57 (78) mm3 (automated < manual) for MCI, and 81 (125) mm3 (automated < manual) for AD subjects. Overall, the mean (SD) of differences in the manual and automated hippocampal volumes was 27 mm3 (120 mm3) (or 1.2% of mean volume) with manual > automated. The largest outlier in terms of difference compared with manual measures is shown in Figure 4. This difference in volume can be largely attributed to the automated region including more of the tail of the hippocampus and more of the medial aspect than was included in the manual segmentations.
Table 5.
Mean (SD) Jaccard index for each stage in the accuracy assessment, separated by groups (control, MCI and AD). Only the Jaccard indices in all three groups (in the “total” column) were used to choose the optimal methods and parameters. Mean (SD) JI of MRF parameters are shown to 3 decimal points to distinguish fine-grained effects.
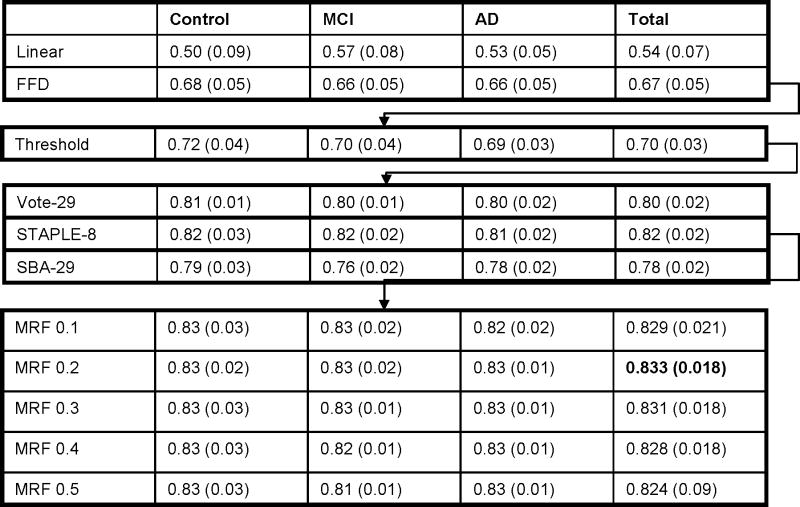 |
Figure 3.
Average Jaccard index of the left hippocampal regions from the baseline images of 15 randomly selected subjects for vote rule, STAPLE and SBA used to assess the optimal number of templates to combine in each method. Error bars denote standard errors.
Table 6.
Mean (SD) of the volumes (in mm3) in the left hippocampus in the baseline images of the subset of 15 subjects used to assess optimal methods and parameters.
| Control (n=5) | MCI (n=5) | AD (n=5) | |
|---|---|---|---|
| Manual | 2525 (529) | 2228 (342) | 1900 (299) |
| Automated | 2581 (625) | 2172 (400) | 1820 (217) |
| manual vs automated | |||
| mean of difference (95% CI) | -56 (-212 to 100) p=0.37 |
57 (-40 to 154) p=0.18 |
81 (-74 to 236) =0.22 |
| SD of differences | 126 | 78 | 125 |
| SD ratio (95% CI) | 0.85 (0.66 to 1.09) p=0.13 |
0.85 (0.66 to 1.10) p=0.15 |
1.38 (0.74 to 2.57) p=0.22 |
Figure 4.
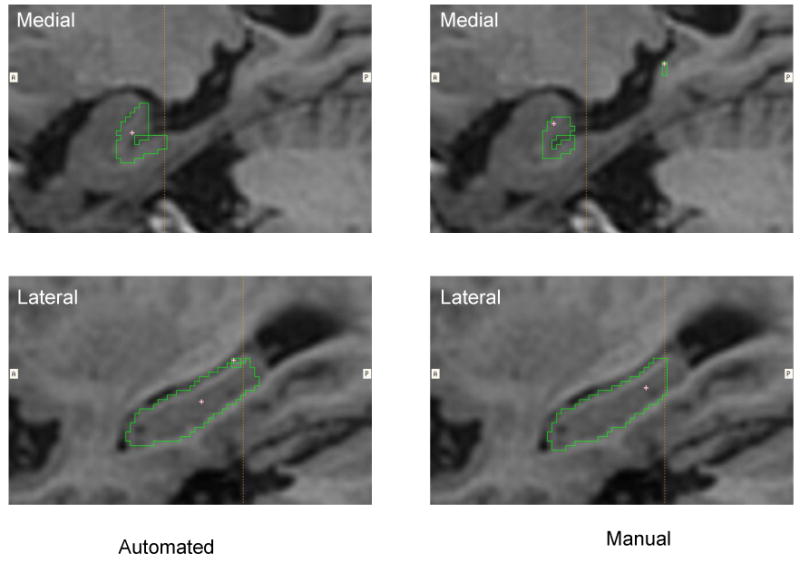
The largest outlier of the automated hippocampal segmentation in the subset of 15 subjects in terms of difference compared with manual measures. This difference in volume can be largely attributed to the automated region including more of the tail of the hippocampus (lower panel) and more of the medial aspect of body and tail (upper panels) than was included in the manual segmentations.
Table 7 shows the mean (SD) rates of atrophy for MAPS-HBSI, manualHBSI and manual methods. The SD of the difference between manualHBSI and manual rates was 3.33 % /year for controls, 1.65 % /year for MCI, and 1.01 % /year for AD subjects. The analogous statistics for the difference between MAPS-HBSI and manual rates was 3.10% /year for controls, 1.50% /year for MCI, and 1.62 % /year for AD subjects.
Table 7.
Mean (SD) annualised percentage rates of atrophy in the left hippocampus in the baseline images of the subset of 15 subjects used to assess optimal methods and parameters.
| Control (n=5) | MCI (n=5) | AD (n=5) | |
|---|---|---|---|
| manual | 1.53 (2.27) | 4.73 (2.02) | 5.44 (1.90) |
| manualHBSI | 1.28 (4.19) | 4.89 (2.81) | 6.97 (1.40) |
| MAPS-HBSI | 1.49 (3.87) | 5.18 (2.80) | 6.48 (1.06) |
| manualHBSI vs manual | |||
| difference in means (95% CI) |
-0.26 (-4.36 to 3.85), p=0.87 |
0.16 (-1.89 to 2.2) p=0.84 |
1.53 (0.27 to 2.79) p=0.03 |
| SD of the difference | 3.33 | 1.65 | 1.01 |
| SD ratio (95% CI) | 1.85 (0.58 to 5.91) p=0.25 |
1.39 (0.55 to 3.51) p=0.39 |
0.76 (0.31 to 1.72) p=0.38 |
| MAPS-HBSI vs manual | |||
| difference in means (95% CI) |
-0.04 (-3.90 to 3.81) p=0.98 |
0.46 (-1.41 to 2.32) p=0.53 |
1.05 (-0.97 to 3.06) p=0.22 |
| SD of the difference | 3.10 | 1.50 | 1.62 |
| SD ratio (95% CI) | 1.71 (0.52 to 5.55) p=0.31 |
1.38 (0.59 to 3.24) p=0.35 |
0.56 (0.16 to 1.91) p=0.30 |
| MAPS-HBSI vs manualHBSI | |||
| difference in means (95% CI) | 0.21 (-0.28 to 0.70) p=0.30 |
0.30 (-0.61 to 1.20) p=0.41 |
-0.48 (-1.94 to 0.97) p=0.41 |
| SD of the difference | 0.40 | 0.73 | 1.17 |
| SD ratio (95% CI) | 0.92 (0.83 to 1.03) p=0.10 |
0.99 (0.63 to 1.57) p=0.97 |
0.76 (0.23 to 2.51) p=0.59 |
The estimated intra-rater ICC (95% CI) of the expert segmentor S1 for the manually segmented baseline hippocampal volumes was 0.993 (0.980 to 0.998), suggesting very high reliability for a population in which there are equal numbers of controls, MCI subjects, and AD subjects. For the repeat volumes the estimated intra-rater ICC was 0.997 (0.992 to 0.999). The mean (SD) JI between the two different manual segmentations by the same segmentor S1 was 0.91 (0.02) for AD, 0.92 (0.01) for MCI, 0.92 (0.02) for controls and 0.92 (0.02) for all three groups. The estimated intra-rater ICC of change in volumes using manual methods was 0.761 (0.444 to 0.910), and for volumes found using manualHBSI was 0.985 (0.957 to 0.995).
The estimated inter-rater ICC (95% CI) between the expert segmentors S1 and S2 for the manually-segmented baseline hippocampal volumes was 0.953 (0.616 to 0.988), suggesting high reliability for a population in which there are equal numbers of controls, MCI subjects, and AD subjects. The mean (SD) JI between the different manual segmentations delineated by the expert segmentors S1 and S2 was 0.86 (0.03) for AD, 0.88 (0.02) for MCI, 0.87 (0.02) for controls and 0.87 (0.03) for all three groups. The estimated inter-rater ICC of change in volumes using manual methods was 0.762 (0.438 to 0.912), and for volumes found using manualHBSI was 0.953 (0.786 to 0.986).
Furthermore, we found that the mean (SD) JIs between the automated left hippocampal regions and manually regions delineated by the expert segmentor S2 in the baseline images to be 0.81 (0.03).
Method validation using a manually-segmented subset of 30 subjects
The mean (SD) JIs of the left hippocampus in the baseline images were 0.80 (0.03) for controls, 0.81 (0.03) for MCI, 0.79 (0.0.05) for AD and 0.80 (0.04) across the 3 groups. Table 8 shows the means (SD) of the of the manual and automated hippocampal volumes. The mean (SD) of differences in the manual and automated hippocampal volumes by baseline diagnostic group were 37 (168) mm3 for controls, 82 (93) mm3 for MCI, and 182 (133) mm3 for AD subjects with automated volumes lower than manual volumes in all the 3 groups. Overall, the mean (SD) of differences in the manual and automated hippocampal volumes was 101 mm3 (144 mm3) (or 4.4% of mean volume) with manual > automated.
Table 8.
Mean (SD) of the volumes (in mm3) in the left hippocampus in the baseline images of the subset of 30 subjects for method validation.
| Control (n=10) | MCI (n=10) | AD (n=10) | |
|---|---|---|---|
| Manual | 2563 (358) | 2331 (410) | 1994 (478) |
| Automated | 2526 (304) | 2249 (371) | 1813 (444) |
| manual vs automated | |||
| mean of difference (95% CI) | 37 (-83 to 157) p=0.25 |
82 (16 to 149) p=0.01 |
182 (87 to 277) p=0.001 |
| SD of differences | 168 | 93 | 133 |
| SD ratio (95% CI) | 1.18 (0.81 to 1.71) p=0.35 |
1.10 (0.93 to 1.31) p=0.23 |
1.08 (0.86 to 1.34) p=0.48 |
Analysis of the full dataset
Visual assessment
No large segmentation errors were found in the automated baseline hippocampal regions. The most common errors were: a) inclusion of extra hippocampal tissue in the temporal lobe (either white or grey matter) or b) exclusion of some hippocampal tissue probably due to noisy images and thresholds being too extreme for that specific image (Figure 5). No manual editing was taken to correct these segmentation errors.
Figure 5.
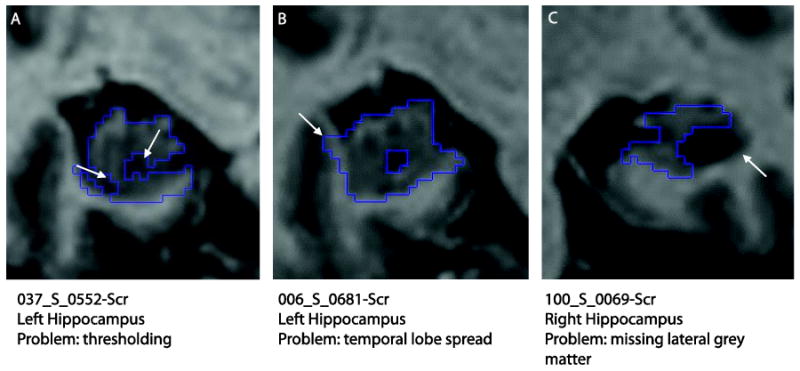
Automated hippocampal segmentation errors. (A) Thresholding excluding hippocampal tissue, (B) extra-hippocampal tissue included (white and grey matter of the temporal lobe) and (C) exclusion of lateral hippocampal grey matter due to large hippocampal cyst and lack of temporal horn.
Comparison between controls, MCI and AD
Table 9 shows the mean (SD) unadjusted total (left + right) hippocampal volumes and atrophy rates in the full dataset, together with adjusted mean differences between groups defined by baseline diagnosis. These are graphically depicted in Figures 6a (volumes) and 6b (rates) using box plots. We have visually assessed the baseline and registered repeat images of the outliers with negative atrophy rates in Figure 6(b), and found that two of them had slightly larger hippocampi in the repeat images and one had motion artifacts. After adjustment for age, gender and TIV, the estimated mean volume (95% CI) in the MCI group was 802mm3 (673 to 932, p<0.001) lower than in the control group, while the adjusted mean in the AD group was 437mm3 (294 to 579, p<0.001) lower than the MCI group. Adjusting for age and gender, the mean atrophy rate in the MCI group was 1.66 percentage points (1.25 to 2.07, p<0.001) greater than in the control group, while the adjusted mean rate in the AD group was 1.68 percentage points (1.23 to 2.13, p<0.001) higher than in the MCI group. The volumes and atrophy rates in the full dataset for the left or right hippocampus can be found in the Table SP1 in the supplementary data.
Table 9.
Mean (SD) unadjusted total (left+right) hippocampal volumes and annualised percentage atrophy rates in whole dataset. Group comparisons show the estimated mean difference in volumes/rates (95% CI), adjusted for TIV (volumes only), age, and gender.
| Control (n=200) | MCI (n=335) | AD (n=147) | MCI vs Control difference in means (95% CI) |
AD vs Control difference in means (95% CI) |
AD vs MCI difference in means (95% CI) |
|
|---|---|---|---|---|---|---|
| Automated total volumes, mm3 | 5251 (659) | 4450 (766) | 3984 (784) | -802 (-932 to -673), p<0.001 | -1239 (-1395 to -1083), p<0.001 | -437 (-579 to -294), p<0.001 |
| MAPS-HBSI atrophy rate, % | 1.10 (1.89) | 2.68 (2.45) | 4.43 (2.59) | 1.66 (1.25 to 2.07), p<0.001 | 3.34 (2.85 to 3.83), p<0.001 | 1.68 (1.23 to 2.13), p<0.001 |
Figure 6.
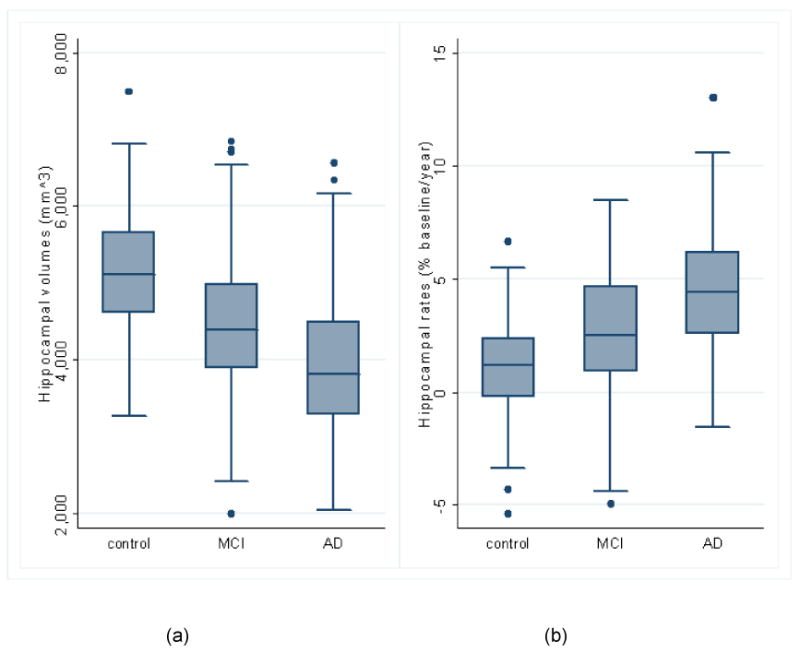
Group comparisons using baseline diagnosis (control (n=200), MCI (n=335), AD (n=147)) in box plots*. (a) Unadjusted total (left+right) hippocampal volumes; (b) Unadjusted atrophy rates from automated MAPS-HBSI.
*The horizontal line in the box represents the median value, and the box represents the interquartile range (IQR). The whiskers represent the upper and lower adjacent values, which are the highest value not greater than 75th percentile + 1.5 times IQR and the lowest value not less than 25th percentile - 1.5 times IQR. Values outside the whiskers are marked as dots.
Comparison between MCI subgroups
Mean (SD) unadjusted total (left+right) hippocampal volumes and atrophy rates, together with the adjusted mean differences are shown in Table 10 for the MCI subgroups. The corresponding box plots are Figures 7a (volumes) and 7b (rates). The adjusted (for age, gender and TIV) mean volume in reverters (95% CI) was 328mm3 (193 lower to 850 higher, p=0.217) and 737mm3 (210 to 1265, p=0.006) higher than in the stable and converter groups respectively. The adjusted mean volume was 409mm3 (244 to 574, p<0.001) lower in converters compared to stable MCI subjects. Adjusting for age and gender, the mean atrophy rate in reverters was 0.43 percentage points (-1.24 to 2.10, p=0.61) lower than stable subjects and 1.84 percentage points (0.15 to 3.53, p=0.03) lower than converters, while the adjusted mean rate was 1.41 percentage points (0.89 to 1.94, p<0.001) higher in converters compared to stable subjects. The volumes and atrophy rates of the MCI subgroups for the left or right hippocampus can be found in the Table SP2 in the supplementary data.
Table 10.
Mean (SD) total (left+right) hippocampal volumes and annualised percentage atrophy rates in MCI subjects, by MCI subgroup. Group comparisons show the estimated mean difference in volumes/rates (95% CI), adjusted for TIV (volumes only), age, and gender.
| MCI reverters (n=8) | MCI stable (n=204) | MCI converters (n=123) | Stable vs reverters difference in means (95% CI) |
Converters vs reverters difference in means (95% CI) |
Converters vs stable difference in means (95% CI) |
|
|---|---|---|---|---|---|---|
| Automated total volumes, mm3 | 5134 (489) | 4580 (839) | 4182 (855) | -328 (-850 to 193), p=0.217 | -737 (-1265 to -210), p=0.006 | -409 (-574 to -244), p<0.001 |
| MAPS-HBSI atrophy rate, % | 1.73 (2.35) | 2.17 (2.22) | 3.60 (2.57) | 0.43 (-1.24 to 2.10), p=0.61 | 1.84 (0.15 to 3.53), p=0.03 | 1.41 (0.89 to 1.94), p<0.001 |
Figure 7.
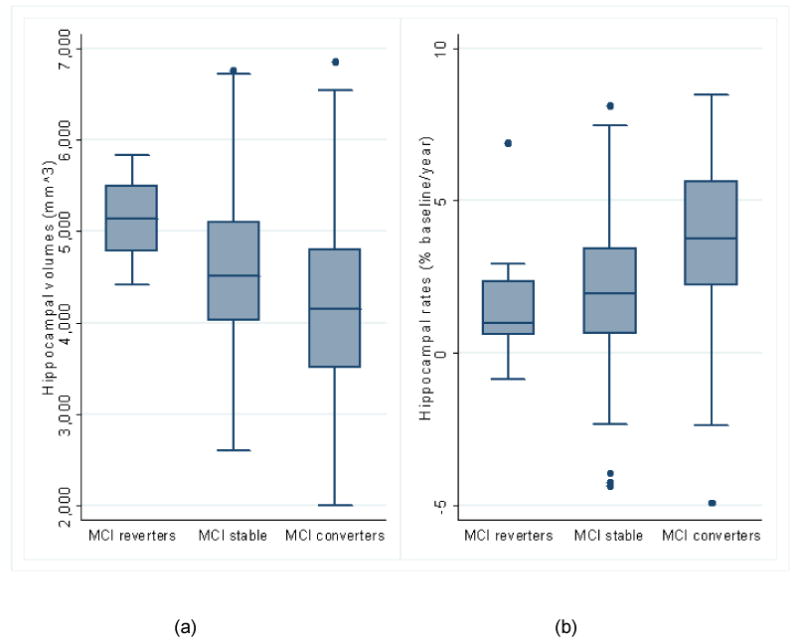
MCI subgroup comparisons (reverters (n=8), stable (n=204), converters (n=123)) in box plots. (a) Unadjusted total (left+right) hippocampal volumes; (b) Unadjusted atrophy rates from automated MAPS-HBSI.
Comparison between MAPS and SNT
We found no evidence of differences between the subset of subjects having an SNT measure and the full subset in the distributions of age, gender, baseline MMSE, and TIV. Table 11 shows the results comparing MAPS with SNT. The means and SDs of the baseline volumes using MAPS were larger than SNT in all three groups. There was no evidence that the mean indirect atrophy rate obtained using MAPS differs from SNT in controls (p=0.26), but in MCI subjects the mean rate (95% CI) using SNT was 0.95 percentage point (0.03 to 1.87, p=0.02) lower than using MAPS, and in AD subjects the mean rate using SNT was 1.55 percentage points (0.30 to 2.81, p=0.01) higher than using MAPS. The SDs of the indirect atrophy rates using MAPS were smaller than SNT in all three groups. For an AD trial powered on hippocampal indirect atrophy rates, the estimated sample size (95% CI) using MAPS was 12% (36% lower to 90% higher, p>0.05) higher than SNT when using atrophy rates from AD subjects alone, and the estimated sample size (95% CI) using MAPS was 71% (42% to 88%, p<0.05) lower than SNT when using atrophy rates from MCI subjects alone. In MCI subjects, there was no evidence of correlation between baseline hippocampal volume and logical memory score, measured either with MAPS (correlation (95% CI) 0.14 (-0.02 to 0.30), p=0.09), or SNT (0.15 (-0.02 to 0.30), p=0.08), whereas in AD subjects, there was evidence of positive correlation: 0.38 (0.14 to 0.57, p=0.003) (MAPS), 0.27 (0.03 to 0.49, p=0.03) SNT. In MCI subjects baseline volume was positively correlated with AVLT score, using both MAPS (0.24 (0.08 to 0.39, p=0.004)) and SNT (0.17 (0.00 to 0.32, p=0.05)), whereas in AD subjects there was no evidence of correlation. In MCI and AD subjects there was no evidence of correlation between change in AVLT score and indirect atrophy rate, measured either using MAPS or SNT.
Table 11.
Comparison between our results and those reported by Schuff et al. (2009) obtained from the ADNI website. Three subjects were excluded from the indirect atrophy rate comparison due to missing SNT volumes at 12-month follow-up (2 MCI subjects (031_S_0568 and 002_S_0954) and 1 AD subject (141_S_0852)). Three more subjects were excluded from the correlation analysis because: the baseline AVLT score of 1 AD subject (099_S_0372) was -1 in Trial V, the 12-month repeat AVLT scores of 1 AD subject (073_S_0565) were all -1 in Trials I-V and the 12-month repeat AVLT scores of 1 MCI subject (067_S_0607) were -1 in Trials IV and V.
| MAPS | SNT | MAPS vs SNT | ||
|---|---|---|---|---|
| Mean (SD) baseline total (left+right) hippocampal volume, mm3 | Differences in mean (95% CI) | SD ratio (95% CI) | ||
| Controls (n=98) | 5080 (718) | 4260 (623) | 821 (742 to 900), p<0.001 | 1.15 (1.03 to 1.30), p=0.01 |
| MCI (n=143) | 4393 (841) | 3670 (705) | 723 (662 to 785), p<0.001 | 1.19 (1.11 to 1.28), p<0.001 |
| AD (n=62) | 3946 (921) | 3300 (770) | 648 (556 to 740), p<0.001 | 1.27 (1.16 to 1.39), p<0.001 |
| Mean (SD) indirect atrophy rate, % | Differences in mean (95% CI) | SD ratio (95% CI) | ||
| Controls (n=98) | 1.40 (3.11) | 1.04 (5.41) | 0.37 (-0.74 to 1.47), p=0.26 | 0.58 (0.47 to 0.70), p<0.001 |
| MCI (n=141) | 3.68 (3.92) | 2.73 (5.40) | 0.95 (0.03 to 1.87), p=0.02 | 0.73 (0.62 to 0.85), p<0.001 |
| AD (n=61) | 4.57 (3.76) | 6.12 (4.77) | -1.55 (-2.81 to -0.30), p=0.01 | 0.79 (0.62 to 1.00), p=0.05 |
| Sample size (95% CI) | Sample size ratio (95% CI) | |||
| AD rate alone | 170 (99 to 486) | 152 (93 to 364) | 1.12 (0.64 to 1.90), p>0.05 | |
| Controlling for aging | 354 (169 to 1415) | 221 (114 to 643) | 1.60 (0.69 to 3.83), p>0.05 | |
| Sample size (95% CI) | Sample size ratio (95% CI) | |||
| MCI rate alone | 285 (193 to 470) | 981 (522 to 2501) | 0.29 (0.12 to 0.58), p<0.05 | |
| Controlling for aging | 742 (367 to 2143) | 2545 (710 to 63884) | 0.29 (0.01 to 1.20), p>0.05 | |
| Correlation between baseline volume and logical memory score (95% CI) | Differences in correlation (95% CI) | |||
| MCI (n = 143) | 0.14 (-0.02 to 0.30) p=0.09 |
0.15 (-0.02 to 0.30) p=0.08 |
0.00 (-0.07 to 0.06), p=0.90 | |
| AD (n = 62) | 0.38 (0.14 to 0.57) p=0.003 |
0.27 (0.03 to 0.49) p=0.03 |
0.10 (-0.01 to 0.21), p=0.07 | |
| Correlation between baseline volume and AVLT (95% CI) | Differences in correlation (95% CI) | |||
| MCI (n = 143) | 0.24 (0.08 to 0.39) p=0.004 |
0.17 (0.00 to 0.32) p=0.05 |
0.07 (-0.01 to 0.15), p=0.08 | |
| AD (n = 61) | 0.12 (-0.14 to 0.36) p=0.36 |
0.05 (-0.20 to 0.30) p=0.70 |
0.07 (-0.04 to 0.19), p=0.24 | |
| Correlation between atrophy rate and change in AVLT (95% CI) | Differences in correlation (95% CI) | |||
| MCI (n=140) | -0.15 (-0.31 to 0.02) p=0.08 |
-0.02 (-0.19 to 0.14) p=0.78 |
-0.13 (-0.35 to 0.09), p=0.26 | |
| AD (n = 59) | 0.13 (-0.13 to 0.37) p=0.33 |
0.05 (-0.21 to 0.30) p=0.71 |
0.08 (-0.22 to 0.38), p=0.60 | |
Comparison between MAPS indirect and direct atrophy rates
Table 12 shows the results comparing the indirect and direct methods of calculating hippocampal atrophy rates (MAPS indirect atrophy rate and MAPS-HBSI respectively). There was no evidence that the MAPS indirect atrophy rate differs from MAPS-HBSI in controls (p=0.13) and AD (p=0.49), but in MCI subjects the mean MAPS indirect atrophy rate (95% CI) was 0.88 percentage point (0.38 to 1.37, p<0.001) higher than using MAPS-HBSI. The SDs of the MAPS indirect atrophy rate were larger than MAPS-HBSI in all three groups. For an AD trial powered on hippocampal atrophy rates, the estimated sample size (95% CI) using MAPS indirect atrophy rate was 151% (43% to 466%, p<0.05) higher than MAPS-HBSI when using atrophy rates from AD subjects alone, and the estimated sample size (95% CI) using MAPS indirect atrophy rate was 50% (9% to 121%, p<0.05) higher than MAPS-HBSI when using atrophy rates from MCI subjects alone. In MCI subjects, MAPS-HBSI was negatively correlated with change in AVLT score (-0.22 (-0.38 to -0.06, p=0.008)), but there was no evidence of correlation with MAPS indirect atrophy rate (-0.15 (-0.31 to 0.02, p=0.08)). In AD subjects there was no evidence of correlation between change in AVLT score and either MAPS indirect atrophy rate or MAPS-HBSI.
Table 12.
Comparison between MAPS indirect atrophy rate and MAPS-HBSI. Three subjects were excluded from the indirect atrophy rate comparison due to missing SNT volumes at 12-month follow-up (2 MCI subjects (031_S_0568 and 002_S_0954) and 1 AD subject (141_S_0852)). Three subjects were excluded from the correlation analysis because: the baseline AVLT score of 1 AD subject (099_S_0372) was -1 in Trial V, the 12-month repeat AVLT scores of 1 AD subject (073_S_0565) were all -1 in Trials I-V and the 12-month repeat AVLT scores of 1 MCI subject (067_S_0607) were -1 in Trials IV and V.
| MAPS indirect atrophy rate | MAPS-HBSI | MAPS indirect atrophy rate vs MAPS-HBSI | ||
|---|---|---|---|---|
| Mean (SD) atrophy rate, % | Differences in mean (95% CI) | SD ratio (95% CI) | ||
| Controls (n=98) | 1.40 (3.11) | 1.15 (2.02) | 0.25 (-0.19 to 0.69), p=0.13 | 1.54 (1.33 to 1.78), p<0.001 |
| MCI (n=141) | 3.68 (3.92) | 2.80 (2.43) | 0.88 (0.38 to 1.37), p<0.001 | 1.61 (1.42 to 1.83), p<0.001 |
| AD (n=61) | 4.57 (3.76) | 4.57 (2.38) | -0.01 (-0.83 to 0.81), p=0.49 | 1.58 (1.27 to 1.97), p<0.001 |
| Sample size (95% CI) | Sample size ratio (95% CI) | |||
| AD rate alone | 170 (99 to 488) | 68 (48 to 99) | 2.51 (1.43 to 5.66), p<0.05 | |
| Controlling for aging | 354 (169 to 1419) | 121 (77 to 206) | 2.92 (1.45 to 8.64), p<0.05 | |
| Sample size (95% CI) | Sample size ratio (95% CI) | |||
| MCI rate alone | 285 (193 to 471) | 189 (139 to 289) | 1.50 (1.09 to 2.21), p<0.05 | |
| Controlling for aging | 742 (367 to 2147) | 545 (296 to 1331) | 1.36 (0.73 to 2.80), p>0.05 | |
| Correlation between atrophy rate and change in AVLT (95% CI) | Differences in correlation (95% CI) | |||
| MCI (n=140) | -0.15 (-0.31 to 0.02) p=0.08 |
-0.22 (-0.38 to -0.06) p=0.008 |
0.07 (-0.03 to 0.18 to 0.03), p=0.18 | |
| AD (n = 59) | 0.13 (-0.13 to 0.37) p=0.33 |
0.04 (-0.22 to 0.20) p=0.77 |
0.09 (-0.23 to 0.41), p=0.58 | |
Discussion
Based on a training sample of 15 subjects, we found that the best method for generating a baseline hippocampal volume with our template library utilised non-linear registration (FFD) together with intensity thresholding and combining the best matched eight segmentations using STAPLE to which a Markov random field filter of 0.2 weighting was applied. This generated volumes whose means and SDs were similar to those produced using manual segmentation, with the largest difference being in the AD group with automated volumes on average being lower than manual by 81 mm3 which corresponds to about 4% of the manual AD hippocampal volume. Overall, the mean difference between our automated volumes and the manual measurements was 27mm3 or just over 1% of the mean of all volumes. The automated regions also agreed well (average JI = 0.81) with the manual regions delineated by a second expert segmentor S2 who did not generate the manual regions in the template library. Using an HBSI measure on these regions, we were able to produce a rate of atrophy similar to manual atrophy rates, with the largest difference in means being in the AD group (5.4% manual vs. 6.5% automated) and the overall mean difference in rates being just under 0.5% /year.
We found that the accuracy of MAPS on unseen data was very high, having achieved a mean (SD) JI of 0.80 (0.04) when comparing the automated baseline hippocampal regions with manual regions delineated by the expert segmentor S1 from a set of 30 subjects (10 AD, 10 MCI and 10 controls). The SD of the automated volumes was similar to the manual volumes in all three groups. The means of automated volumes were smaller than manual volumes in all three groups, with the largest difference being in the AD group, with automated volumes on average being lower than manual by 182 mm3 which corresponds to about 10% of the manual AD hippocampal volume. Overall, the mean (SD) of differences in the manual and automated hippocampal volumes was 101 mm3 (144 mm3) (or 4.4% of mean volume) with manual > automated.
Application of MAPS and MAPS-HBSI to a large dataset showed the expected pattern of hippocampal volumes (AD<MCI<controls) and atrophy rates (AD>MCI>controls). Further to this, we found differences across MCI subgroups based on follow-up diagnosis determined up to 36 months from baseline, with hippocampal volumes statistically significantly lower in those subjects who progressed to a diagnosis of dementia at some point in the study compared to those who remained stable or “reverted” to normal. Atrophy rates from MAPS-HBSI also showed the expected pattern (MCI reverters < MCI stable < MCI converters), with converters having a hippocampal atrophy rate that was statistically significantly higher than the other groups and twice as high as the reverters. Although MCI reverters had higher mean volumes and lower rates than the MCI stable group these differences did not reach statistical significance, which is likely due to the small size of the reverter group (n=8).
The comparison of our volumes and indirect atrophy rates with those calculated using SNT (previously published by Schuff et al. (2009) and treated as the gold standard in the work of Wolz et al. (2010) and Lötjönen, J. et al. (2010)) revealed that there was a marked difference in volumes, with MAPS having larger volumes than SNT. The difference in absolute volume is not surprising given that the conventions of anatomical boundaries of the hippocampus differ slightly between MAPS and SNT (Konrad et al., 2009), e.g. the alveus was included in our hippocampal regions while it was not included in regions from SNT. The mean indirect atrophy rate using MAPS was 0.95 percentage point higher than the mean using SNT in the MCI group and 1.55 percentage points lower than the mean using SNT in the AD group; however the MAPS indirect atrophy rates had lower SDs in all three groups. The estimated sample sizes calculated based on MCI atrophy rates using MAPS was 285, which was 71% smaller than using SNT (981).
Our final investigation of a comparison of MAPS indirect atrophy rate and MAPS-HBSI showed that the mean MAPS-HBSI was 0.88 percentage point lower than MAPS indirect atrophy rate in the MCI group. More importantly, the MAPS-HBSI has markedly lower SDs in all three groups. Possible reasons for the lower SD in rates include the use of a registration-based method to detect boundary shift of the hippocampus (HBSI) compared with segmentation of the hippocampus at the two time-points, as has been previously demonstrated in whole brain analysis (Frost et al., 2004). We have also shown previously that the use of HBSI with a manual baseline region results in lower SDs of rates in control groups compared with rates generated from our “gold standard” manual segmentation of baseline and repeat scans (Barnes et al., 2004; Barnes et al., 2007). Consequently, the estimated sample size calculated based on AD subjects alone using MAPS-HBSI was 68, which was 60% smaller than using MAPS indirect atrophy rates. There was evidence of correlation between MAPS-HBSI and 1-year change in AVLT score in MCI subjects, whereas no evidence of correlations was found between the 1-year change in AVLT score and MAPS (or SNT) indirect atrophy rates.
Our technical findings relate well to those of other research groups. We found non-linear registration to perform better than linear registration as this has been previously shown (Woods et al., 1998b). A combination of labels has also been shown to be useful (Rohlfing et al., 2004; Warfield et al., 2004) so our finding of overall improvement by including those best matches was expected. Overall, the different methods of combinations of labels (vote rule, SBA and STAPLE) produced similarly good results. It is interesting that the results of vote rule and STAPLE were more similar than those of SBA. STAPLE had slightly better accuracy than vote rule, which is consistent with a previous publication (Rohlfing et al., 2004). SBA had slightly lower accuracy than vote rule. This was consistent with previous results showing that the vote rule had slightly better accuracy when combining more than five segmentations (see Figure 7a of (Rohlfing and Maurer, Jr., 2007)).
The optimal number of segmentations for vote rule, SBA and STAPLE were 29, 29 and 8. Figure 3 shows that segmentation accuracy increases first and approaches a plateau when more segmentations are combined. A similar plateau effect in the range of 20 – 30 segmentations was reported by Aljabar et al. (2009) and Collins et al. (2009) when fusing hippocampal segmentations using the vote rule. It should also be noted that the less accurate (i.e. lower rank) segmentations may introduce bias into the combined segmentation in all three methods if the segmentation errors are not randomly distributed. Aljabar et al. (2009) showed a gradual decrease in accuracy for hippocampal segmentation when more than about 30 ranked atlases are combined.
Overall, our technique is most similar to that reported by Aljabar et al. (Aljabar et al., 2009). It differs in the following ways: (1) Aljabar et al. used vote rule to fuse the segmentations, whereas we used STAPLE with MRF to combine the segmentations. Furthermore, Table 5 shows that STAPLE with MRF performed slightly better than vote rule when used to fuse the segmentations from our technique; (2) Aljabar et al. ranked the atlases after nonrigidly registering all the images to the Montreal Neurological Institute (MNI) BrainWeb single subject simulated T1-weighted MR image, whereas we ranked the atlases after affinely registering all the images to a single control subject in the template library. Note that Klein et al. (2008) mentioned the need to initialise the STAPLE algorithm using a probabilistic segmentation; in the STAPLE implementation given to us by Warfield, this is performed internally by averaging the input segmentations to provide a global prior.
We have obtained one of the best accuracies reported to-date for automated hippocampal segmentation when compared with gold standard manual segmentations from a set of 30 randomly chosen subjects (10 AD, 10 MCI and 10 controls) from ADNI. Expressing our JI (of 0.80 from the independent test data) as a Dice score5 equates to 0.89, with the previous highest Dice scores (N = number of hippocampi in the study) being 0.81(N=100) (Pohl et al., 2007), 0.83 (N=550) (Aljabar et al., 2009), 0.83 (N=60) (Heckemann et al., 2006), 0.86 (N=54) (Barnes et al., 2008a), 0.86 (N=40) (Morra et al., 2008), 0.86 (N=14) (Fischl et al., 2002), 0.85 (N=30) (Powell et al., 2008), 0.86 (N=40) (van der Lijn et al., 2008), 0.87 (N=30) (Chupin et al., 2008), 0.88 (N=5) (Gousias et al., 2008) (from a cohort of 2 year old children), 0.85 (N=364) (Wolz et al., 2010), 0.89 (N=120) (Lötjönen et al., 2010) and 0.89 (N=160) (Collins and Pruessner, 2009). Note that our inter- and intra-rater JI values correspond to Dice scores of 0.93 and 0.96 respectively. Comparing these to the results from using our automatic method with different training and test data (0.89) or with the same training data segmented by a different rater (0.90) or the same rater (0.91), suggests that the method has not been over-trained, and that there is potential to improve it further, ideally to approach the upper bounds of inter- or intra-rater agreement.
A large number of studies have shown AD subjects to have lower hippocampal volumes than controls with MCI subjects having intermediate volumes (AD<MCI<control) (Chupin et al., 2009a; Henneman et al., 2009; Schuff et al., 2009; Shi et al., 2009). MCI converters have also been shown to have a lower baseline hippocampal volume compared with non-converters or stable MCI subjects (Chupin et al., 2009a; Devanand et al., 2007; Jack et al., 2000).
Hippocampal atrophy rates using automated baseline regions and HBSI of 4.4% per year for AD (mean age = 75) and 1.1% per year for controls (mean age = 76) are similar to those reported in the literature. A recent meta-analysis of studies prior to ADNI estimated rates to be approximately 4.7% per year in AD subjects (mean age = 73) and controls 1.4% per year in healthy controls (mean age = 78) (Barnes et al., 2009). Mean hippocampal atrophy rates in ADNI have been reported to be 0.8% per year in controls (mean age = 76) and 4.4% per year in AD (mean age = 76) (Schuff et al., 2009). MCI atrophy rates have been shown to be between those of AD and controls, with those progressing to a diagnosis of dementia having higher rates than those remaining stable: median rates being 4.3% vs. 3.0% per year (mean age = 71 in MCI) (Henneman et al., 2009), 3.3% vs 1.8% per year (mean age = 77 in converters and 76 in stable subjects) (Jack et al., 2004).
The strengths of this study include the large and multi-site nature of data collection (though training and testing subsets were relatively small) and the availability of follow-up on all subjects enabling assessment of clinical change from baseline. A notable difference between the outcomes in ADNI and previous studies relates to MCI conversion to AD. One recent meta-analysis showed that in studies adhering to the Mayo clinic definition, allowing for dementia type, the conversion rate from MCI to AD was 8.1% /year (95% CI = 6.3–10.0%) (Mitchell and Shiri-Feshki, 2009). However, the higher rate of conversion in ADNI (∼16% /year) is likely to due to the stringent criteria used to recruit subjects with MCI meaning they were likely to be further down the clinical spectrum (i.e. closer to an AD diagnosis) compared with other studies (Petersen et al., 2009).
We acknowledge a number of limitations to this study. We only performed inter-subject registration using ratio image uniformity as the cost function rather than evaluating several possible cost functions. Also, we used the cross-correlation to choose the best-matched images from the template library rather than evaluating other image similarity measures, since cross-correlation has been shown to be a good measure for the hippocampus (Aljabar et al., 2009). Although there is longitudinal follow-up on all subjects there is no pathological confirmation of disease which would provide diagnostic certainty. However, this is the setting in which clinical trials must be conducted. Not only did hippocampal atrophy rates differ between AD and the other groups, but MCI subjects who progressed clinically also had higher rates than those who did not.
In general, the use of a template library in a multi-atlas method depends on a number of factors, such as the quality of template images (e.g. signal-to-noise ratio and contrast-to-noise ratio), anatomical differences between the subjects in the template library and target group, and the manual segmentation protocol. Although the template library used in this study was from a different cohort and included a spectrum of hippocampal volumes (both AD and controls) it did not contain subjects with MCI. However, we saw no evidence that the MCI subjects were more poorly segmented than the control or AD groups, as they had similar JIs compared with control and AD groups. This would be expected given the overlap in hippocampal volumes and morphology across the control-MCI-AD spectrum. Finally, we did not assess whether our algorithm differed in ability to segment hippocampi or detect change over time according to imaging site or field strength. However, one previous study reported no evidence of differing variability across sites (Schuff et al., 2009) and our estimation of HBSI parameters was performed on a subject by subject basis which allows for some differences across the scanning sites.
In addition to this, the number of MCI reverters in this study was low at only 8 subjects, which was small when compared to the number of MCI stable and converters. The MCI reverters were possibly subjects with small test-retest fluctuations in performance or genuine changes in cognitive ability. However, they did meet criteria for MCI at baseline and after this time did not. One would hypothesise that these subjects would have larger hippocampal volumes and lower rates and with MAPS and MAPS-HBSI we find this was so (albeit differences were not significant when reverters and stable MCI subjects were compared).
We conclude that MAPS has a high level of accuracy for segmentation of the hippocampus and is robust to multi-site data. Our automatically obtained regions can be used to measure hippocampal volume change over time using boundary shift measures (MAPS-HBSI). These methods show expected patterns of volume difference AD<MCI<control, and atrophy rate AD>MCI>control, and show differences in volume and rate in MCI groups according to clinical follow-up with MCI converters< MCI stable< MCI reverters for volumes, and MCI converters> MCI stable> MCI reverters for atrophy rates. MAPS and MAPS-HBSI may be useful in large-scale multi-centre trials to assess both baseline characteristics and disease progression.
Supplementary Material
Acknowledgments
The implementations of vote rule and SBA use the Insight Segmentation and Registration Toolkit (ITK), an open source software developed as an initiative of the U.S. National Library of Medicine and available at www.itk.org. We thank Simon Warfield for kindly providing us with the source code of STAPLE. The research of STAPLE was supported in part by NIH R01 RR021885 from the National Center For Research Resources, and by an award from the Neuroscience Blueprint I/C through R01 EB008015 from the National Institute of Biomedical Imaging and Bioengineering. The Foundation for the National Institutes of Health (www.fnih.org) coordinates the private sector participation of the $60 million ADNI public-private partnership that was begun by the National Institute on Aging (NIA) and supported by the National Institutes of Health. To date, more than $27 million has been provided to the Foundation for NIH by Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly and Co., Merck & Co., Inc., Novartis AG, Pfizer Inc., F. Hoffmann-La Roche, Schering-Plough, Synarc Inc., and Wyeth, as well as non-profit partners the Alzheimer's Association and the Institute for the Study of Aging. This work was undertaken at UCL/UCLH which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. The Dementia Research Centre is an Alzheimer's Research Trust Co-ordinating centre. KKL and MC are supported by TSB grant M1638A, NCF is funded by the Medical Research Council (UK). JB is supported by an Alzheimer's Research Trust (ART, UK) Research Fellowship partly supported by the Kirby Laing Foundation. The authors would like to thank the ADNI study subjects and investigators for their participation.
Appendix
Template library creation
The subjects in the template library included 36 subjects with clinically probable AD and 19 controls who had a mean age of approximately 70 years. In brief, all segmentations were performed using MIDAS software which allows segmentation and viewing of 3D images in coronal, axial and sagittal planes (Freeborough et al., 1997). Whole brain segmentations were performed in native space. Hippocampal segmentations were performed using manual delineation on images rigidly registered (Woods et al., 1998a) to the MNI 305 template (Mazziotta et al., 1995).
The anatomy in our hippocampal segmentation protocol included dentate gyrus, the hippocampus proper, the subiculum and the alveus. Measurements were taken from every coronal slice from the posterior to anterior boundaries using a standard neuroanatomical atlas (Duvernoy, 1998). The posterior limit of the hippocampus was defined as the coronal slice where the longest length of the crus of the fornix was seen (Watson et al., 1992). The hippocampus was bounded superiorly, medially and laterally by cerebrospinal fluid (CSF) and inferiorly by the white matter subjacent to the subiculum. The head of the hippocampus was delineated from the amygdala by inclusion of the alveus, which was best seen as a band of high signal intensity on the sagittal sections. Intensity thresholds restricting hippocampal voxels to lie within 70 to 110% of mean brain intensity were used to improve consistency by excluding white matter and CSF voxels.
A library of images was generated for the hippocampus by choosing a 76-year-old male control subject with MMSE 30/30 (not included in the 19 mentioned above) whose hippocampal volume was close to the mean hippocampal volume of the whole group. To adjust for the difference in head position and size, all images were registered using 12 degrees of freedom (dof) brain-to-brain assessing the cost function (ratio image uniformity) over the segmented brain regions (Woods et al., 1998a). The hippocampi were then rigidly aligned using only the single subject control hippocampus regions to assess the cost function. This local rigid registration was used in order to improve overlap of hippocampal areas. Without distorting the size and shape of the hippocampus, hippocampal regions for each subject were transformed using the registration parameters of these steps and dilated by two voxels to provide an area over which similarity between images could be measured. Note that the dilated regions were only used for the template selection step.
BSI double intensity window approach
The BSI double intensity window approach was previously described (Hobbs et al., 2009). A double intensity window was included for the HBSI calculation in order to capture boundary shift at both the hippocampus–CSF border, and the hippocampus–WM border. The optimal intensity window parameters were chosen using an automatic intensity window selection method. In order to capture most of the tissue-type change between the hippocampus and CSF (or hippocampus and WM), it was desirable to ignore changes within the same tissue type, and to maximize changes between different tissue types. Therefore, the lower intensity window for capturing CSF and hippocampus change was chosen to be (ICSF mean + ICSF std, Ihippo mean − Ihippo std), and the higher intensity window for capturing hippocampus and WM change was chosen to be (Ihippo mean + Ihippo std, IWM mean − IWM std) where ICSF mean, ICSF std, Ihippo mean, Ihippo std, IWM mean and IWM std were the mean and standard deviation of CSF, hippocampus and WM intensities. Ihippo mean and Ihippo std were estimated over the baseline hippocampal region, and ICSF mean, ICSF std, IWM mean and IWM std were estimated using a k-means clustering method over a dilated (by three voxels) hippocampal region.
Footnotes
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. ADNI investigators included (complete listing available at www.loni.ucla.edu/ADNI/Collaboration/ADNI_Citatation.shtml).
TIV estimation will be evaluated in greater detail elsewhere, but in brief, using 67 subjects with manually derived TIV (according to the protocol of (Whitwell et al., 2001) values from SPM8's new segmentation toolbox had a correlation of 0.989, which exceeded values from SPM5 and SPM2.
Downloaded on 17th September 2009.
Dice score (D) is related to the Jaccard index (J) by the equation D = 2J/(1+J).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46:726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ashton EA, Parker KJ, Berg MJ, Chen CW. A novel volumetric feature extraction technique with applications to MR images. IEEE Trans Med Imaging. 1997;16:365–371. doi: 10.1109/42.611343. [DOI] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. A meta-analysis of hippocampal atrophy rates in Alzheimer's disease. Neurobiol Aging. 2009;30:1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008a;40:1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Barnes J, Lewis EB, Scahill RI, Bartlett JW, Frost C, Schott JM, Rossor MN, Fox NC. Automated measurement of hippocampal atrophy rates using fluid-registered serial MRI in AD and controls. JCAT. 2007;31:581–587. doi: 10.1097/rct.0b013e31802f4139. [DOI] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Boyes RG, Frost C, Lewis EB, Rossor CL, Rossor MN, Fox NC. Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates. Neuroimage. 2004;23:574–581. doi: 10.1016/j.neuroimage.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Frost C, Schott JM, Rossor MN, Fox NC. Increased hippocampal atrophy rates in AD over 6 months using serial MR imaging. Neurobiol Aging. 2008b;29:1199–1203. doi: 10.1016/j.neurobiolaging.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathologica. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Aizenstein HA, Davis SW, Becker JT, Thompson PM, Meltzer CC, Liu Y. Atlas-based hippocampus segmentation in Alzheimer's disease and mild cognitive impairment. Neuroimage. 2005;27:979–990. doi: 10.1016/j.neuroimage.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M, Chetelat G, Lemieux L, Dubois B, Garnero L, Benali H, Eustache F, Lehericy S, Desgranges B, Colliot O. Fully automatic hippocampus segmentation discriminates between early Alzheimer's disease and normal aging. 5th IEEE International Symposium on Biomedical Imaging Computer; 2008; 2008. pp. 97–100. [Google Scholar]
- Chupin M, Gerardin E, Cuingnet R, Boutet C, Lemieux L, Lehericy S, Benali H, Garnero L, Colliot O. Fully automatic hippocampus segmentation and classification in Alzheimer's disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009a;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M, Hammers A, Liu RS, Colliot O, Burdett J, Bardinet E, Duncan JS, Garnero L, Lemieux L. Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation 2. Neuroimage. 2009b;46:749–761. doi: 10.1016/j.neuroimage.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DL, Alex PZ, Wim FCB, Alan CE. ANIMAL+INSECT: Improved Cortical Structure Segmentation. Proceedings of the 16th International Conference on Information Processing in Medical Imaging; 1999. pp. 210–223. [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D Model-Based Neuroanatomical Segmentation. Hum Brain Mapp. 1996;3:190–208. [Google Scholar]
- Collins DL, Pruessner JC. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI. Medical Image Computing and Computer-Assisted Intervention Part II. 2009:592–600. doi: 10.1007/978-3-642-04271-3_72. [DOI] [PubMed] [Google Scholar]
- Crum WR, Scahill RI, Fox NC. Automated hippocampal segmentation by regional fluid registration of serial MRI: validation and application in Alzheimer's disease. Neuroimage. 2001;13:847–855. doi: 10.1006/nimg.2001.0744. [DOI] [PubMed] [Google Scholar]
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Honig LS, Mayeux R, Stern Y, Tabert MH, de Leon MJ. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger-Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, Meguro K, O'Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Stern Y, Visser PJ, Scheltens P. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- Duchesne S, Pruessner JC, Collins DL. Appearance-Based Segmentation of Medial Temporal Lobe Structures. Neuroimage. 2002;17:515–531. [PubMed] [Google Scholar]
- Duvernoy HM. The Human Hippocampus. Springer-Verlag; Heidelberg: 1998. [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der KA, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fox NC, Black RS, Gilman S, Rossor MN, Griffith SG, Jenkins L, Koller M. Effects of A beta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005;64:1563–1572. doi: 10.1212/01.WNL.0000159743.08996.99. [DOI] [PubMed] [Google Scholar]
- Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. Presymptomatic hippocampal atrophy in Alzheimer's disease: a longitudinal MRI study. Brain. 1996;119:2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging. 1997;16:623–629. doi: 10.1109/42.640753. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC, Kitney RI. Interactive algorithms for the segmentation and quantitation of 3-D MRI brain scans. Computer Methods and Programs in Biomedicine. 1997;53:15–25. doi: 10.1016/s0169-2607(97)01803-8. [DOI] [PubMed] [Google Scholar]
- Frost C, Kenward MG, Fox NC. The analysis of repeated ‘direct’ measures of change illustrated with an application in longitudinal imaging. Stat Med. 2004;23:3275–3286. doi: 10.1002/sim.1909. [DOI] [PubMed] [Google Scholar]
- Gosche KM, Mortimer JA, Smith CD, Markesbery WR, Snowdon DA. An automated technique for measuring hippocampal volumes from MR imaging studies. AJNR Am J Neuroradiol. 2001;22:1686–1689. [PMC free article] [PubMed] [Google Scholar]
- Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40:672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Gunter JL, Bernstein MA, Borowski BJ, Felmlee JP, Blezek DJ, Mallozzi R, Levy JR, Schuff N, Jack CR. Validation Testing of the MRI Calibration Phantom for the Alzheimer's Disease Neuroimaging Initiative Study. ISMRM 2006 [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, Gado M, Joshi SC, Miller MI, Sheline YI, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomical atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers A, Heckemann R, Koepp MJ, Duncan JS, Hajnal JV, Rueckert D, Aljabar P. Automatic detection and quantification of hippocampal atrophy on MRI in temporal lobe epilepsy: a proof-of-principle study. Neuroimage. 2007;36:38–47. doi: 10.1016/j.neuroimage.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Hammers A, Koepp MJ, Free SL, Brett M, Richardson MP, Labbe C, Cunningham VJ, Brooks DJ, Duncan J. Implementation and application of a brain template for multiple volumes of interest. Hum Brain Mapp. 2002;15:165–174. doi: 10.1002/hbm.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Kazui H, Matsumoto K, Nakano Y, Yasuda M, Mori E. Does donepezil treatment slow the progression of hippocampal atrophy in patients with Alzheimer's disease? American Journal of Psychiatry. 2005;162:676–682. doi: 10.1176/appi.ajp.162.4.676. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Henneman WJ, Sluimer JD, Barnes J, van der Flier WM, Sluimer IC, Fox NC, Scheltens P, Vrenken H, Barkhof F. Hippocampal atrophy rates in Alzheimer disease: added value over whole brain volume measures. Neurology. 2009;72:999–1007. doi: 10.1212/01.wnl.0000344568.09360.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs NZ, Henley SM, Wild EJ, Leung KK, Frost C, Barker RA, Scahill RI, Barnes J, Tabrizi SJ, Fox NC. Automated quantification of caudate atrophy by local registration of serial MRI: evaluation and application in Huntington's disease. Neuroimage. 2009;47:1659–1665. doi: 10.1016/j.neuroimage.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, Weiner MW. Comparison of Automated and Manual MRI Volumetry of Hippocampus in Normal Aging and Dementia. J Magn Reson Imaging. 2002;16:305–310. doi: 10.1002/jmri.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. La distribution de la flore dans la zone alpine. Rev Gen Sci Pures Appl. 1907;18:961–967. [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell L, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu Y, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Tangalos EG, Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Weigand SD, O'Brien PC, Gunter JL, Boeve BF, Knopman DS, Smith GE, Ivnik RJ, Tangalos EG, Petersen RC. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI. Neurology. 2005;65:1227–1231. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O'Brien PC, Cha R, Knopman D, Petersen RC. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60:253–260. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Weigand SD, Shiung MM, Przybelski SA, O'Brien PC, Gunter JL, Knopman DS, Boeve BF, Smith GE, Petersen RC. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008b;70:1740–1752. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CRJ, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der KA, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kelemen A, Szekely G, Gerig G. Elastic model-based segmentation of 3-D neuroradiological data sets. IEEE Trans Med Imaging. 1999;18:828–839. doi: 10.1109/42.811260. [DOI] [PubMed] [Google Scholar]
- Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JP. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys. 2008;35:1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Lancaster J, Thompson P, Toga AW, Brewer P, Hardies J, Fox P. An optimized individual target brain in the Talairach coordinate system. Neuroimage. 2002;17:922–927. [PubMed] [Google Scholar]
- Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. Defining the human hippocampus in cerebral magnetic resonance images--an overview of current segmentation protocols. Neuroimage. 2009;47:1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likeman M, Anderson VM, Stevens JM, Waldman AD, Godbolt AK, Frost C, Rossor MN, Fox NC. Visual assessment of atrophy on magnetic resonance imaging in the diagnosis of pathologically confirmed young-onset dementias. Archives of Neurology. 2005;62:1410–1415. doi: 10.1001/archneur.62.9.1410. [DOI] [PubMed] [Google Scholar]
- Lötjönen J, Wolz R, Koikkalainen JR, Thurfjell L, Waldemar G, Soininen H, Rueckert D. Fast and robust multi-atlas segmentation of brain magnetic resonance images. Neuroimage. 2010;49:2352. doi: 10.1016/j.neuroimage.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: Theory and rationale for its development. Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Mitchell AJ, Shiri-Feshki M. Rate of progression of mild cognitive impairment to dementia--meta-analysis of 41 robust inception cohort studies. Acta Psychiatr Scand. 2009;119:252–265. doi: 10.1111/j.1600-0447.2008.01326.x. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack J, Weiner MW, Thompson PM. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana PA, Brey WW, Kulkarni V, Sievenpiper CL. Compensation for surface coil sensitivity variation in magnetic resonance imaging. Magnetic Resonance Imaging. 1998;6:271–274. doi: 10.1016/0730-725x(88)90401-8. [DOI] [PubMed] [Google Scholar]
- Patenaude B, Smith S, Kennedy D, Jenkinson M. FIRST - FMRIB's Integrated Registration and Segmentation Tool. 13th Annual Meeting of the Organization for Human Brain Mapping.2007. [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW. Alzheimer's Disease Neuroimaging Initiative (ADNI). Clinical characterization. Neurology. 2009 doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitiot A, Delingette H, Thompson PM, Ayache N. Expert knowledge-guided segmentation system for brain MRI. Neuroimage. 2004;23 1:S85–S96. doi: 10.1016/j.neuroimage.2004.07.040. [DOI] [PubMed] [Google Scholar]
- Pitman EJG. A Note on Normal Correlation. Biometrika. 1939;31:9–12. [Google Scholar]
- Pohl KM, Bouix S, Nakamura M, Rohlfing T, McCarley RW, Kikinis R, Grimson WE, Shenton ME, Wells WM. A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging. 2007;26:1201–1212. doi: 10.1109/TMI.2007.901433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey A. L'examen clinique en psychologie. Presses Universitaires de France; 1964. [Google Scholar]
- Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, Fox NC. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol. 2006;5:828–834. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Maurer CR., Jr Shape-based averaging. IEEE Trans Image Process. 2007;16:153–161. doi: 10.1109/tip.2006.884936. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Russakoff DB, Maurer CR., Jr Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans Med Imaging. 2004;23:983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Moffat S, Resnick SM, Davatzikos C. Measuring size and shape of the hippocampus in MR images using a deformable shape model. Neuroimage. 2002;15:422–434. doi: 10.1006/nimg.2001.0987. [DOI] [PubMed] [Google Scholar]
- Shi F, Liu B, Zhou Y, Yu C, Jiang T. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer's disease: Meta-analyses of MRI studies. Hippocampus. 2009 doi: 10.1002/hipo.20573. [DOI] [PubMed] [Google Scholar]
- Shuter B, Yeh IB, Graham S, Au C, Wang SC. Reproducibility of brain tissue volumes in longitudinal studies: effects of changes in signal-to-noise ratio and scanner software. Neuroimage. 2008;41:371–379. doi: 10.1016/j.neuroimage.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- van der Lijn F, den Heijer T, Breteler MM, Niessen WJ. Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. Neuroimage. 2008;43:708–720. doi: 10.1016/j.neuroimage.2008.07.058. [DOI] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20:667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42:1743–1750. doi: 10.1212/wnl.42.9.1743. [DOI] [PubMed] [Google Scholar]
- Webb J, Guimond A, Eldridge P, Chadwick D, Meunier J, Thirion JP, Roberts N. Automatic detection of hippocampal atrophy on magnetic resonance images. Magn Reson Imaging. 1999;17:1149–1161. doi: 10.1016/s0730-725x(99)00044-2. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Manual for the Wechsler Adult Intelligence Scale Revised. Psychological Corporation; New York: 1981. [Google Scholar]
- Whitwell JL, Crum WR, Watt HC, Fox NC. Normalization of cerebral volumes by use of intracranial volume: implications for longitudinal quantitative MR imaging. AJNR Am J Neuroradiol. 2001;22:1483–1489. [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D. LEAP: learning embeddings for atlas propagation. Neuroimage. 2010;49:1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998a;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998b;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.