SUMMARY
The major capsid protein, VP1, of the human Polyomavirus BK (BKV) is structurally divided into five outer loops, referred to as BC, DE, EF, GH, and HI. The BC loop includes a short region, named the BKV subtyping region, spanning nucleotides 1744–1812 and characterized by non-synonymous nucleotide polymorphisms that have been used to classify different strains of BKV into four subtypes. The aim of this study was to determine if the nucleotide changes clustered within the BKV subtyping region may influence the in vitro growth efficiency of the virus. We therefore infected the African Green Monkey kidney cell line Vero with four different viral strains (named BKV I, II, III, and IV) that contained the nucleotide sequences of the BKV subtypes within the same genomic background. Infected cells were followed for 59 days and viral replication was assessed at different time points by Quantitative Real Time PCR (Q-PCR). BKV I, II, and IV were successfully propagated over time in Vero cells, whereas BKV III viral loads progressively decreased during the infection course, demonstrating that the non-synonymous nucleotide polymorphisms of subtype III confer a strong disadvantage for viral replication. Since subtype III differs from all the other subtypes at position 68 of the VP1, where Leu is replaced by Gln, we created viral strains bearing Gln at this position together with the polymorphisms of subtypes I, II, IV and tested their growth in Vero cells. Our results demonstrate that this amino acid substitution does not lower the replication efficiency of subtypes I, II, and IV. In conclusion, this study provides further insights to the importance of the BC loop of BKV in the virus life cycle. In addition, given the effect of the amino acid substitutions of the four BKV subtypes on infectious spread of the virus, our results suggest the need to investigate their potential association with BKV related complications.
Keywords: BK virus, subtype, BC loop, subtyping region
1. Introduction
The human polyomavirus BK (BKV) belongs to the Polyomaviridae family and was first isolated from the urine of a renal allograft recipient (Gardner et al., 1971). The virus is ubiquitous in the human population worldwide and primary infection generally occurs during early childhood, being followed by a lifelong and usually asymptomatic persistence in the kidney (reviewed in Knowles, 2006). The onset of BKV related pathological conditions mainly depends on the immune status of the patient. For example, symptomatic replication of BKV in renal tubular epithelial cells may occur in renal transplant patients as a consequence of immunosuppressive therapy and cause polyomavirus associated nephropathy (PVAN) (Hirsch et al., 2002).
The circular, double-stranded BKV genome encodes six viral proteins: two early regulatory proteins (small and large T antigens), three capsid proteins (VP1, VP2, VP3) and a late auxilliary protein, named Agnoprotein. The origin of replication and the promoter region including binding sites for transcription factors are located in a region of approximately 400 bp, referred to as non-coding control region (NCCR), which may easily undergo rearrangements during passages of virus in cell cultures (Johnsen et al., 1995; Rinaldo et al., 2005; Rubinstein et al., 1991). The major BKV capsid protein, VP1, can be divided, as well as the VP1 of other Polyomaviruses, into five outer domains or loops, known as BC, DE, EF, GH, and HI, that connect the different β strands of the polypeptide (Griffith et al., 1992; Liddington et al., 1991). The importance of the VP1 loops of Polyomaviruses in mediating capsid assembly and attachment to the host cell receptor has been widely demonstrated (Dugan et al., 2007; Gee et al., 2004; Stehle and Harrison, 1994). In particular, it has been predicted that the receptor binding site of BKV may be located in a shallow groove formed by the BC and HI loops of VP1 (Dugan et al., 2007). The BC loop of BKV contains a 69 bp region, named VP1 subtyping region, that spans nucleotides 1744–1812 (aa 61–83) (numbering according to BKV Dunlop strain; Seif et al., 1979), that has been used to identify four main viral subtypes on the basis of nucleotide polymorphisms: subtype I (including for example the MM, PT, JL and Dunlop strain), subtype II (SB strain), subtype III (AS strain), and subtype IV (IV and MG strains) (Jin et al., 1993). These nucleotide polymorphisms result in amino acid changes that are responsible for the existence of four viral antigenic subtypes (Jin et al., 1993; Knowles et al., 1989) (Table 1). The four BKV subtypes are differently distributed in the human population: subtype I is widespread in the world, subtype IV is mainly distributed in east Asia and Europe, whereas subtypes II and III are rarely detected worldwide (reviewed in Yogo et al., 2009).
Table 1.
Nucleotide polymorphisms and consequent amino acid substitutions within the BKV subtyping region, used by Jin et al. (1993) to classify the different BKV isolates into four viral subtypes. Non-discriminatory nucleotides have been omitted for simplicity.
| BKV subtype (viral strain) | Nucleotides at indicated positionsa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1744–46 | 1747 | 1760 | 1765–67 | 1768–70 | 1775 | 1784 | 1787 | 1792–94 | 1809 | 1811 | |
| I (DUN) | G.A | A | T | .TA | AAG | G | A | A | AGC | G | G |
| II (SB) | G.T | A | A | .TA | AAG | C | A | C | GAC | C | A |
| III (AS) | G.T | A | A | .AG | CAC | G | A | C | GAG | C | G |
| IV (IV) | A.T | G | A | .TA | AGA | C | C | C | GAG | C | G |
| Amino acids at indicated positions | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 61 | 62 | 66 | 68 | 69 | 71 | 74 | 75 | 77 | 82 | 83 | |
| I (DUN) | Glu | Asn | Phe | Leu | Lys | Ser | Asn | Asp | Ser | Glu | Arg |
| II (SB) | Asp | Asn | Tyr | Leu | Lys | Thr | Asn | Ala | Asp | Asp | Lys |
| III (AS) | Asp | Asn | Tyr | Gln | His | Ser | Asn | Ala | Glu | Asp | Arg |
| IV (IV) | Asn | Asp | Tyr | Leu | Arg | Thr | Thr | Ala | Glu | Asp | Arg |
Numbering according to Seif et al., 1979
The aim of this study was to determine if the nucleotide polymorphisms of the four subtypes of BKV may influence the in vitro growth ability of the virus. We addressed this issue using an in vitro system, that allowed the infection and replication of recombinant viral genomes containing the nucleotide polymorphisms of subtypes I, II, III, and IV in the African Green Monkey kidney cell line, Vero. In addition, since the in vitro propagation of BKV isolates could introduce rearrangements within their NCCR, we assessed the stability of this genomic region by direct sequencing. To our knowledge, this is the first study aimed to investigate and compare the effect of the nucleotide polymorphisms and the resulting amino acid changes of subtypes I, II, III, and IV on the viral in vitro replication ability.
2. Materials and methods
2.1. Cell cultures
Vero and COS 7 cells (Amercian Type Culture Collection; ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Euroclone, Italy) supplemented with 2% or 10% heat inactivated fetal bovine serum (FBS, Euroclone, Italy), antibiotics (penicillin, 100 U; streptomycin, 100 μg/ml, Euroclone, Italy) and L-Glutamine (2mM, Euroclone, Italy), in a humified environment with 5% CO2 at 37°C.
2.2. Amplification of the VP1 subtyping region of subtypes II, III, IV, I Gln 68, II Gln 68 and IV Gln 68, and the NCCR by standard PCR
VP1 subtyping region of subtypes II, III, and IV
The VP1 subtyping region of subtypes II, III, and IV was amplified by using primers BKSubF (5′-CAGAAATGGGGGATCCAGA-3′, nt 1724–1742) and BKSubR (5′-GGAATTCTTGCTGTGCTGTAA-3′, nt 1847-1827), which contained the restriction sites for the endonucleases BamHI and EcoRI, respectively (the restriction sites are underscored in the primer sequence). The length of the amplified fragment was 123 bp.
As template DNA we used plasmids that had been previously obtained in a BKV subtyping study performed at the Center for Neurovirology at Temple University. These plasmids had been created by cloning into the pCR2.1 vector the 1633–2009 nucleotide region of VP1 amplified from renal transplant patients shedding the virus in their urine. We chose, as template, plasmids containing the VP1 region amplified from patients harboring subtypes II (SB strain), III (AS strain), and IV (IV strain).
The amplifications were carried out in a total volume of 50 μl containing 100 ng of template DNA, 500 nM of each primer, 0.6 mM dNTPs, 1.5 mM MgCl2 and 2U of EuroTaq DNA Polymerase (EuroClone, Italy) in the presence of 1X Reaction Buffer supplied by the manufacturer. The amplification was performed running the following protocol on a GeneAmp PCR System 9700 (Applied Biosystems, USA): an initial denaturation at 94°C for 5 min, followed by 35 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 64°C and 30 sec extension at 72°C, and a final extension step at 72°C for 7 min. The products of amplification were analyzed by means of 3% agarose gel electrophoresis.
VP1 subtyping region of subtypes I Gln 68, II Gln 68 and IV Gln 68
The VP1 subtyping region of subtypes I Gln 68, II Gln 68 and IV Gln 68 was amplified by using three different forward primers (BKV I Gln, BKV II Gln, BKV IV Gln, respectively) and the same reverse primer BKVSubR. BKV I Gln, BKV II Gln, and BKV IV Gln contain two nucleotide mismatches at position 1766–1767 in comparison to the template DNA, which was represented respectively by plasmids pucDun, puc II, and puc IV, which will be described later in section 2.3. The three forward primers also contain the restriction site for BamHI (Table 2). The length of the amplified fragment was 117 bp.
Table 2.
Sequence of primers BKV I Gln, BKV II Gln, and BKV IV Gln. The three primers span nucleotide position 1731–1775 of the VP1.
| Name | Sequence |
|---|---|
| BKV I Gln | 5′-GGGGGATCCaAGATGAAAACCTTAGGGGCTTTAGTCAGbAAGCTAAG-3′ |
| BKV II Gln | 5′-GGGGGATCCaAGATGATAACCTTAGGGGCTATAGTCAGbAAGCTAAC-3′ |
| BKV IV Gln | 5′-GGGGGATCCaAGATAATGACCTTAGGGGCTATAGTCAGbAGACTAAC-3′ |
BamHI restriction site;
nucleotide mismatches (positions 1766–1767) between the forward primers and the template DNA (pucDun, puc II, and puc IV, respectively), which bears nucleotides TA at that genomic position.
The amplification was performed carrying out the same protocol used for the amplification of the VP1 subtyping region of subtypes II, III, and IV, and running the following cycles on a GeneAmp PCR System 9700 (Applied Biosystems, USA): an initial denaturation at 94°C for 5 min, followed by 35 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 60°C and 30 sec extension at 72°C, and a final extension step at 72°C for 7 min. The products of amplification were analyzed by means of 3% agarose gel electrophoresis.
NCCR region
The NCCR of the viral strains collected at different time points of infection was amplified by using primers BKTT-1 and BKTT-2 (Flaegstad et al., 1991), which are expected to generate a fragment of 705 bp when the BKV Dunlop strain is used as template in the reaction. The amplifications were carried out in a total volume of 50 μl containing 5 μl of viral DNA extracted from culture medium, 500 nM of each primer, 0.6 mM dNTPs, 1.5 mM MgCl2 and 2U of EuroTaq DNA Polymerase (Euroclone, Italy) in the presence of 1X Reaction Buffer supplied by the manufacturer. The amplification was performed running the following protocol in a GeneAmp PCR System 9700 (Applied Biosystems, USA): an initial denaturation at 94°C for 5 min, followed by 35 cycles of 30 sec denaturation at 94°C, 40 sec annealing at 57°C and 30 sec extension at 72°C, and a final extension step at 72°C for 7 min. The products of amplification were analyzed by means of 1% agarose gel electrophoresis.
2.3. Construction of recombinant plasmids containing the nucleotide polymorphisms of subtypes I, II, III, IV, I Gln 68, II Gln 68 and IV Gln 68
Nucleotide polyomorphisms for each subtype were introduced into the BKV Dunlop backbone by replacing the region encoding 1734–1846 with the corresponding subtype sequence using unique native BamHI and EcoRI sites flanking the VP1 subtyping region at positions 1734–39 and 1841–46 of the BKV Dunlop genome, respectively. To accomplish this, the entire BKV Dunlop genome was excised from pBKV plasmid (American Type Culture Collection; ATCC) and cloned into the PstI site of pUC19, from which its BamHI and EcoRI restriction sites were previously removed. The resulting vector, pUCDun, was digested with BamHI and EcoRI (Roche, USA) and excised region was replaced with PCR fragments amplified with primers BKSubF and BKSubR. The resulting plasmids, containing the nucleotide polymorphisms of subtypes II, III, and IV, were referred to as pUC II, pUC III and pUC IV. In addition, in order to obtain recombinant plasmids containing the nucleotide polymorphisms of subtypes I Gln 68, II Gln 68 and IV Gln 68, the pUCDun 1734–1846 excised region was replaced with PCR fragments amplified with primers BKV I Gln/BKVSubR, BKV II Gln/BKVSubR, BKV IV Gln/BKVSubR, respectively. The resulting plasmids were referred to as pUC I Gln, pUC II Gln, and pUC IV Gln.
For high copy expression of plasmids, the INVαF′ strain of Escherichia Coli (Invitrogen, USA) was transformed with pUCDun, pUC II, pUC III, pUC IV, pUC I Gln, pUC II Gln, and pUC IV Gln. Plasmid DNA was extracted from bacterial cells using the QIAGEN Plasmid Minikit (QIAGEN, Germany), following the manifacturer’s instruction. Plasmids were sequenced to confirm the presence of the insert and to rule out the possibility that any mutations or insertions were introduced during genetic manipulation.
2.4. Transfections and virion production
Plasmids were digested with PstI (Roche, USA) in order to separate the complete BKV genome from the plasmid backbone. 5 to 8 μg of digested viral DNA were then transfected in COS 7 cells using FuGENE 6 (Roche, USA), according to the manifacturer’s instructions (FuGENE: DNA ratio = 3:2). Transfected cells were cultured in growth medium and harvested when evident cytopathic effects were visible (approximately 2 weeks). Collected cells were subjected to three freeze-thaw cycles (dry ice/37°C) and then incubated in 0.2 U/ml neuraminidase for 30 min at 37°C, to release virus bound to cell membranes. Neuraminidase was then inactivated by incubation at 56°C for 30 min. Cellular debris were centrifuged at 3200 rpm for 45 min at 4°C. Supernatants containing viral stocks (conveniently named BKV I, II, III, IV, I Gln 68, II Gln 68, IV Gln 68) were collected, aliquoted and stored at −80°C. For quantification of viral genomes, viruses were heated at 95°C for 10 min and diluted 1:1000 before performing Q-PCR analysis as described later in section 2.7. A molecular characterization of the VP1 region of collected viral strains was performed by direct sequencing, in order to rule out the presence of nucleotide substitutions.
2.5. Viral infections
4 × 105 Vero cells were plated in T25 flasks. The following day, cells at approximately 80% confluence were infected with 109 copies of each viral stock. The infections were performed in triplicate by adsorption in serum free medium for 90 min at 37°C. The inoculum was then removed, cells were washed twice with phosphate buffer saline (PBS) to eliminate unbound virus, and DMEM containing 2% FBS was added. Mock infected cells, that were used as negative controls, were given the same treatment, but without the addition of virus. After infection with BKV I, BKV II, BKV III, BKV IV, and infection with BKV I Gln 68, BKV II Gln 68, BKV IV Gln 68 together with the corresponding subtypes without the Leu to Gln substitution, cells were continuously cultured in 2% FBS DMEM for 59 and 46 days, respectively. In both experiments, cells were transferred at split ratio of 1:6 once every 6 to 8 days. Therefore, a total of 8 and 6 subculturing passages were required to maintain cells up to day 59 and day 46, respectively. 1ml of fresh medium was added to each flask 3 to 4 days after subculturing. It is worth noting that the cells infected with the different viral strains were simultaneously subjected to medium changing and subculturing, therefore all the cells underwent the same treatment during the infection course. In addition, cells were inspected for cytopathic effects daily. Aliquots of culture medium and cells infected with BKV I, BKV II, BKV III, and BKV IV were collected at days 5, 11, 17, 24, 31, 47, 59 post-infection (P.I.), whereas aliquots of culture medium and cells infected with BKV I Gln 68, II Gln 68, IV Gln 68, I, II, and IV were collected at days 5, 12, 18, 25, 32, 39, 46 P.I.. Viral genomic copies were then quantified in both collected cells and culture medium.
2.6. DNA extraction
Viral DNA was extracted from culture medium and Vero cell pellets using the Nucleospin RNA Virus Kit (Macherey-Nagel, Germany) and the QIAamp DNA Blood Minikit (QIAGEN, Germany), respectively, according to the manufacturer’s instruction. Extracted viral DNA was diluted 1:1000 before Quantitative Real-Time PCR (Q-PCR) analysis.
2.7. Quantification of viral load
Q-PCR was performed using TaqMan chemistry and the 7300 Real Time PCR System (Applied Biosystems, USA). The following primers and probe were used for amplification and detection of a conserved region of the VP1 gene: BKVPf (5′-AGTGGATGGGCAGCCTATGTA-3′, nt 2511–2531), BKVPr (5′-TCATATCTGGGTCCCCTGGA-3′, nt 2605-2586), and MGB probe BKVPp (5′FAM- AGGTAGAAGAGGTTAGGGTGTTTGATGGCACAG-3′MGB, nt 2578-2546). The reactions were performed in a final volume of 25 μl containing a 1X Taqman Universal PCR Master Mix (Applied Biosytems, USA), 0.4 μM BKVPf, 0.9 μM BKVPr, 0.2 μM BKVPp and 5 μl of diluted viral nucleic acid. Thermal cycling was carried out according to the following steps: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, at the end of which the fluorescence was read. A standard curve for the quantification of BKV viral load was obtained after serial dilution of the plasmid pBKV, which contained the whole genome of the BKV Dunlop strain, with 10 ng of pBKV corresponding to 109 copies of the viral genome (dilutions range: 102 to 106 plasmid copies). Negative and positive controls were included in each run. Each sample, standard and control was tested in triplicate. The detection limit of the assay was determined at 5 copies/reaction. Data were expressed as copies of viral DNA per mL of cell medium or per μg of DNA extracted from the cell pellets.
2.8. DNA sequencing and analysis
DNA sequencing was performed by external facilities (Kimmel Cancer Center Institute - Nucleic Acid Facility, Philadelphia; Primm, Milan). Sequences were analyzed by using BLAST at NCBI.
3. Results and discussion
3.1. Replication rates of BKV I, II, III, and IV in Vero cells
The first step of the study consisted of creating recombinant viral genomes that contained the nucleotide polymorphisms of the four BKV subtypes within the same genomic background. The recombinant viral genomes were then transfected into COS 7 cells in order to produce complete viral particles. The COS 7 cell line was chosen for the production of viral stocks since it expresses high levels of SV40 T-Ag, and is therefore suitable to support the rapid propagation of the closely related BK virus in vitro. Viruses were then harvested and viral titer determined by Q-PCR. A molecular characterization of the VP1 region of each viral strain was performed, showing that no nucleotide changes were introduced during growth in COS 7 cells (data not shown). Infections of Vero cells were then performed. Vero cells were chosen for the infection since they are able to sustain productive replication of BKV (Acott et al., 2006) and have been widely used for basic research purposes, providing important data regarding virus binding to its receptor, the infectivity profile of different BKV strains and the effect of specific drugs on viral replication (Acott et al., 2006; Sinibaldi et al., 1987, 1990). Cells infected with the same starting number of genomic copies for each viral strain were allowed to grow for 59 days with occasional subculturing every 6 to 8 days. The replication rates of BKV I, II, III, and IV were assayed at days 5, 11, 17, 24, 31, 47, 59 P.I. by Q-PCR performed on aliquots of culture medium and cells. In addition, the VP1 region of viral strains collected at day 59 P.I. was amplified and sequenced but no nucleotide substitutions were detected (data not shown).
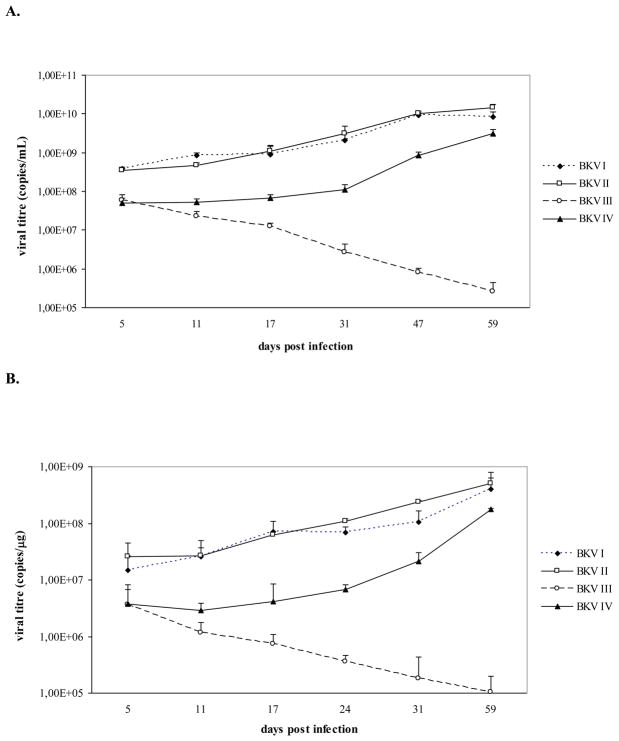
As shown in figure 1, viral genomes corresponding to BKV I, II, III, and IV were all detected throughout the course of infection. The variation of BKV I, II, III, and IV viral titres detected in the culture medium samples overlapped with the viral titres detected in the cell pellets. At the first time point of sample collection (day 5 P.I.), the four BKV strains exhibited similar viral loads. However, as the infection course progressed, BKV I, II, and IV were detected at progressively higher viral loads that peaked at day 59 P.I., whereas BKV III viral titres gradually decreased in both cell pellets and culture medium samples. Therefore, differences in viral load between subtypes I, II, IV and subtype III progressively increased and peaked at the last time point of samples collection (at day 59 P.I., subtype I, II, IV viral loads were approximately 4-log and 3-log higher than subtype III viral load in the culture medium and in the cell pellets, respectively). In addition, BKV I and II exhibited the highest viral loads throughout the course of infection. BKV IV showed the same replication pattern as BKV I and II but was detected with slightly lower viral titres throughout the infection course (Fig. 1). Clear cytopathic effects, represented by detachment of cells from the surface of the flasks, were observed in the last phase of infection (starting from approximately day 40 P.I.) only in cell cultures infected with BKV I, II, and IV (data not shown).
Fig. 1. Nucleotide polymorphisms of the four BKV subtypes alter the virus growth efficiency in Vero cells.
Vero cells were infected with BKV strains (BKV I, II, III, and IV) containing the nucleotide polymorphisms of the four BKV subtypes. During the infection course, aliquots of culture medium and cell pellets were collected and viral replication was assessed by Q-PCR. A. the graph shows the number of genomic copies of BKV I, II, III, and IV detected in the culture medium samples collected at days 5, 11, 17, 31, 47, 59 P.I. (data were expressed as viral copies per ml of culture medium). Day 24 P.I. is not indicated in the graph since aliquots of culture medium were not collected at this time point. B. the graph shows the number of copies of BKV I, II, III, and IV detected in aliquots of Vero cells collected at days 5, 11, 17, 24, 31, and 59 P.I. (data were expressed as viral copies per μg of DNA extracted from cell pellets). Day 47 P.I. is not indicated in the graph since aliquots of Vero cells were not collected at this time point. Both graphs indicate that BKV I, II, and IV share similar replication patterns. On the contrary, the polymorphisms of subtype III negatively affect the in vitro propagation of the virus, since viral titres of BKV III detected in both culture medium and cell pellets progressively decreased during the course of infection.
On the whole, our data showed that the polymorphisms of the four BKV subtypes strongly influence the growth efficiency of the virus in Vero cells. In particular, BKV III was associated with the lowest in vitro replication ability which suggests that the nucleotide polymorphisms and the resulting amino acid changes of subtype III may confer a strong disadvantage for its in vitro propagation. In addition, the nucleotide polymorphisms of subtype IV seem to slightly influence the BKV growth efficiency, even if they do not affect virus lifecycle as the polymorphisms of subtype III. In fact, even if BKV IV was detected with lower viral titres than BKV I and BKV II during the course of the infection, this viral strain was rapidly propagated in cell culture and was characterized by the same replication pattern of BKV I and II.
To our knowledge, this is the first study that investigated and compared the in vitro growth efficiency of viral strains bearing the nucleotide polymorphisms of the four BKV subtypes. Previously, Nukuzuma et al. tested the in vitro replication rates of full-length subtypes I and IV BKV strains, demonstrating that subtype I was able to grow whereas subtype IV was mostly not detected throughout cultivation (Nukuzuma et al., 2005). The differences between our results and the results reported by Nukuzuma et al. may be explained by the different cell lines employed and variations in methods used for viral propagation and detection. Nukuzuma et al. also suggested that the different in vitro growth properties of subtypes I and IV may potentially determine the proportion of these two subtypes in the human population. However, the results of our study suggest that differences in in vitro replication rates due to amino acid substitutions within the BKV subtyping region can not fully explain the distribution of these subtypes within the human population. In fact, the amino acid polymorphisms of subtypes III, which has a markedly lower distribution than subtype I, result in a lower in vitro growth efficiency. On the other hand, the amino acid polymorphisms of subtype II, which is also rarely detected in the human populations, confer similar replication properties to subtype I, the predominant subtype. Therefore, other factors which remain to be determined are likely to contribute to the peculiar distribution of the four BKV subtypes within the human population. We may also hypothesize that, in addition to its possible association to the virus distribution within the human population, the different replication ability of the four BKV subtypes may be related to distinct clinical outcomes. In a previous work (Tremolada et al., 2010), we failed to detect an association between a specific subtype and the onset of PVAN, but the number of patients included was too low to draw any conclusion regarding this issue. Therefore, further studies are needed to determine if BKV subtypes characterized by a lower in vitro replication ability may result in a milder clinical course in infected patients.
The amino acid changes discriminating the four subtypes of BKV are located within the BC loop of the virus. Therefore, our results demonstrate that the BC loop of BKV could play a crucial role in the virus lifecycle. It has been shown that mutations within this loop of VP1 may affect the ability of the virus to interact with its receptor on the host cell (Stehle and Harrison, 1996). Subtype III differs from the other subtypes at position 68, where leucine, a highly hydrophobic residue which is characteristic of subtypes I, II, and IV, is replaced by glutamine, which is highly hydrophilic. We may therefore hypothesize that this difference may alter the structure of the BC loop or directly affect the ability of subtype III to interact with molecular moieties on the cellular receptor.
Similarly, subtype IV bears asparagine, a neutral residue, at position 61, while the other subtypes bear glutamic or aspartic acids, which are both negatively charged. This change of charge may slightly affect the ability of the virus to spread in cell culture and may explain why BKV IV was detected with lower viral titres than BKV I and II. Moreover, it is possible that amino acid substitutions within the BC loop of BKV may also affect other aspects of the virus lifecycle, such as capsid assembly or DNA packaging, as have been previously shown (Dugan et al., 2007).
3.2. Replication rates of BKV I Gln 68, II Gln 68, and IV Gln 68 in Vero cells
In order to verify the hypothesis that the above mentioned Leu to Gln amino acid change at position 68, that is characteristic of subtype III, could be responsible of the reduced replication capacity of this viral strain, we performed further cloning experiments and in vitro infections. Nucleotides TA at positions 1766–1767 of subtypes I, II and IV were replaced by nucleotides AG of subtype III, in order to create recombinant viral constructs coding a Gln instead of a Leu at position 68. The resulting plasmids were named puc I Gln, puc II Gln, puc IV Gln. We then created complete viral particles by transfection-infection of COS 7 cells and sequenced their VP1 in order to rule out the presence of nucleotide substitutions (data not shown). Vero cells were infected using the same starting number of genomic copies of BKV I Gln 68, II Gln 68, and IV Gln 68. Infections were also performed with BKV I, II, and IV, which were used as controls. Cells were allowed to grow for 46 days with occasional subculturing every 6 to 7 days. The replication rates of BKV I Gln 68, II Gln 68, IV Gln 68, I, II, and IV were tested by Q-PCR on aliquots of culture medium and cells collected at days 5, 12, 18, 25, 32, 39, 46 P.I..
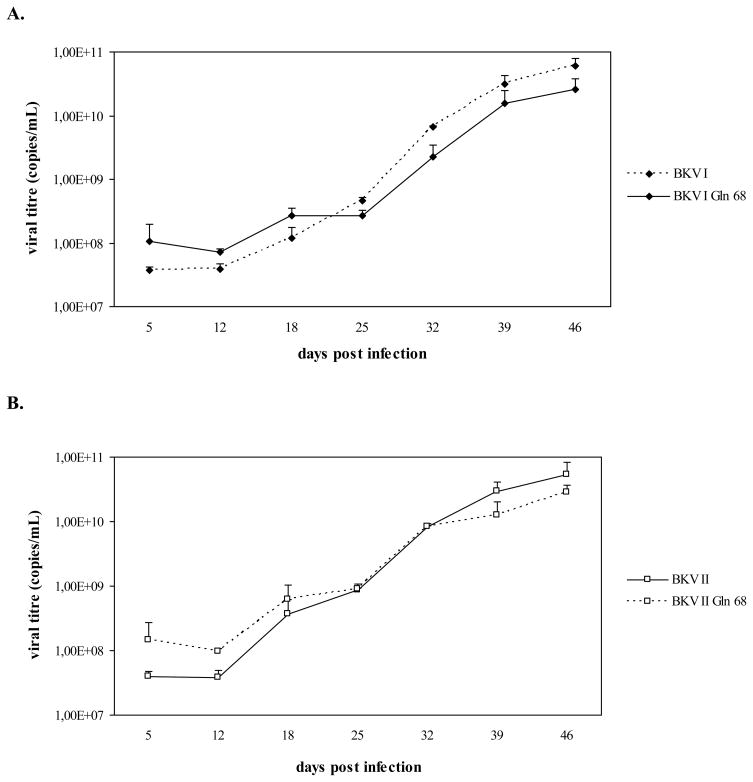
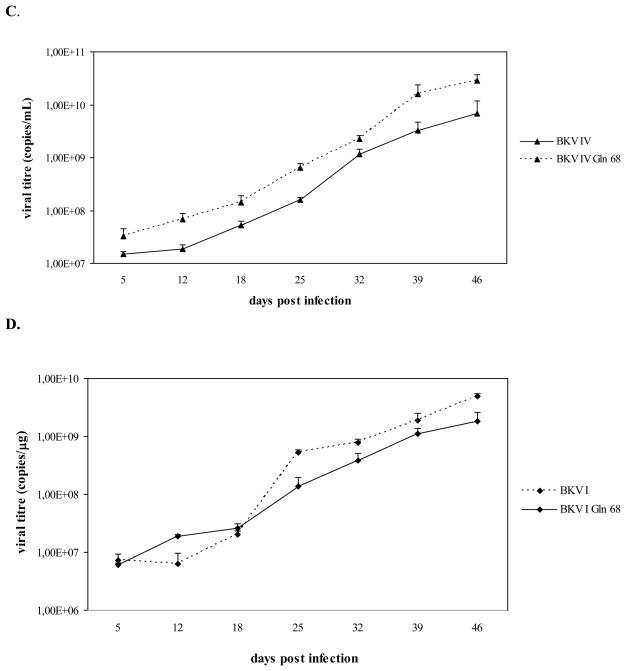
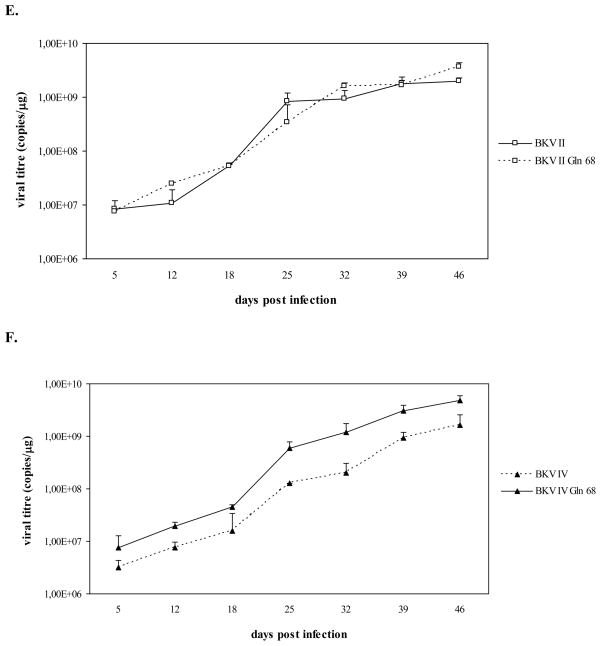
As shown in figure 2, viral genomes corresponding to BKV I Gln 68, II Gln 68, and IV Gln 68 were all detected throughout the course of infection. Similarly to the results obtained with the first infection experiment, the variation of viral titres detected in the culture medium samples overlapped with the viral titres detected in the cell pellets. In addition, the growth curves of BKV I Gln 68, II Gln 68, and IV Gln 68 were similar to those of BKV I, II, and IV, respectively, and all these viral strains were detected with progressively higher viral titres throughout the course of the infection, indicating their successful propagation in Vero cells (Fig. 2). Cytopathic effects, represented by detachment of cells from the surface of the flasks, were observed in the last phase of infection (starting from approximately day 33 P.I.) in all cell cultures (data not shown).
Fig. 2. A Leu to Gln substitution at position 68 of the VP1 does not affect the growth efficiency of subtypes I, II, and IV in Vero cells.
Vero cells were infected with BKV I, II, IV and BKV strains (BKV I Gln 68, II Gln 68, and IV Gln 68) whose VP1 contained the amino acid polymorphisms of subtypes I, II, and IV, with the exception of position 68, where a Leu was substituted by a Gln, as in subtype III. During the infection course, aliquots of culture medium and cell pellets were collected and viral replication was assessed by Q-PCR. A, B, C. the graphs compare the number of genomic copies of BKV I and I Gln 68, BKV II and II Gln 68, BKV IV and BKV IV Gln 68, respectively, detected in the culture medium samples collected at days 5, 12, 18, 25, 32, 39, 46 P.I. (data were expressed as viral copies per ml of culture medium). D, E, F. the graphs compare the number of genomic copies of BKV I and I Gln 68, BKV II and II Gln 68, BKV IV and BKV IV Gln 68, respectively, detected in aliquots of Vero cells collected at days 5, 12, 18, 25, 32, 39, 46 P.I. (data were expressed as viral copies per μg of DNA extracted from cell pellets). On the whole, all the viral strains were detected with progressively higher viral titres during the infection course, thus indicating that the presence of a Gln instead of a Leu at position 68 of VP1 does not affect the replication efficiency of subtypes I, II, and IV.
On the whole, the presence of a Gln instead of a Leu at position 68 of the VP1 region did not seem to influence the replication efficiency of subtypes I, II, and IV. Therefore, these results do not confirm the role of this amino acid substitution in determining the lower growth capacity of subtype III. However, it should be pointed out that the final structure of a protein is determined by the mutual interaction between the different amino acids that constitute the molecule. Therefore, it is possible that the critical effect of the presence of a Gln instead of a Leu at position 68 may mostly result as a consequence of the interaction between this amino acid and the peculiar polymorphisms of subtype III.
3.3. Molecular characterization of the NCCR during the infection course
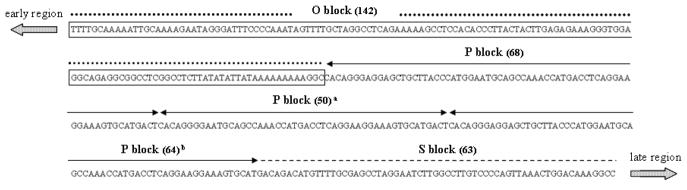
It has been reported that the BKV NCCR frequently undergoes rearrangement during passage in cell culture (Johnsen et al., 1995; Rinaldo et al., 2005; Rubinstein et al., 1991). These genomic modifications may affect the virus in vitro growth efficiency (Moens et al., 1995; Rubinstein et al., 1987), the host cell permissivity and the transforming ability of the virus (Johnsen et al., 1995). Therefore, an important step in our study consisted of determining the molecular characterization of the NCCR of the viral strains isolated during infection of Vero cells with BKV I, II, III, and IV, in order to assess the stability of this genomic region. The BKV strains used in this study all shared the same NCCR organization since they were created by molecular cloning of the VP1 subtyping region within the Dunlop strain background. Therefore, the NCCRs of BKV I, II, III, and IV were PCR-amplified from the culture medium on days 5, 17, 31, and 59 P.I., gel-purified, sequenced and compared to the Dunlop NCCR sequence (Fig. 3).
Fig. 3. Nucleotide sequence of the BKV NCCR of the Dunlop strain (GenBank ID: V01108).
The NCCR of BKV has been divided into sequence blocks, in order to help visualize rearrangements of the different NCCR variants (Markowitz and Dynan, 1988). The O block contains the origin of replication and TATA box, while the other blocks contain the promoter, the enhancer and the binding sites for transcription factors. In the Dunlop NCCR genomic organization, blocks Q and R, which are usually present in the archetypal organization, are missing, whereas the P block is repeated three times (two of the three P blocks are partially deleted). All the BKV strains used in our study were characterized by a Dunlop NCCR organization. The numbers in brackets indicate the length in nucleotide of each block. a This P block bears a deletion spanning nucleotides 8–25. b This P block bears a deletion spanning nucleotides 65–68.
The results obtained indicated that all the viral strains analyzed were characterized by a Dunlop NCCR organization, and no deletions, duplications or insertions were found in the clones analyzed. Only a single nucleotide substitution at position 308 (A→C) within the NCCR of the BKV IV strain isolated at day 59 P.I. was detected. On the whole, however, the NCCRs of the BKV strains isolated were conserved during their growth in Vero cells and shared 100% homology with the Dunlop strain NCCR (GenBank ID: V01108). Based on the data obtained, we could rule out the possibility that the different replication patterns of BKV I, II, III and IV could have been related to the onset of rearrangements within the NCCR during the course of infection.
The stability of the NCCR emerging from our study is not consistent with most of the previously published data, reporting that this genomic region is frequently subjected to deletions, duplications or insertions. However, it is worth mentioning that most studies that investigated stability of the NCCR during passage in cell culture have involved viral strains bearing an archetypal genomic organization. The absence of NCCR rearrangements found in our study could may be therefore related to the viral strain used for the infection. In addition, we cannot exclude the possibility that structural modifications within the NCCR may have been detected if the viral strains had been monitored for longer than 59 days in culture (Rinaldo et al., 2005). Moreover, the onset of rearrangements within the NCCR may also depend on the cell line used for the in vitro propagation of the virus. Acott et al. showed that changes within the NCCR of several BKV isolates propagated in Vero cells started to appear at around 50 days P.I., suggesting that genomic modifications may become frequent only after a prolonged infection course (Acott et al., 2006).
4. Conclusions
In this study we demonstrated that the polymorphisms of the four BKV subtypes influence their replication rates in Vero cells. These results were confirmed by Q-PCR, monitoring the cytopathic effects and by confirming the absence of regulatory region rearrangements during the infection course. Therefore, our data support the crucial role of the VP1 BC loop in the life cycle of BKV. In addition, the effect of the amino acid changes of subtypes I, II, III, and IV on virus infectivity suggests the need to investigate the potential pathogenic role of these amino acid substitutions and the potential association between one or more subtypes and BKV related pathologies. Our results did not confirm the role of a Leu to Gln substitution at position 68 of the VP1 region in determining the different replication properties of the BKV subtypes. Therefore, other amino acid positions as well as other hypotheses need to be evaluated in order to explain the lower replication capacity of subtype III in comparison to that of the other BKV subtypes. The information concerning the virus viability and growth may be useful to support, in the future, the development of new clinical tools for the treatment of complications related to BKV reactivation.
Acknowledgments
This work was partially supported by NIMH, grant R01-MH07258 to PF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sara Tremolada, Email: sara.tremolada@unimi.it.
Serena Delbue, Email: serena.delbue@unimi.it.
Sara Larocca, Email: sara.larocca@studenti.unimi.it.
Camilla Carloni, Email: camilla.carloni@studenti.unimi.it.
Francesca Elia, Email: francesca.elia@studenti.unimi.it.
Kamel Khalili, Email: kamel.khalili@temple.edu.
Jennifer Gordon, Email: jennifer.gordon@temple.edu.
Pasquale Ferrante, Email: pasquale.ferrante@unimi.it.
References
- Acott PD, O’Regan PA, Lee SH, Crocker JF. Utilization of vero cells for primary and chronic BK virus infection. Transplant Proc. 2006;38:3502–3505. doi: 10.1016/j.transproceed.2006.10.163. [DOI] [PubMed] [Google Scholar]
- Dugan A, Gasparovic ML, Tsomaia N, Mierke DF, O’Hara BA, Manley K, Atwood WJ. Identification of amino acid residues in BK virus VP1 critical for viability and growth. J Virol. 2007;81:11798–11808. doi: 10.1128/JVI.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaegstad T, Sundsfjord A, Arthur RR, Pedersen M, Traavik T, Subramani S. Amplification and sequencing of the control regions of BK and JC virus from human urine by polymerase chain reaction. Virology. 1991;180:553–560. doi: 10.1016/0042-6822(91)90069-n. [DOI] [PubMed] [Google Scholar]
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Gee GV, Tsomaia N, Mierke DF, Atwood WJ. Modeling a sialic acid binding pocket in the external loops of JC virus VP1. J Biol Chem. 2004;279:49172–49176. doi: 10.1074/jbc.M409326200. [DOI] [PubMed] [Google Scholar]
- Griffith JP, Griffith DL, Rayment I, Murakami WT, Caspar DL. Inside polyomavirus at 25-A resolution. Nature. 1992;355:652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Steiger J. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002;347:488–496. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- Jin L, Gibson PE, Knowles WA, Clewley JP. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J Med Virol. 1993;39:50–56. doi: 10.1002/jmv.1890390110. [DOI] [PubMed] [Google Scholar]
- Johnsen JI, Seternes OM, Johansen T, Moens U, Mäntyjärvi R, Traavik T. Subpopulations of non-coding control region variants within a cell culture-passaged stock of BK virus: Sequence comparisons and biological characteristics. J Gen Virol. 1995;76:1571–1581. doi: 10.1099/0022-1317-76-7-1571. [DOI] [PubMed] [Google Scholar]
- Knowles WA. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV) Adv Exp Med Biol. 2006;577:19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- Knowles WA, Gibson PE, Gardner SD. Serological typing scheme for BK-like isolates of human polyomavirus. J Med Virol. 1989;28:118–123. doi: 10.1002/jmv.1890280212. [DOI] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Markowitz RB, Dynan WS. Binding of cellular proteins to the regulatory region of BK virus DNA. J Virol. 1988;62:3388–3398. doi: 10.1128/jvi.62.9.3388-3398.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens U, Johansen T, Johnsen JI, Seternes OM, Traavik T. Noncoding control region of naturally occurring BK virus variants: Sequence comparison and functional analysis. Virus Genes. 1995;10:261–275. doi: 10.1007/BF01701816. [DOI] [PubMed] [Google Scholar]
- Nukuzuma S, Takasaka T, Zheng HY, Zhong S, Chen Q, Kitamura T, Yogo Y. Subtype I BK polyomavirus strains grow more efficiently in human renal epithelial cells than subtype IV strains. J Gen Virol. 2006;87:1893–1901. doi: 10.1099/vir.0.81698-0. [DOI] [PubMed] [Google Scholar]
- Rinaldo CH, Hansen H, Traavik T. Human endothelial cells allow passage of an archetypal BK virus (BKV) strain – a tool for cultivation and functional studies of natural BKV strains. Arch Virol. 2005;150:1449–1458. doi: 10.1007/s00705-005-0511-3. [DOI] [PubMed] [Google Scholar]
- Rubinstein R, Pare N, Harley EH. Structure and function of the transcriptional control region of nonpassaged BK virus. J Virol. 1987;61:1747–1750. doi: 10.1128/jvi.61.5.1747-1750.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R, Schoonakker A, Harley EH. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J Virol. 1991;65:1600–1604. doi: 10.1128/jvi.65.3.1600-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I, Khoury G, Dhar R. The genome of human papovavirus BKV. Cell. 1979;18:963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Sinibaldi L, Goldoni P, Pietropaolo V, Longhi C, Orsi N. Involvement of gangliosides in the interaction between BK virus and Vero cells. Arch Virol. 1990;113:291–296. doi: 10.1007/BF01316682. [DOI] [PubMed] [Google Scholar]
- Sinibaldi L, Viti D, Goldoni P, Cavallo G, Caroni C, Orsi N. Inhibition of BK virus haemagglutination by gangliosides. J Gen Virol. 1987;68:879–883. doi: 10.1099/0022-1317-68-3-879. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Tremolada S, Delbue S, Castagnoli L, Allegroni S, Miglio U, Boldorini R, Elia F, Gordon J, Ferrante P. Mutations in the external loops of BK virus VP1 and urine viral load in renal transplant recipients. J Cell Physiol. 2010;222:195–199. doi: 10.1002/jcp.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Sugimoto C, Zhong S, Homma Y. Evolution of the BK polyomavirus: epidemiological, anthropological and clinical implications. Rev Med Virol. 2009;19:185–199. doi: 10.1002/rmv.613. [DOI] [PubMed] [Google Scholar]