Summary
Accurate chromosome segregation requires carefully regulated interactions between kinetochores and microtubules, but how plasticity is achieved to correct diverse attachment defects remains unclear. Here, we demonstrate that Aurora B kinase phosphorylates three spatially distinct targets within the conserved outer kinetochore KNL1/Mis12 complex/Ndc80 complex (KMN) network, the key player in kinetochore-microtubule attachments. The combinatorial phosphorylation of the KMN network generates graded levels of microtubule binding activity, with full phosphorylation severely compromising microtubule binding. Altering the phosphorylation state of each protein causes corresponding chromosome segregation defects. Importantly, the spatial distribution of these targets along the kinetochore axis leads to their differential phosphorylation in response to changes in tension and attachment state. In total, rather than generating exclusively binary changes in microtubule binding, our results suggest a mechanism for the tension-dependent fine tuning of kinetochore-microtubule interactions.
Keywords: Mitosis, Kinetochore, Microtubule, Phosphorylation, Chromosome Segregation
Introduction
To ensure the accurate and equal distribution of genetic material to the daughter cells during mitosis, chromosomes bi-orient on the mitotic spindle under tension from dynamic spindle microtubules. The kinetochore links microtubules to centromeric DNA and forms a large macromolecular assembly that is essential for chromosome segregation (Welburn and Cheeseman, 2008). Proteomic studies have shown that the kinetochore is organized into discrete sub-complexes that play distinct functions within the kinetochore (Cheeseman and Desai, 2008), but which must ultimately be integrated to mediate chromosome segregation. Beyond its role as a structural linker between the centromere and microtubules, the kinetochore also couples the energy from depolymerizing microtubules to chromosome movement and monitors the fidelity of microtubule-kinetochore attachments until bi-orientation is achieved.
The rapid and reversible regulation of kinetochore function by post-translational modifications is critical to monitor and correct kinetochore-microtubule attachments. In particular, Aurora B kinase, a component of the conserved chromosomal passenger complex (Ruchaud et al., 2007), is a key player in regulating kinetochore function. In budding yeast, Aurora B has been proposed to generate unattached kinetochores in response to a lack of inter-kinetochore tension (Pinsky et al., 2006; Tanaka et al., 2002). In contrast, rather than becoming fully unattached, mal-oriented vertebrate kinetochore-microtubule attachments appear to become destabilized upon Aurora B activation allowing chromosomes to move closer to the spindle poles before bi-orientation occurs (Lampson et al., 2004). During the course of a normal cell division and in response to mitotic errors, Aurora B must potentially correct a range of attachment defects. However, it remains to be determined whether Aurora B acts as a switch to generate binary on/off changes in microtubule binding state or whether it can function to modulate kinetochore-microtubule attachments.
To control kinetochore-microtubule attachments, Aurora B must ultimately regulate the key components of the outer kinetochore. The core of the kinetochore-microtubule interface is provided by the conserved 9 subunit KNL1/Mis12 complex/Ndc80 complex (KMN) network (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006; Wei et al., 2007). Recent work has demonstrated that the microtubule binding activity of the Ndc80 complex is controlled by Aurora B phosphorylation of an N-terminal 100 amino acid unstructured region (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006). This phosphorylation likely acts by generating negative charges that disrupt the electrostatic interactions between Ndc80 and the acidic C-terminal tails of microtubules (Guimaraes et al., 2008; Miller et al., 2008). While previous work has concentrated primarily on the regulation of the Ndc80 complex, the microtubule binding activity of the KMN network also requires important contributions from KNL1 and the Mis12 complex (Cheeseman et al., 2006).
Here, we demonstrate that Aurora B phosphorylates multiple subunits of the KMN network that additionally includes the Mis12 complex subunit Dsn1 and an N-terminal microtubule binding domain of KNL1. Phosphorylation of all three targets is necessary to inactivate the microtubule binding activity of the KMN network, while phosphorylation of a subset of proteins results in intermediate activity states. Disrupting these phosphorylation events in vertebrate cells causes corresponding defects in chromosome segregation. Importantly, we demonstrate that Aurora B phosphorylation of the KMN network responds to kinetochore attachment state, and that the phosphorylation of these spatially distinct targets occurs differentially during chromosome alignment. These results provide a mechanism to modulate the function of the kinetochore-microtubule interface by generating multiple activity states, rather than simply a binary on/off situation, and provide for the fine tuning of chromosome segregation by Aurora B.
Results
Aurora B targets multiple proteins within the KMN network
The core of the kinetochore-microtubule interface is provided by the conserved 9 subunit KNL1/Mis12 complex/Ndc80 complex (KMN) network (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006; Wei et al., 2007). While previous work has focused exclusively on the regulation of the Ndc80 complex by Aurora B (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006; Guimaraes et al., 2008; Miller et al., 2008), KNL1 and the Mis12 complex also contribute to the synergistic microtubule binding activity of the KMN network (Cheeseman et al., 2006). Thus, it is critical to analyze the regulation of the complete KMN network. To determine whether Aurora B has additional KMN network substrates, we used two complementary strategies in combination with mass spectrometry. First, to define the physiologically relevant phosphorylation sites, we isolated the KMN network by immunoprecipitation from HeLa cells. Second, to identify sites that are specific to Aurora B kinase, we phosphorylated the reconstituted human Mis12 complex (Kline et al., 2006) and the C. elegans KNL-1/MIS-12 complex (Cheeseman et al., 2006) in vitro with the budding yeast Aurora B kinase Ipl1. In each case, we analyzed the samples by mass spectrometry both with and without phosphopeptide enrichment (Supplemental Fig. 1A). These approaches identified four phosphorylation sites in the Mis12 complex subunit hDsn1, two of which are conserved in the C. elegans Dsn1 counterpart KNL-3, and two conserved sites at the N-terminus of KNL-1 as targets of Aurora B (Supplemental Fig. 1B and C). In vitro phosphorylation of the human KNL1 N-terminus (for which we are unable to purify full length protein) also identified similar Aurora B phosphorylation sites. We confirmed these sites by conducting kinase assays with non-phosphorylatable mutants (denoted as ProteinSA) of Dsn1/KNL-3 or KNL-1 (Supplemental Fig. 1D). In combination with the results described below, these data demonstrate that KNL1 and Dsn1 are conserved targets of Aurora B.
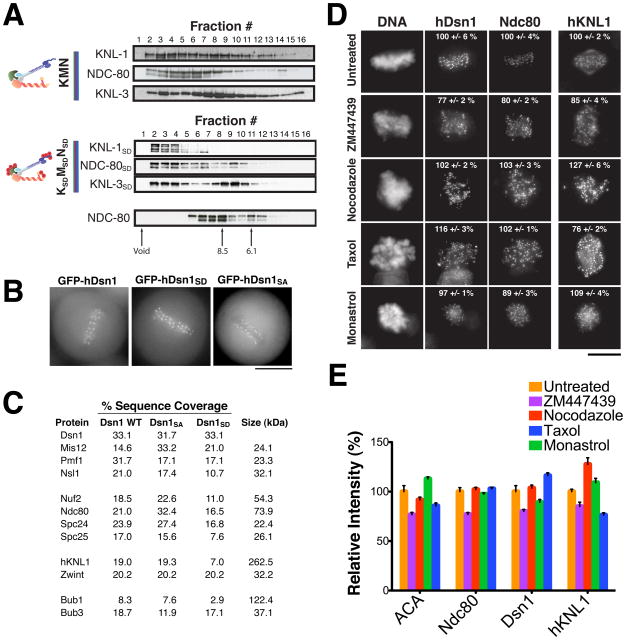
Aurora B phosphorylation does not affect the associations within the KMN network or its assembly at kinetochores
Phosphorylation of these additional Aurora B targets may regulate the kinetochore-microtubule interface directly. Alternatively, as suggested by recent work in Xenopus extracts (Emanuele et al., 2008), phosphorylation may contribute to kinetochore assembly by altering protein-protein interactions. To assess the role of phosphorylation on KMN network assembly, we mutated the complete complement of phosphorylation sites in each Aurora B target to aspartate to mimic constitutive phosphorylation (denoted as ProteinSD). We were able to reconstitute and assemble the complete KMN network in vitro regardless of its phosphorylation state (Fig. 1A; Supplemental Fig. 2A; (Cheeseman et al., 2006). Since the Mis12 complex has an established role in kinetochore assembly (Kline et al., 2006), we tested the specific effect of hDsn1 phosphorylation on its localization and associations. Clonal cell lines stably expressing GFP-hDsn1 Aurora B phosphomimetic and non-phosphorylatable mutants localized to kinetochores identically to wild type hDsn1 (Fig. 1B). We note that this contrasts with previous results in which transiently expressed hDsn1 Ser100/Ser109 to alanine double mutants did not localize to kinetochores (Yang et al., 2008). In addition to showing similar localization, one step immunoprecipitations of GFPLAP-tagged hDsn1, hDsn1SA and hDsn1SD each isolated the complete KMN network as well as the outer kinetochore proteins Zwint, Bub1, and Bub3 (Fig. 1C). We also found that conditional replacement Dsn1 phosphorylation site mutants in chicken DT40 cells did not affect kinetochore assembly (see below). Finally, treatment with the Aurora B inhibitor ZM447439 only modestly affected the kinetochore localization of the representative KMN network subunits hKNL1, hDsn1 and Ndc80/HEC1 (Fig. 1D and E) highlighting that the outer kinetochore can still maintain a supramolecular assembly in the absence of Aurora B activity. In contrast, treatment with ZM447439 completely eliminated phospho-Histone H3 staining, an established Aurora B substrate (Supplemental Fig. 2B). In total, our analysis suggests that Aurora B phosphorylation of the KMN network does not play a significant role in the assembly or maintenance of the outer kinetochore in human cells.
Figure 1. Phosphorylation of the KMN network by Aurora B does not affect kinetochore assembly.
(A) In vitro assembly properties of the C. elegans KMN and KSDMSDNSD network. The KNL-1 and MIS-12 complex were co-purified and incubated with the NDC80 complex, followed by gel filtration chromatography of the sample. Elution profiles of the KMN network (top), the KSDMSDNSD network (middle), and the NDC-80 complex alone (bottom) are shown. The KNL-3, KNL-1 and NDC-80 components were detected by Western blotting. Also see Figure S2. (B) Images of live mitotic cells from clonal human cell lines stably expressing moderate amounts of GFP-hDsn1, GFP-hDsn1SA and GFP-hDsn1SD. Each fusion protein localizes to kinetochores throughout mitosis. Scale bar, 10 μm. (C) Purification of GFP-hDsn1, GFP-hDsn1SA and GFP-hDsn1SD and their associated proteins using a one-step immunoprecipitation to directly isolate the tagged hDsn1 from stable clonal cell lines. Percent sequence coverage from the mass spectrometric analysis of these samples for each KMN network component and associated spindle checkpoint components is indicated. (D) Immunofluorescence images acquired using antibodies against Hec1 (hNdc80), hKNL1 and hDsn1 in HeLa cells under control conditions or after 3 hr treatment with nocodazole, taxol, monastrol, or ZM447439. Scale bar, 10 μm. Also see Figure S2. (E) Bar graph showing the average ratio of kinetochore fluorescence intensities (percent relative to control) for the indicated antibodies/conditions. Error bars represent standard error. The fluorescence intensity of 30–80 kinetochores per cell was measured for 5 to 10 cells per condition.
Phosphorylation of the KMN network results in graded changes in microtubule binding activity
We next sought to define the contribution of the identified phosphorylation sites in the KMN network to regulating its microtubule binding activity in vitro. For these experiments, we utilized C. elegans KMN network, in which the human hDsn1 subunit of the Mis12 complex corresponds to C. elegans KNL-3 (Fig. 2A), and which can be reconstituted in vitro (Cheeseman et al., 2006) allowing us to dissect the biochemical properties of the complete KMN network. To date, it has not been possible to reconstitute full length human hKNL1, which is greater than 265 kDa. However, where possible, we confirmed these results using the corresponding human proteins. Due to the established cooperative microtubule binding behavior of the KMN network constituents (Cheeseman et al., 2006), assays were conducted with 50 nM input protein to maximize any differences in binding behavior.
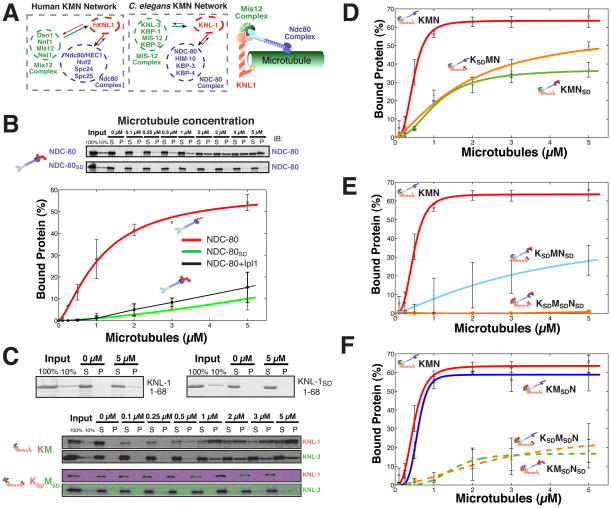
Figure 2. Combinatorial phosphorylation of the KMN network generates multiple levels of microtubule binding activity.
(A) Left, diagrams and cartoon model showing the components and organization of the human and C. elegans KMN networks. (B) Top, gel showing microtubule co-sedimentation of 50 nM NDC-80 complex and NDC-80SD complex. Bottom, graph showing the average percent protein bound relative to microtubule concentration for the wild type NDC-80 complex, NDC-80 complex phosphorylated by Ipl1 (data from (Cheeseman et al., 2006)), and the NDC-80SD complex. (C) Co-sedimentation assays with the N-terminus of C. elegans KNL-1 and a KNL-1SD and the full length KNL-1/MIS-12 complex and KNL-1SD/MIS-12SD complex. (D, E, and F) Graphs showing the microtubule binding activity for 50 nM input protein of the indicated KMN network mutants. The points were fitted with MATLAB using a modified Hill equation. Error bars represent the standard deviation. Representative gels for the data shown in these graphs and microtubule binding data for a KSAMSANSA mutant is included in Supplemental Fig. 3.
We previously demonstrated that in vitro phosphorylation of the Ndc80 complex by Aurora B significantly reduced its microtubule binding affinity (Cheeseman et al., 2006). Importantly, the affinity of phosphomimetic mutant NDC-80SD complex for microtubules is virtually identical to that of NDC-80 complex that has been phosphorylated in vitro by Aurora B (Fig. 2B), providing an important tool to dissect the function of this phosphorylation. However, while phosphorylated Ndc80 complex displays negligible microtubule binding activity on its own, this phosphorylation is not sufficient to prevent binding of the KMN network to microtubules. The KMNSD network containing NDC-80SD has an apparent microtubule binding affinity of 1 μM, nearly half that of wild type KMN network (KD = 0.6 μM; Fig. 2D). Thus, while Ndc80 phosphorylation reduces binding of the KMN network to microtubules, this affinity remains physiologically relevant when compared to other established microtubule interacting proteins.
Within the KMN network, KNL-1 also contributes to microtubule binding, but the region responsible for this activity was unknown. We found that the 68 amino acids at the N-terminus of KNL-1 are necessary (not shown) and sufficient (Fig. 2C) for binding to microtubules. Interestingly, the Aurora B phosphorylation sites that we identified in KNL1 are present within this region suggesting that they may regulate microtubule interactions. Indeed, phosphomimetic mutants abolished the binding of the C. elegans KNL-1 (Fig. 2C) or human hKNL1 (Supplemental Fig. 3A) N-terminal fragments, or full length C. elegans KNL-1 (Fig. 2C; Supplemental Fig. 3C) to microtubules. However, as with the Ndc80 complex, while phosphorylation of KNL-1 abolished its affinity for microtubules on its own, the KSDMN network containing KNL-1SD displayed only a moderate reduction in microtubule binding (KD = 2 μM; Fig. 2D).
Since individual Ndc80 or KNL-1 phosphorylation did not eliminate microtubule binding, we next tested whether Aurora B phosphorylation of multiple proteins within KMN network could abrogate binding. Surprisingly, the KSDMNSD network in which both established microtubule binding components are perturbed, still binds to microtubules, albeit with low apparent affinity (KD = 4.9 μM; Fig. 2E). Finally, since the MIS-12 complex is also phosphorylated by Aurora B, we tested the fully phosphomimetic KMN network. The KSDMSDNSD network, despite its capacity to assemble normally (see Fig. 1), did not associate with microtubules under the conditions analyzed (Fig. 2E). Importantly, the KSAMSANSA network in which each phosphorylation site was mutated to alanine did not affect microtubule binding (Supplemental Fig. 3D, E) demonstrating that simply changing these residues to another amino acid does not affect KMN network activity in vitro.
The effect of MIS-12 complex phosphorylation on the microtubule binding activity of the KMN network is surprising given that this complex does not associate directly with microtubules (Cheeseman et al., 2006). To define the contribution of MIS-12 complex phosphorylation, we tested a series of mutant complexes. The KMSDN network, in which only the KNL-3 subunit of the MIS-12 complex is mutated to mimic constitutive phosphorylation, displayed a similar binding affinity for microtubules as the wild type KMN network (Fig. 2F). However, a significant reduction in affinity was observed when the MIS-12SD complex was combined with either KNL-1SD or the NDC-80SD complex (KSDMSDN and KMSDNSD have respective KD = 5.3 and 4.6 μM, compared to 2.1 and 1 μM for KSDMN and KMNSD; Fig. 2F). Therefore, phosphorylation of the MIS-12 complex sensitizes the microtubule binding activity of the KMN network such that the additional phosphorylation of at least one microtubule binding component creates a dramatic change in activity. In total, this biochemical analysis reveals important contributions from the phosphorylation of Ndc80, Dsn1, and KNL1 to regulating microtubule interactions. These results provide a mechanism to severely compromise the activity of the complete KMN network through the combined phosphorylation of each subunit, but also provide a way to generate graded changes in microtubule binding activity through phosphorylation of a subset of proteins.
Phosphorylation of the KMN network is critical for regulating chromosome segregation in vivo
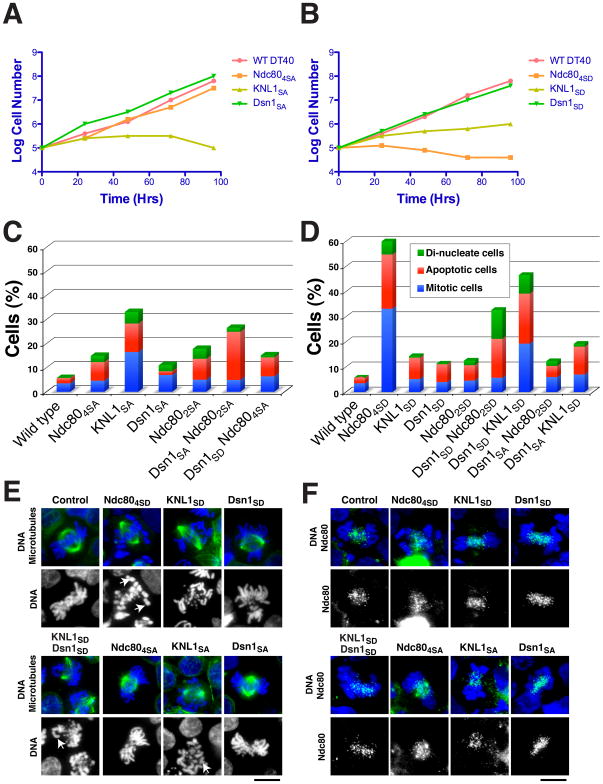
To determine whether the combinatorial regulation of the KMN network by Aurora B observed in vitro is critical to controlling kinetochore-microtubule attachments in vivo, we next investigated the effect of the phosphomimetic (to aspartate) and non-phosphorylatable (to alanine) mutants of Dsn1, KNL1 and Ndc80 in vertebrate cells. We predicted both types of mutations in critical phosphorylation sites would affect chromosome segregation since preventing the inactivation of inappropriately attached kinetochores by Aurora B (in the non-phosphorylatable mutant), or constitutively inactivating this attachment (in the phosphomimetic mutant) would both result in severe consequences. For these experiments, we used chicken DT40 cells that are well suited to such studies due to high rates of homologous recombination (Fukagawa et al., 2001), allowing us to generate conditional replacement alleles. Consistent with previous work (DeLuca et al., 2006; Guimaraes et al., 2008), mutation of multiple phosphorylation sites in ggNdc80 (Ser5, Ser15, Ser16, Ser44) resulted in mitotic defects (Fig. 3). The viability of the phosphomimetic Ndc80 mutant was severely compromised (Fig. 3B), while the non-phosphorylatable mutant was still viable (Fig. 3A), but showed an increase in apoptotic cells (Fig. 3C). Thus, while altering the regulation of Ndc80 affects proper chromosome segregation, these phenotypes suggest that additional regulation occurs downstream of Aurora B. Consistent with this, previous work on budding yeast Ndc80 phosphorylation demonstrated that S to A mutations of the equivalent sites was non-essential (Akiyoshi et al., 2009; Kemmler et al., 2009) arguing for the existence of other key targets.
Figure 3. The phosphorylation of the KMN network has synergistic effects in vivo.
(A and B) Growth curve analysis of cellular viability for the non-phosphorylatable (A) or phosphomimetic (B) KMN mutants in chicken DT40 cells. Tetracycline was added at time 0 to the culture to eliminate expression of the wild type protein. The number of cells not stained with trypan blue was counted. (C and D) Summary of phenotypes for complementation experiments in Ndc80, KNL1 or Dsn1 deficient DT40 cells expressing the non-phosphorylatable (C) or phosphomimetic (D) mutants. In cell lines that express Dsn1 in combination with KNL1 or Ndc80 phospho-mutants, the endogenous Dsn1 is also present. 500–1000 cells were counted for each cell line. (E) Chromosome morphology and mitotic spindle shown by DNA staining using DAPI (blue) and α-tubulin staining (green) in the Ndc80, KNL1 or Dsn1 deficient DT40 cells expressing the non-phosphorylatable or phosphomimetic mutants. White arrowheads show kinetochore attachment defects. (F) Outer kinetochore assembly state in the phospho-mutants shown by DNA staining using DAPI (blue) and Ndc80 staining (green). Scale bars: 10 μm.
The ggKNL1SD and ggKNL1SA mutants both displayed a severe decrease in viability (Fig. 3A, B) and an accumulation of cells in mitosis or undergoing apoptosis (Fig. 3C, D). In contrast, consistent with our biochemical analysis (Fig. 2), ggDsn1 phosphorylation site mutants were viable (Fig. 3A, B) and displayed only mild defects (Fig. 3C, D). In addition, none of the phosphomimetic or phosphoinhibitory mutants in Ndc80/Hec1, KNL1, or Dsn1 altered kinetochore assembly based on the localization of Ndc80 (Fig. 3F). Thus, disrupting the phosphorylation of individual KMN network subunits results in defects that are consistent with the roles in regulating kinetochore-microtubule interactions that we defined biochemically. However, while there are not dramatic changes in kinetochore assembly in these mutants, we note that in cells it is possible that these phosphorylation sites affect multiple aspects of KMN network function in addition to microtubule interactions. This is particularly true for KNL1, which has been reported to interact with multiple binding partners (see Cheeseman and Desai, 2008).
Although the ggDsn1 mutants did not show dramatic defects in vivo, based on our biochemical analysis we predicted that these would synergize with KNL1 or Ndc80 phosphorylation site mutants. We therefore analyzed ggKNL1SD and ggNdc80SD mutants in the presence of exogenously expressed ggDsn1SD mutant, or ggNdc80SA in combination with ggDsn1SA to assemble doubly mutant KMN network. In each case, we observed a significant increase in chromosome alignment defects (Fig. 3E) and an accumulation of cells undergoing a mitotic arrest or apoptosis (Fig. 3C, D) relative to cells containing the Ndc80 mutants or KNL1 mutant alone. In contrast, we did not observe a similar increase in defective cells using ggDsn1SA in combination with phosphomimetic ggKNL1SD or ggNdc80SD mutants, or vice versa (Fig. 3C, D). These results imply that Dsn1 has a key role in coordinating the effect of Aurora B phosphorylation of the microtubule binding Ndc80 and KNL1 subunits to regulate the KMN network.
Tension-dependent differential phosphorylation of the KMN network by Aurora B
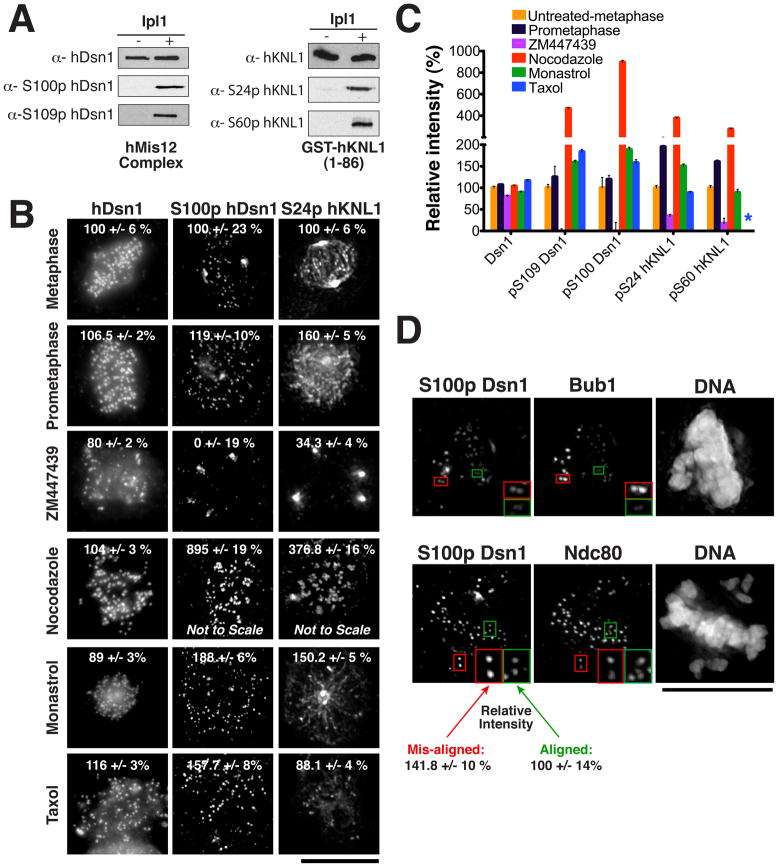
The biochemical and functional analyses described above demonstrate that phosphorylation of each KMN network subunit contributes to regulating microtubule binding and chromosome segregation. Current models suggest that Aurora B phosphorylates its targets when inter-kinetochore tension is low, but this has not been demonstrated for relevant endogenous outer kinetochore substrates. To define the response KMN network phosphorylation, we generated antibodies that specifically recognize the phosphorylated form of Ser100 or Ser109 in hDsn1 and Ser24 or Ser60 in hKNL1 (Fig. 4A). Immunofluorescence using these antibodies in HeLa cells revealed kinetochore staining that was severely reduced by treatment with the Aurora B inhibitor ZM447439 (Fig. 4B and C). In contrast, antibodies that detect Ndc80, hDsn1, and hKNL1 regardless of phosphorylation state displayed only minor changes in levels under all conditions analyzed (Fig. 4B, C; Fig. 1).
Figure 4. Aurora B phosphorylation of the KMN network occurs under conditions where attachments must be corrected.
(A) Western blots validating the phosphospecific hDsn1 and hKNL1 antibodies with recombinant protein using the indicated conditions. (B) Immunofluorescence images acquired using the indicated antibodies in untreated HeLa cells or in cells treated with nocodazole, taxol, monastrol or ZM447439 for 3 hrs. Numbers indicate the average percent quantitated fluorescence (+/− sem) relative to control treated metaphase cells. Images are scaled equivalently except for the nocodazole treated samples. Phospho-antibodies show spindle pole background staining that is not eliminated by ZM447439 treatment. (C) Ratio of fluorescence intensities for the images in (B) expressed as a percentage relative to controls. Asterix indicates that intensity in taxol treated cells for S60p hKNL1 was not determined due to extensive spindle background in this condition. (D) Immunofluorescence images acquired using the indicated antibodies in HeLa cells treated with 6 ng/ml Nocodazole to generate cells with both aligned (green boxes) and mis-aligned (red boxes) chromosomes. The checkpoint protein Bub1 localizes preferentially to unattached kinetochores or those that are not under tension. Phospho-Dsn1 shows a statistically significant increase (95% confidence interval in a T-test) on mis-aligned chromosomes (125 kinetochores analyzed) relative to aligned chromosomes (240 kinetochores analyzed). Scale bars, 10 μm.
To evaluate the targeting of hDsn1 and hKNL1 phosphorylation, we next used phosphospecific antibodies to examine cells with distinct kinetochore-microtubule attachment states. In cells with chromosomes aligned at the metaphase plate, we detected low hDsn1 and hKNL1 phosphorylation at all kinetochores. Cells in prometaphase showed slightly elevated and uniform hKNL1, and to a lesser extent hDsn1, phosphorylation. However, when cells were treated with nocodazole to depolymerize microtubules and create unattached kinetochores, we observed a dramatic increase in both hDsn1 and hKNL1 phosphorylation (Fig. 4B, C). This increased phosphorylation was dependent upon Aurora B since treatment with both nocodazole and ZM447439 eliminated this phosphorylation (not shown). When cells were treated with taxol, to stabilize microtubules and reduce inter-kinetochore tension, or the Eg5 inhibitor monastrol, to create mono-orientated sister chromatids, Aurora B phosphorylation of hDsn1 was also increased, but not to the same extent as nocodazole treatment (Fig. 4B, C). Under these conditions, hKNL1 phosphorylation was only modestly increased (Fig. 4B, C). Finally, we treated cells with low levels of nocodazole to generate cells with only a few mis-aligned chromosomes. In these cases, we observed a statistically significant increase (>95% confidence based on a independent T test) in the amount of phospho-Dsn1 staining on the mis-aligned chromosomes relative to the aligned chromosomes (Fig. 4D). The increase was comparable to the level of phosphorylation observed on prometaphase chromosomes (Fig. 4B, C). Therefore, phosphorylation of hDsn1 and hKNL1 by Aurora B occurs at basal levels during mitosis, but increases in response to defects in kinetochore-microtubule attachments and intra-kinetochore tension.
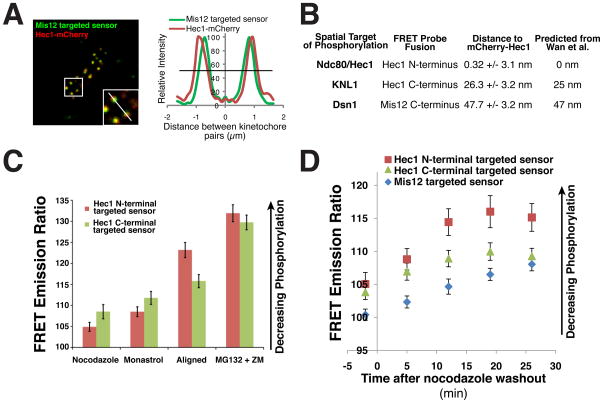
The phospho-specific antibodies allowed us to define the behavior of the endogenous phosphorylation events for Dsn1 and KNL1, but do not allow a direct comparison between the phosphorylation of each protein. hDsn1, hKNL1, and Ndc80/HEC1 are physically associated within the KMN network, but they are positioned at spatially distinct regions within the kinetochore (Wan et al., 2009). Current models suggest that tension decreases phosphorylation by Aurora B by separating the kinase, which is targeted to the inner centromere, from its targets at kinetochores (Liu et al., 2009). However, it was unclear whether small spatial differences within the kinetochore could result in distinct phosphorylation by Aurora B. Because the KMN network covers a distance of ~50 nm along the inner-outer kinetochore axis, distinct substrates may be differentially phosphorylated in response to changes in tension. To test this, we used the established FRET probe (Liu et al., 2009) to monitor the Aurora B phosphorylation at positions within the kinetochore that correspond to the phosphorylation sites we defined in Dsn1 (fusion with Mis12), KNL1 (fusion to the C-terminus of Hec1), and Ndc80 (fusion to the N-terminal microtubule binding domain of Hec1). Analysis of the relative spatial position of these probes demonstrated that they were indeed positioned as expected based on previous work (Fig. 5A and 5B; (Wan et al., 2009)). Each probe displayed increased phosphorylation (as measured by a lower FRET emission ratio) in nocodazole treated cells compared to cells with metaphase aligned chromosomes (Fig. 5C and 5D). However, similar to the analysis using phospho-specific antibodies, residual phosphorylation was still observed on aligned chromosomes as treatment with ZM447439 further decreased phosphorylation (Fig. 5C and 5D). Importantly, the N-terminal HEC1 targeted FRET probe displayed much lower phosphorylation on aligned chromosomes than the C-terminal targeted FRET probe (which mimics KNL1 position), suggesting that Ndc80 phosphorylation is more strongly reduced as tension is established relative to hKNL1 and hDsn1 phosphorylation. Time-lapse analysis following nocodazole washout demonstrated that the position of each FRET probe affected the time required to reach maximal dephosphorylation with the N-terminal Ndc80 targeted probe changing most quickly and the Mis12 targeted probe changing the most slowly (Fig. 5D). Thus, although Dsn1, KNL1, and Ndc80 are all targets of Aurora B, due to their distinct spatial positions within the kinetochore, they are likely to be differentially regulated in a manner that is dependent on the kinetochore-microtubule attachment state.
Figure 5. Differential phosphorylation of the KMN network under distinct kinetochore attachment states modulates outer kinetochore microtubule binding activity.
(A) Image and linescan showing position of mCherry-Hec1 relative to a Mis12 targeted FRET sensor. (B) Table showing the average relative position of an N-terminally tagged mCherry-Hec1 (+/− SEM) to the indicated FRET probe tested as illustrated in (A). At least 5 different cells were imaged for each probe with at least 10 kinetochore pairs per cell. These positions agree closely with those determined by (Wan et al., 2009). (C) Average FRET emission ratio (+/− SEM) for an artificial Aurora B substrate FRET probe (Liu et al., 2009) targeted to kinetochores using either the Ndc80/Hec1 N-terminus or C-terminus in the indicated conditions (n > 20 cells per condition). (D) Graph as in (C) for FRET probe targeted with hMis12 or the Ndc80/HEC1 N or C-terminus for time points following the wash out of 30 ng/ml nocodazole. For each time point, >20 kinetochores were measured in >20 cells.
Overall, these results demonstrate that Dsn1, KNL1 and Ndc80 are key targets of Aurora B at the outer kinetochore and that phosphorylation of these Aurora B sites is necessary for correct chromosome alignment and segregation. Altering the phosphorylation state of each individual protein has different effects on microtubule binding activity and chromosome segregation, and these phosphorylation events occur distinctly in response to defects in kinetochore tension providing a mechanism to differentially regulate the outer kinetochore.
Discussion
Aurora B phosphorylates multiple targets in the KMN network to fully inactivate its microtubule-binding activity
Aurora B kinase is required to correct microtubule-kinetochore attachment defects (Hauf et al., 2003; Pinsky et al., 2006), but the precise mechanism of this regulation was unclear. Prior to our studies, the N-terminus of Ndc80 was identified as the major conserved substrate of Aurora B at the outer kinetochore (Cheeseman et al., 2006; Ciferri et al., 2008; DeLuca et al., 2006). While Ndc80 is an important component of the kinetochore-microtubule interface, we demonstrate that phosphorylation of Ndc80 is not sufficient to inactivate the microtubule binding activity of the KMN network in vitro. Similarly, non-phosphorylatable mutants of Ndc80 in chicken cells are viable. In both cases, this suggests that regulation of the kinetochore-microtubule interface by Aurora B requires additional phosphorylation events.
We have identified the microtubule binding domain of KNL-1 and the Dsn1/KNL-3 subunit of the Mis12 complex as important additional substrates of Aurora B. Phosphorylation of these three KMN network components does not disrupt protein-protein interactions within the KMN network or prevent kinetochore assembly. In contrast, individual phosphorylation of Ndc80, KNL1, or Dsn1/KNL-3 results in distinct changes in the microtubule binding affinity of the KMN network, while simultaneous phosphorylation of all three subunits inactivates microtubule binding. These results provide a mechanism to completely inactivate the microtubule binding activity of the KMN network, the core conserved component of the kinetochore-microtubule interface, but also provide a way to modulate its microtubule binding properties to destabilize attachments without completely eliminating them.
Phosphorylation of Ndc80 and KNL1 by Aurora B specifically targets the microtubule binding domains of these proteins. In contrast, the Mis12 complex does not interact directly with the microtubules, but is required to synergize the microtubule binding of the Ndc80 complex and KNL-1 (Cheeseman et al., 2006). Phosphorylation of the Mis12 complex subunit Dsn1 alone does not affect the microtubule binding activity of the KMN network. However, phosphorylation of the Mis12 complex has a previously unidentified allosteric effect on KMN network activity in vitro when the KMN network is also phosphorylated on one of the microtubule-binding subunits. Similarly, in vertebrate cells, the Dsn1 phosphorylation sites are not essential, but cause increased defects when combined with mutations in KNL1 or Ndc80. Thus, Dsn1 phosphorylation sensitizes the KMN network to the phosphorylation of its microtubule binding subunits to dramatically reduce KMN network activity.
Fine tuning kinetochore-microtubule attachments
Previous work demonstrated that Aurora B activity is critical for the correction of defective kinetochore-microtubule attachments, including those that occur during the course of a normal cell division. Models for the function of Aurora B have proposed two distinct states of kinetochore activity; an “on”-state where the kinetochore is dephosphorylated and is able to attach to microtubules, and an “off”-state where Aurora B phosphorylates kinetochore substrates to inactivate kinetochore-microtubule attachments. Previous work on the role of phosphorylation on protein function, including the regulation of Sic1 by CDK (Nash et al., 2001), indicates that multiple phosphorylation sites within a single protein function to generate a molecular switch-like behavior. The nature of the microtubule binding interface, the diverse errors in kinetochore-microtubule attachments that must be corrected, and the mechanisms by which this correction occurs would benefit from the ability to generate a switch-like behavior as well as a more graded response.
Our work supports the existence of an “off” state for KMN network function in which Ndc80, KNL1, and Dsn1/KNL-3 are all phosphorylated (Fig. 5). However, our work also provides for additional regulation states that would allow for the modulation of kinetochore-microtubule attachments depending on which subunits are phosphorylated. In vivo, this differential regulation could result in a distinct threshold of KMN network activation in response to various cellular stimuli. In addition to full activity and partial or complete inactivation of the KMN network, our work has suggested the potential existence of an additional state where only the Mis12 complex subunit Dsn1 is phosphorylated (Fig. 5). In this case, phosphorylation would not alter kinetochore-microtubule attachments, but the KMN network would be “primed” such that additional phosphorylation of either KNL1 or Ndc80 would result in a dramatic decrease in microtubule binding activity.
Recent work has suggested a spatial model for Aurora B function in which the proximity of a substrate to the kinase controls its phosphorylation state (Liu et al., 2009). Under conditions where there is a strong defect in intra-kinetochore tension, Aurora B would phosphorylate multiple substrates (including the entire KMN network) that are now spatially closer to the inner centromere where Aurora B resides (Fig. 5). In this case, complete phosphorylation of the KMN network would fully inactivate it and create an unattached kinetochore. As chromosome alignment is reestablished, the outer kinetochore is spatially repositioned by attachment of incoming microtubules to generate tension. Ndc80 is located the most distally from the inner centromere (Wan et al., 2009) and based on this position, it would be the first KMN network component to be dephosphorylated as tension is re-established. Consistent with this, we found that small spatial differences within the kinetochore result in differences in the dephosphorylation of an artificial Aurora B substrate. Dephosphorylation of Ndc80 would result in a KMN network where only KNL1 and Dsn1 were phosphorylated causing a partial, but not complete, reduction in microtubule binding activity. In total, these results provide a model that would allow the differential regulation of kinetochore-microtubule interactions by the coordinate regulation of spatially distinct targets at the outer kinetochore.
Experimental procedures
Protein Purification
The 4 subunit human Mis12 complex was co-expressed in BL21 E. coli and isolated using an established purification procedure (Kline et al., 2006). The C. elegans NDC-80 complex and the MIS-12 complex/KNL-1 were expressed separately and assembled in vitro as described (Cheeseman et al., 2006). GST fusions to the C. elegans KNL-1 (amino acids 1-68) and human hKNL1 (1-86) N-termini were expressed in pGEX-6P-1. Site-directed mutagenesis was performed using Quickchange (Stratagene).
Identification of phosphorylation sites
To identify endogenous phosphorylation sites, GFPLAP tagged Mis12 was isolated from HeLa cells in the presence of phosphatase inhibitors (20 mM b-glycerophosphate, 10 mM Na pyrophosphate, 5 mM Na azide, 10 mM NaF, 0.4 mM Na orthovanadate). To identify sites directly phosphorylated by Aurora B in vitro, 1 mg recombinant yeast GST-Ipl1 was incubated with 2–4 mg of recombinant C. elegans MIS-12 complex and KNL-1, human Mis12 complex, or the hKNL1 N-terminus for 30 minutes in presence of 100 nM ATP. For radioactivity kinase assays, reactions were performed with the addition of 1 mCi ATP-gP32. Phosphorylation sites within the KMN network were identified by mass spectrometry using an LTQ XL Ion trap mass spectrometer (Thermo) using MudPIT and SEQUEST software as described previously (Washburn et al., 2001). Titanium oxide and IMAC procedures were also used to enrich for phosphopeptides (Cantin and Yates, 2004).
Microtubule binding assays
Microtubule binding assays were performed as described (Cheeseman et al., 2006). MATLAB and least square refinement was used to fit the data with the modified Hill equation (Miller et al., 2008).
Immunofluorescence
HeLa cells were incubated for 3 hours with drugs at the following concentrations: monastrol: 100 μM, nocodazole: 0.2 μg/ml, ZM447439: 2 μM, taxol: 1 μM. For treatment of the cells with low levels of nocodazole, 6ng/ml was used. Immunofluorescence in human cells was conducted as previously described (Kline et al., 2006) using antibodies against microtubules (DM1α; Sigma), phospho histone H3 (Upstate), mouse anti-HEC1 (9G3, Abcam), rabbit α-hDsn1 and α-hKNL1 (Kline et al., 2006), and human anti-centromere antibodies (ACA antibodies, Inc., Davis, CA). The phospho-specific antibodies against Ser100 and Ser109 in hDsn1 and Ser24 and Ser60 in hKNL1 were generated by Abgent against immunizing phosphorylated peptides 95SWRRA Sp MKETN105 and 104TNRRK Sp LHPIH114 for Dsn1 and 48NALRNKKNSRRV Sp FADTIKVFQT70 and 11TNRRKERPVRRRH Sp SILKPPRSPLQDHC38 for hKNL1. All rabbit antibodies were used at 1 μg/ml. DNA was visualized using 10 μg/ml Hoechst. Images were acquired on a DeltaVision Core deconvolution microscope (Applied Precision) equipped with a CoolSnap HQ2 CCD camera. 30 to 40 z-sections were acquired at 0.2 μm steps using a 100x, 1.3 NA Olympus U-PlanApo objective without binning. Equivalent exposure conditions were used between controls and drug treated cells. To quantitate fluorescent intensity, individual kinetochores were selected from projections (selected based on co-localization with Ndc80/HEC1) and the integrated intensity was determined after subtracting the background fluorescence measured from adjacent regions of the cell using Metamorph. Fluorescent levels at kinetochores was normalized with respect to control cells. At least 5–10 cells were examined for each condition and antibody.
Analysis of phosphorylation site mutants in chicken DT40 cells
DT40 cells were cultured and transfected as described previously (Fukagawa et al., 2001). The corresponding Aurora B phosphorylation sites in the chicken proteins were selected based on the mapping of the endogenous phosphorylated sites in DT40 cells (our unpublished work) and the conservation of the Aurora B phosphorylation sites in humans and chicken. For Dsn1, these included Ser26, Ser30, Ser62, Ser79, Ser102 and Ser111. For KNL1, we selected Ser24 and Ser60. For Ndc80, we selected Ser5, Ser15, Ser16, and Ser44. Wild-type cDNAs were cloned under the CMV promoter. Each mutant cDNA was introduced into the corresponding conditional knockout DT40 cell line for Ndc80 (Hori et al., 2003), KNL-1 (Cheeseman et al., 2008), or Dsn1 (this study). Expression of the mutant proteins was assessed by Western blot in the presence of tetracycline (to deplete the endogenous protein).
To observe the phenotypes for the phosphorylation site mutants, 2 μg/ml tetracycline (Sigma) was added to deplete the endogenous protein. At 24 h following tetracycline addition, cells were collected and stained with anti-α-tubulin antibodies. Cell cycle state and chromosome morphology were observed and collected with a cooled EM CCD camera (Quntem, Roper scientific) mounted on an Olympus IX71 inverted microscope with a 100X objective lens. Subsequent analysis and processing of images were performed using Metamorph software (Roper scientific).
Phosphorylation sensor FRET Assays
Cells were incubated in a low concentration of nocodazole (30 ng/ml) in growth medium for 2–3 hours to release tension between sister kinetochores. To remove nocodazole, cells were washed three times with fresh L15 medium. The time of the first wash was recorded as time zero. One image was acquired before washing out nocodazole, and after the washout one image was acquired every 7 min, starting at t=5 min, to minimize photobleaching. For the FRET sensors, image acquisition and analysis was performed as described previously (Liu et al., 2009).
Supplementary Material
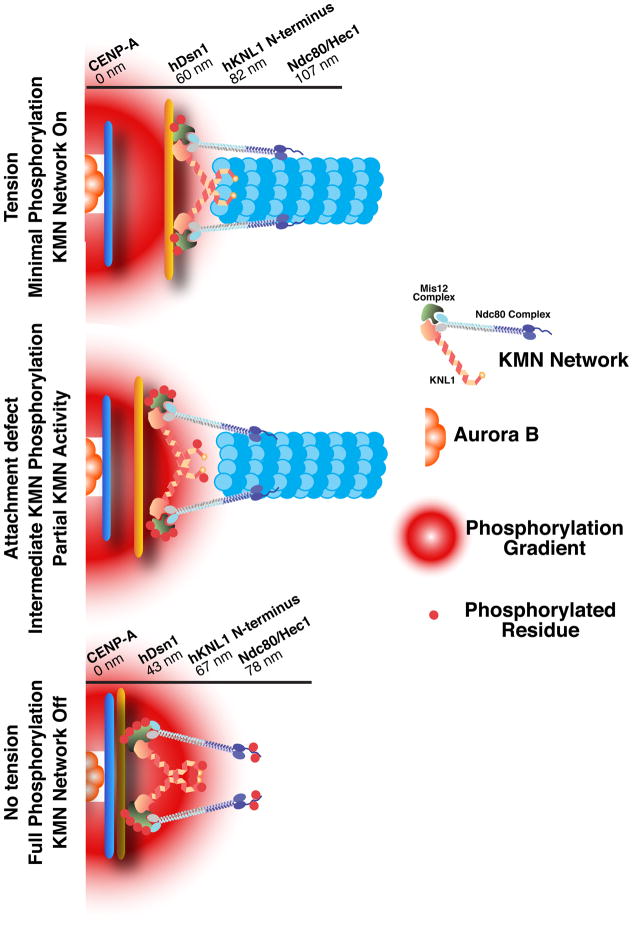
Figure 6. Differential phosphorylation of the KMN network under distinct kinetochore attachment states modulates outer kinetochore microtubule binding activity.
Cartoon depiction of the KMN network at kinetochores under attached (with tension between sister chromatids), intermediate (with partial tension), and unattached (with no tension) states. A gradient of Aurora B phosphorylation emanates from the inner centromere. When tension is present, the kinetochore is stretched relative to its resting state such that the Ndc80 complex is positioned distally to the inner centromere. When no tension is present, the Ndc80 complex is more proximal to the inner centromere, resulting in higher levels of phosphorylation. Values for the position of the indicated subunits within the kinetochore are from (Wan et al., 2009).
Acknowledgments
We thank Katya Grishchuk, Gerben Vader, Terry Orr-Weaver, Angelika Amon, Andreas Hochwagen, Defne Yarar, and members of the Cheeseman lab for helpful discussions and critical reading of the manuscript, Mayumi Takahashi for technical assistance, Sebastien Besson for help with MATLAB, Jennifer Deluca for sharing results prior to publication, and Arshad Desai for his generous support during the early stages of this project. This work was supported by awards from the Smith Family Foundation (I.M.C.), the Massachusetts Life Sciences Center (I.M.C.), and the Searle Scholars Program (I.M.C. and M.A.L.), grants from the National Institute Of General Medical Sciences (GM083988 to M.A.L. and GM088313 to I.M.C.), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Science and Technology (MEXT) of Japan to T.F. I.M.C. is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated Phosphorylation of the Ndc80 Kinetochore Protein in Saccharomyces cerevisiae. Genetics. 2009 doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin GT, Yates JR., 3rd Strategies for shotgun identification of post-translational modifications by mass spectrometry. J Chromatogr A. 2004;1053:7–14. doi: 10.1016/j.chroma.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K Complex Coordinately Direct Kinetochore Assembly in Vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing Chromosome Bi-Orientation by Spatial Separation of Aurora B Kinase from Kinetochore Substrates. Science. 2009 doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wu F, Ward T, Yan F, Wu Q, Wang Z, McGlothen T, Peng W, You T, Sun M, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.