SUMMARY
Neuronal activity-regulated gene expression has been suggested to be an important mediator of the ability of experience to effect long-lasting changes in the structure and function of the nervous system, but the activity-dependent component of gene transcription has never been selectively isolated and tested for its biological significance in the nervous system. Here we describe the generation of an animal that is specifically impaired in the neuronal activity-dependent transcription of the Bdnf gene. The introduction of a subtle knock-in mutation into the mouse Bdnf gene that blocks the ability of the activity-regulated factor CREB to bind Bdnf promoter IV results in an animal in which the sensory experience-dependent induction of Bdnf expression is disrupted in the cortex. Neurons from these animals form fewer inhibitory synapses in culture, have fewer spontaneous inhibitory quantal events in acute cortical slices, and exhibit reduced immunostaining for inhibitory presynaptic markers in the cortex. These results indicate a specific requirement for activity-dependent Bdnf expression in the development of inhibition in the cortex and demonstrate that the activation of gene expression in response to experience-driven neuronal activity has important biological consequences for the development and function of the nervous system.
INTRODUCTION
Experience-dependent changes in the nervous system occur throughout the lifetime of an animal. Sensory experience mediates the structural and functional refinement of developing neuronal circuits (Fox and Wong, 2005), and, in the mature brain, use-dependent modification of neuronal circuits underlies important adaptive functions of the nervous system, including learning, memory, and behavior (Kandel, 2001). The discovery that excitatory neurotransmitters can stimulate new gene transcription by triggering an influx of calcium into post-synaptic neurons (Greenberg et al., 1986; Kornhauser et al., 1990; Morgan et al., 1987) suggested a compelling mechanism by which stimulus-evoked neuronal activity might effect long-lasting changes in the structure and function of the nervous system. In the two decades since, significant advances have been made in elucidating many of the key activity-responsive transcriptional regulators, the calcium-dependent signaling pathways that couple extracellular stimuli to their activation, and the programs of gene expression that are activated in response to neuronal activity (Lanahan and Worley, 1998; West et al., 2002).
Activity-dependent gene expression occurs when synaptic activation triggers calcium influx at the membrane through ligand- and voltage-gated calcium channels, thereby initiating calcium-dependent signaling cascades that amplify and carry the signal to activate transcription factors in the nucleus. Several searches for activity-regulated genes have identified approximately 300 genes as being responsive to neuronal activity (Bartel et al., 1989; Lanahan and Worley, 1998; Lin et al., 2008; Nedivi et al., 1993), many of which fall into one of two categories. The first category includes genes that themselves encode for transcriptional regulators, exemplified by the classic immediate early gene c-fos (Greenberg et al., 1986) or the neuronal bHLH/PAS factor npas4 (Lin et al., 2008). The second category includes genes that encode for neuronally enriched proteins that are believed to function directly at the synapse and play important roles in neural development and plasticity, such as Bdnf (Poo, 2001).
There is no doubt that genes whose transcription is regulated by activity are important for the development and function of the brain. However, despite considerable progress in our understanding of the program of neuronal activity-regulated gene expression, direct evidence that the activity-dependent component of transcription per se is specifically important for nervous system development or function has been elusive. This fact is due in part to limitations in our ability to experimentally manipulate the activity-dependent component of transcription independent of the many other genetic programs regulating a given gene’s transcription. For instance, the role of an activity-regulated gene is most often investigated through loss-of-function studies that remove the gene entirely, independent of the stimulus state or cellular context. This approach can often lead to an outcome in which the functions of a gene that are dependent on its activity-dependent expression are conflated with, for instance, its trophic functions in supporting cell health or survival. In the case of one of the best-studied activity-regulated genes, brain-derived neurotrophic factor (Bdnf), studies in mice that completely lack Bdnf have demonstrated that it plays a key role in neuronal survival, differentiation, migration, and dendritic arborization (Ernfors et al., 1994; Jones et al., 1994; Schwartz et al., 1997). In addition, mice heterozygous for the loss of a Bdnf allele show deficits in synaptic development, function, and plasticity, as well as changes in body weight regulation, locomotor activity, and aggression (Abidin et al., 2008; Abidin et al., 2006; Carter et al., 2002; Kernie et al., 2000; Korte et al., 1995; Lyons et al., 1999; Patterson et al., 1996). Many of these diverse functions of Bdnf, in particular its functions in synaptic development, function, and plasticity, are thought to be regulated at least in part by its neuronal activity-dependent expression (Poo, 2001), but direct evidence for this idea is lacking.
In support of the idea that the many functions of Bdnf in the nervous system may be dependent on the tight temporal, spatial, and stimulus-specific regulation of Bdnf expression is the complexity of its gene structure. The Bdnf gene is comprised of at least eight distinct promoters that initiate transcription of multiple distinct mRNA transcripts, each of which contains an alternative 5’ exon spliced to a common 3’ coding exon that contains the entire open reading frame for the BDNF protein (Aid et al., 2007). Through the use of alternative promoters, splice donors, and polyadenylation sites, at least 18 distinct transcripts can be produced from the Bdnf gene (Figure 1A); remarkably, however, each of these Bdnf mRNAs encodes an identical BDNF protein. The functional significance of the transcriptional organization of the Bdnf gene remains cryptic, but one attractive hypothesis is that the production of many mRNAs that all encode the same protein product provides for multiple layers of regulation of BDNF expression, for instance, at the level of alternative promoter usage, differential mRNA stability and translatability, or differential subcellular localization of either the mRNA message(s) or protein. In support of this idea, it is well known that the various Bdnf promoters are differentially responsive to neuronal activity; in particular, in the cortex, promoter IV-dependent Bdnf transcription accounts for the majority of neuronal activity-induced Bdnf expression (Tao et al., 1998; Timmusk et al., 1994; Timmusk et al., 1993). This observation prompted us to ask whether we could take advantage of our understanding of the activity-dependent regulation of the Bdnf gene to generate an animal model that allows us to ask which, if any, of the physiological functions of Bdnf in the nervous system are specifically controlled by the neuronal activity-regulated component of its expression.
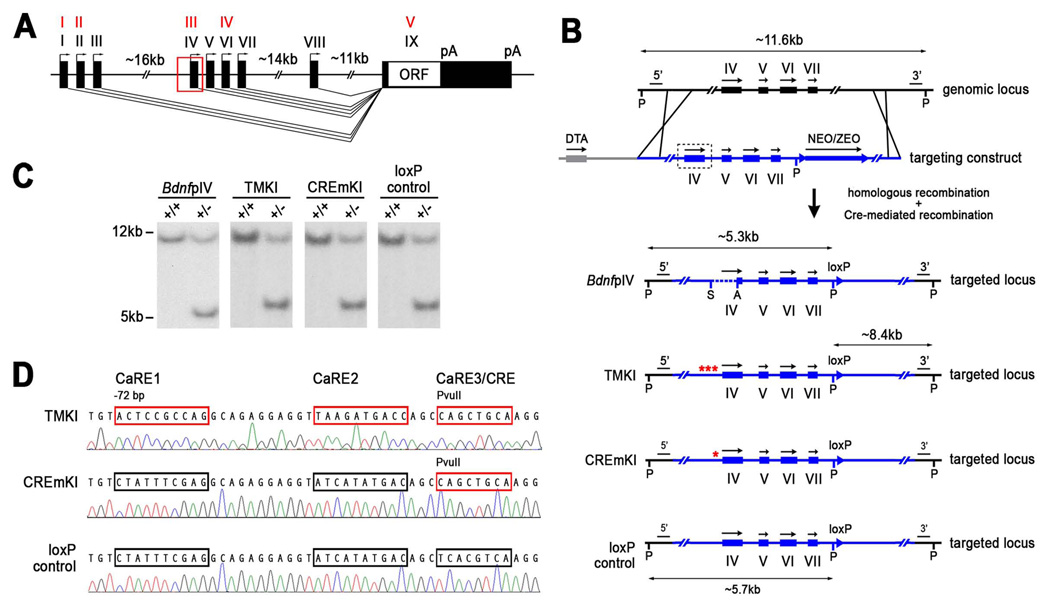
Figure 1.
Generation of Bdnf pIV, TMKI, CREmKI, and loxP control mice. A) Genomic structure of the rodent Bdnf gene adopted from Aid et al., 2007. The discovery of additional exons in the rodent Bdnf gene has resulted in changes in exon nomenclature; the four major transcripts now known as I, II, IV, and VI were initially described as I, II, III, and IV (in red), respectively, in Timmusk et al., 1993. B) Targeting strategy for the introduction of pIV, TMKI, CREmKI, and loxP control mutations into Bdnf promoter IV by homologous recombination. P, PciI; A, AgeI; S, SphI; 5’, 3’ Southern probes; NEO/ZEO, neomycin-zeomycin positive selection cassette; DTA, diphtheria toxin negative selection cassette. C) Southern blot analysis of PciI-digested genomic DNA from targeted ES cells using the 5’ probe indicates correct targeting of each mutation. pIV: WT, 11.6 kb; mut., 5.3 kb; TMKI, CREmKI, loxP control: WT, 11.6 kb; mut., 5.7 kb. D) Direct sequencing of PCR products amplified from genomic DNA isolated from TMKI, CREmKI, or loxP control homozygous mice using primers spanning Bdnf promoter IV.
Our lab and others have extensively investigated the activity-dependent activation of Bdnf promoter IV as a model for understanding the mechanisms mediating calcium-induced neuronal gene expression. Experiments using a plasmid-based reporter to recapitulate promoter IV activity revealed that a cAMP/Ca++-response element-like element (CaRE3/CRE) proximal to the exon IV transcription start site is important for the ability of membrane depolarization to induce promoter IV activation (Shieh et al., 1998; Tao et al., 1998). In vitro studies indicate that this element is bound by the transcription factor CREB, a calcium-regulated factor that has important functions in controlling many adaptive neuronal responses. CREB bound at promoter IV becomes phosphorylated by calcium-regulated kinase cascades in response to neuronal activity and subsequently recruits components of the basal transcriptional machinery to Bdnf promoter IV to activate gene expression (Lonze and Ginty, 2002; West et al., 2001).
Here we report that mutation of the CaRE3/CRE (CREm) at endogenous Bdnf promoter IV by gene targeting results in an animal in which the neuronal activity-dependent component of Bdnf transcription in the cortex is specifically disrupted. CREm knock-in mice exhibit a reduction in the number of inhibitory synapses formed by cortical neurons in culture, a reduction in spontaneous inhibitory quantal transmission measured in acute brain slices, and a reduction in the level of inhibitory presynaptic markers in the cortex. These findings provide a clear demonstration of a biological role for activity-dependent gene expression in the regulation of cortical inhibition, and also indicate a requirement for promoter IV-derived Bdnf expression in this process. Interestingly, our findings point to a previously unappreciated role for CREB in regulating inhibitory synapse development and demonstrate the utility of mice bearing subtle mutations in gene regulatory regions for uncovering novel, target-specific functions for a transcription factor that may not be revealed by traditional loss-of-function studies.
RESULTS
Generation of a mouse model with impaired activity-dependent Bdnf expression
We chose to focus on the Bdnf gene as a model for understanding the functional significance of neuronal activity-regulated gene expression in the brain for several reasons. First, a large body of work has demonstrated a critical role for Bdnf in many diverse processes in nervous system development and function (Poo, 2001), many of which are regulated by neuronal activity. Second, perhaps more than any other activity-regulated gene, the mechanisms by which calcium influx induces Bdnf transcription have been extensively studied, lending insight into the type of genetic lesion that could potentially block activity-dependent expression of the gene (West et al., 2001). In the cortex, for example, promoter IV-derived Bdnf transcripts comprise the majority of neuronal activity-induced Bdnf expression (Tao et al., 1998; Timmusk et al., 1994; Timmusk et al., 1993). In addition, unlike many other prototypical immediate early genes such as c-fos, the induction of Bdnf exon IV-containing transcripts is selectively activated by the influx of extracellular calcium through ligand- and voltage-gated calcium channels; growth factor stimulation, for instance, does not induce robust promoter IV activation. Thus, we reasoned that disruption of exon IV-containing Bdnf transcripts might be sufficient to specifically attenuate activity-dependent expression of Bdnf in the cortex.
Previous studies investigating the mechanisms of Bdnf promoter IV activation have demonstrated that an evolutionarily conserved 170-bp fragment proximal to the exon IV transcriptional start site is sufficient to confer Ca2+-selective responsiveness to a plasmid-based reporter gene when transfected into neurons in vitro. Mutagenesis analysis revealed the existence of three Ca2+-responsive elements (CaREs), CaRE1, CaRE2, and CaRE3/CRE (Figure 1C), each of which, when individually mutated, was sufficient to attenuate activity-dependent induction of the promoter IV reporter (Tao et al., 1998). At the time we began this study, the prevailing view was that CaRE1 is bound by the calcium-responsive transcription factor, CaRF (Tao et al., 2002); CaRE2 contains an E-box element and is bound by the bHLH transcription factors, USF1 and USF2 (Chen et al., 2003b); and CaRE3 is a cAMP/Ca++-response element (CRE)-like element that is bound by CREB (Tao et al., 1998).
Based on the available information at the time, we used homologous recombination in embryonic stem cells to generate three independent lines of mutant mice in which endogenous Bdnf promoter IV is genetically disrupted (Figure 1B). First, we generated a deletion in Bdnf promoter IV (pIV−/−) that removes the previously mapped 170-bp proximal promoter fragment, as well as the transcriptional start site, to completely abrogate transcription from promoter IV. Second, we introduced point mutations into the CREB-binding site CaRE3/CRE element of Bdnf promoter IV (CREmKI, Figure 1B, 1D), which was predicted to block promoter IV induction based on the plasmid reporter assay. Finally, since it is unknown whether mutation of an individual transcriptional control element can independently affect promoter activity in the context of endogenous chromatin, we also generated a third line in which we introduced point mutations into each of the CaRE1, CaRE2, and CaRE3/CRE elements of Bdnf promoter IV in combination (triple-site mutant, TMKI, Figure 1B, 1D). We reasoned that a combination of mutations might function more effectively to block promoter IV activation than mutation of the CaRE3/CRE alone, since previous studies at the c-fos promoter suggest that the concerted action of multiple transcriptional elements is required for gene regulation (Robertson et al., 1995).
For each mutant line, the gene targeting strategy to introduce the mutation into Bdnf promoter IV in embryonic stem cells necessitates the introduction of a 34-bp loxP sequence in the vicinity of the mutation as a by-product of the selection procedure after homologous recombination. To control for the possibility that the presence of the loxP sequence could affect gene expression from the Bdnf locus in unexpected ways, we generated a fourth mouse line in which promoter IV is left unaltered but that differs from a wildtype mouse only by the presence of the loxP sequence in an unconserved region of the large intron between exons VII and VIII (loxP control). Since the loxP site in all three mutants, pIV−/−, CREmKI, and TMKI, is engineered into this identical location (Figure 1B), any differences seen in the mutant lines as compared to the loxP control must therefore stem from their respective mutations at Bdnf promoter IV. Southern blot analysis of genomic DNA isolated from the targeted ES cells, as well as from heterozygous and homozygous mutant mice for all four mutant mouse lines, pIV−/, CREmKI, TMKI, and loxP control, resulted in the predicted mutant allele structure after recombination (Figure 1C, Supplemental Figure 1, data not shown). The CREmKI, TMKI, and loxP control mutations were further verified by direct sequencing of the genomic DNA from homozygous mutants (Figure 1D).
Mutation of the CREB-binding site in Bdnf promoter IV impairs neuronal activity-regulated Bdnf transcription
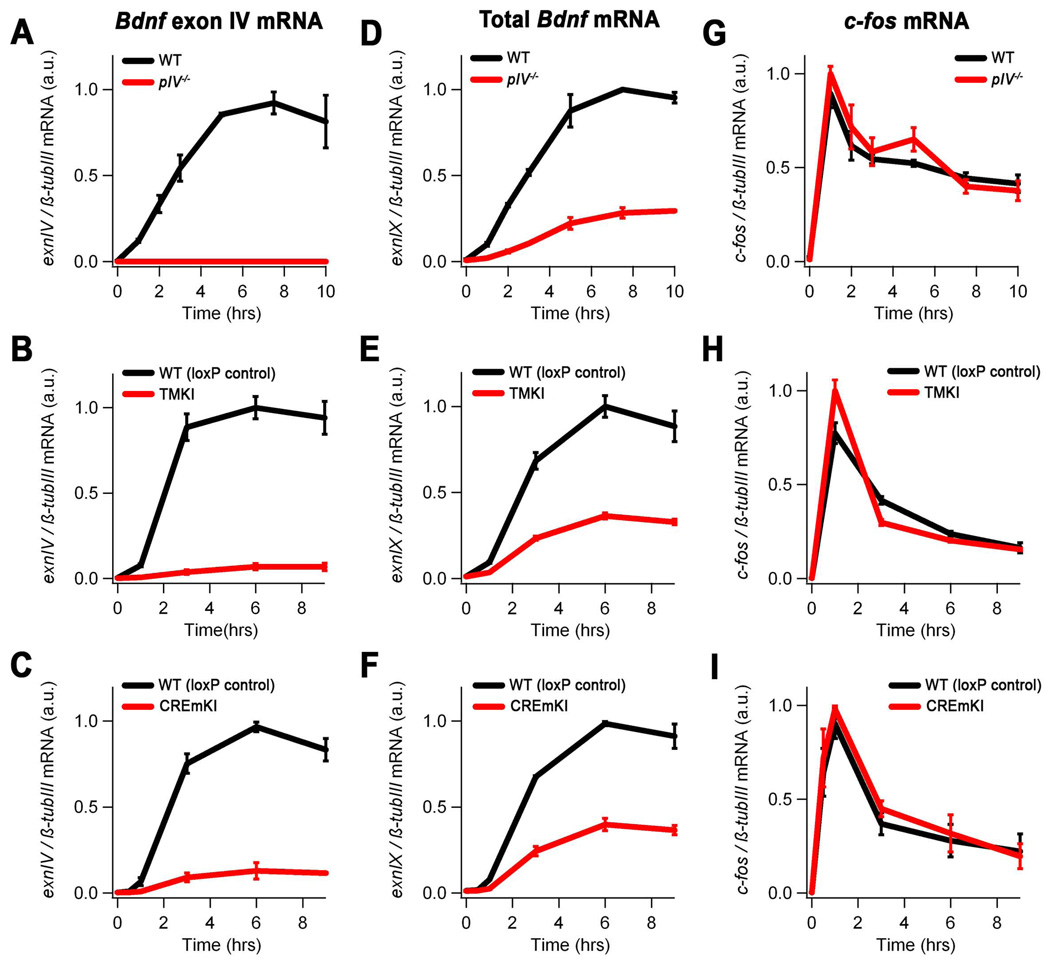
To test whether the introduction of pIV−/−, CREmKI, and TMKI mutations into Bdnf promoter IV blocks neuronal activity-dependent Bdnf expression, we prepared dissociated primary cortical cultures from the brains of homozygous mutant and wildtype or loxP control littermates for each mutant line. Following maintenance in vitro for 5 days (5DIV), neurons were depolarized with high extracellular potassium chloride for varying amounts of time, RNA was extracted, and the levels of Bdnf mRNA transcripts were measured by quantitative real-time PCR (qPCR). As expected, deletion of Bdnf promoter IV results in a complete loss of exon IV-containing Bdnf mRNAs (Figure 2A). Furthermore, mutation of either all three CaREs in combination or of the CaRE3/CRE alone is sufficient to cause an approximately 90% reduction in the level of Bdnf promoter IV induction (Figure 2B, 2C). This failure of Bdnf promoter IV activation does not reflect a general defect in calcium-dependent signaling in these mutants because the induction of other immediate early genes, including c-fos, activity-regulated cytoskeletal-associated protein (Arc), and the other major activity-regulated Bdnf transcript exon I, is unaffected in pIV−/−, CREmKI, or TMKI neurons (Figure 2G–I; Supplemental Figure 2; and data not shown). The impairments in promoter IV activation result in an approximately 70% or 60% reduction in the activity-dependent induction of total Bdnf mRNA in pIV−/− or CREmKI and TMKI cortical neurons, respectively (Figure 2D–F). This impairment in total Bdnf mRNA induction is reflected in an approximately 50% decrease in BDNF protein levels in pIV−/− or CREmKI cortex (Supplemental Figure 3, see also Figure 4F). Thus, all three mutations are sufficient to cause a significant impairment in activity-dependent Bdnf expression in neurons.
Figure 2.
Activity-dependent Bdnf transcription in pIV−/−, TMKI, and CREmKI cortical neurons is impaired in response to membrane depolarization. Levels of Bdnf exon IV (A–C), exon IX (coding exon) (D–F), and c-fos (G–I) mRNA in 5DIV cortical neurons prepared from pIV−/− and wildtype littermates (A,D,G), homozygous TMKI and loxP control littermates (B,E,H), or homozygous CREmKI and loxP control littermates (C,F,I) that were either left untreated or membrane depolarized with high extracellular potassium for the indicated amounts of time. Differences in Bdnf exon IV and total Bdnf mRNA expression are statistically significant for pIV−/−, TMKI, or CREmKI versus their respective controls (P<0.01, repeated-measures ANOVA; P<0.01, pairwise comparisons at each timepoint 1 hr or longer, Bonferroni-Dunn post-hoc test). Data are mean ± SEM from n=3 (pIV−/− and CREmKI) or n=2 (TMKI) independent experiments in which each sample was measured in triplicate.
Figure 4.
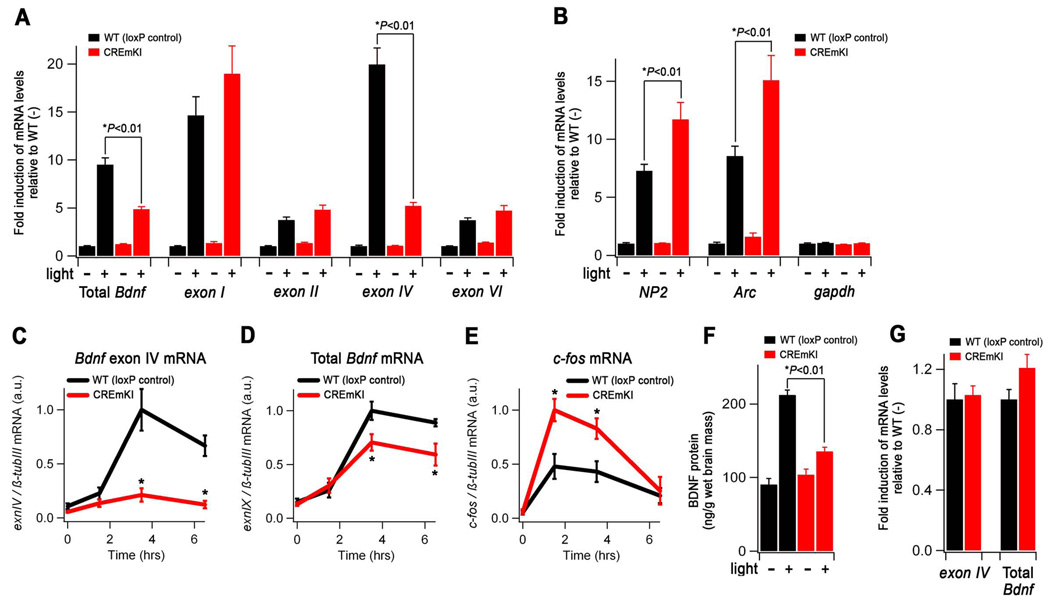
Sensory experience-dependent Bdnf expression is impaired in the intact CREmKI brain. A–B) Levels of the indicated transcripts in the primary visual cortex of adult CREmKI and loxP control mice reared in darkness for 14 days, after which they were either maintained in darkness (−) or exposed to light for 90 min. (+). Within each transcript, mRNA levels are reported as fold induction relative to the loxP control unstimulated (−) condition. In either loxP control or CREmKI visual cortex, light stimulation induced the expression of all mRNAs measured (P<0.01, two-way ANOVA with Bonferroni Dunn post-hoc test), except for gapdh, which served as a non-activity-regulated gene control. C–E) Bdnf exon IV, total Bdnf, and c-fos mRNA levels in the cortex of CREmKI and loxP control mice injected with either saline (time 0) or kainic acid (KA) for the indicated amounts of time. F) Levels of BDNF protein in the primary visual cortex of animals from A–B. G) The unstimulated (−) levels of Bdnf exon IV and total Bdnf in CREmKI and loxP control visual cortex are re-plotted from A) for direct comparison (P>0.05, two-way ANOVA with Bonferroni-Dunn post-hoc test). Data are from n=5–7 animals per condition for both visual stimulation and KA seizure experiments. Each animal was measured in triplicate, and data are presented as mean ± SEM. Asterisks denote P<0.01, two-way (visual simulation) or repeated-measures (KA seizure) ANOVA with Bonferroni-Dunn post-hoc test between indicated pair.
We chose to focus our study on the CREmKI mutant for several reasons. First, the CREmKI mutation represents a subtle change in nucleotide sequence, but is sufficient to cause a substantial impairment in the activity-dependent induction of Bdnf, comparable to that observed in TMKI neurons. Second, the CREmKI mutant provides several significant advantages over the pIV−/− mutant for selectively evaluating the functional significance of both activity-dependent and promoter IV-dependent Bdnf transcription. Whereas the pIV−/− deletion mutant completely removes promoter IV-derived transcript independent of the stimulus state of the neurons, the CREmKI mutant impairs Bdnf promoter IV activation specifically in response to neuronal activity-dependent rises in calcium by eliminating binding of the Ca2+-regulated factor CREB to Bdnf promoter IV (see below, Figure 8). The CREmKI mutation has an additional advantage in that it maintains the endogenous configuration of the Bdnf locus, thereby circumventing potential artifacts that can arise in deletion mutants where the spacing among residual promoter elements and exons is changed. Finally, since previous studies strongly suggest that the CaRE3/CRE element in Bdnf promoter IV is bound by CREB (Tao et al., 1998), the CREmKI mutant also has the potential to provide insight into specific functions of CREB-dependent gene regulation in the nervous system.
Figure 8.
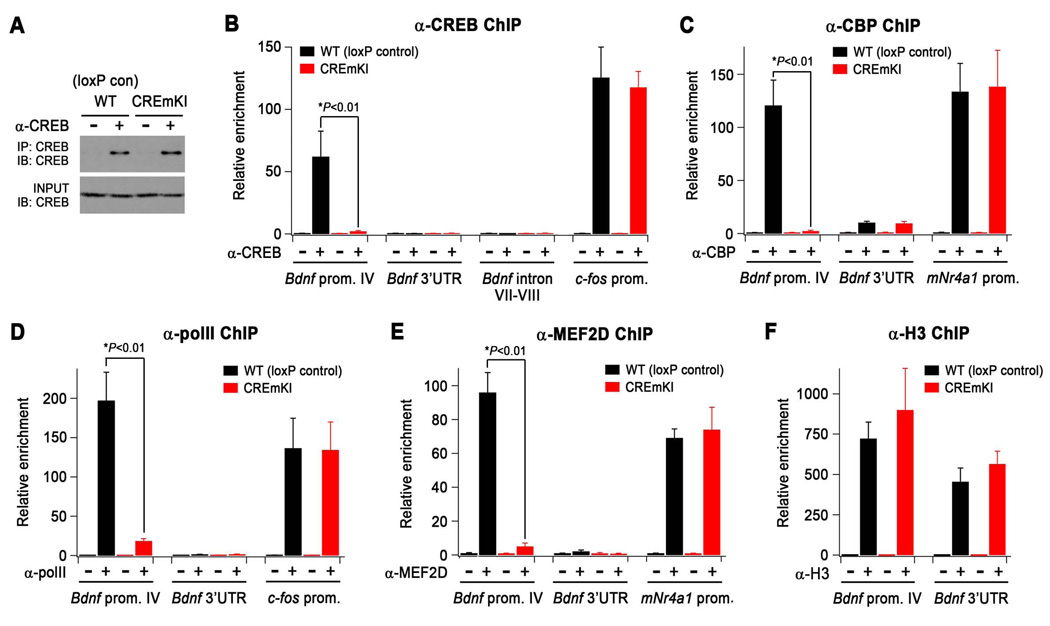
Loss of CREB binding at Bdnf promoter IV in CREmKI brains leads to the disassembly of the transcriptional activating complex at Bdnf promoter IV. A) α-CREB immunoprecipitates were isolated from the cross-linked forebrains of CREmKI and loxP control littermates under conditions identical to those used for chromatin immunoprecipitation (ChIP) and analyzed by Western blotting with an independent antibody against CREB. B–F) qPCR measurement of the levels of the indicated genomic DNA regions in α-CREB (B), α-CBP(C), α-polymerase II (D), α-MEF2D (E), and α-histone H3 (F) chromatin immunoprecipitates prepared from the forebrains of CREmKI and loxP control littermates. Asterisk denotes P<0.01, two-way ANOVA with pairwise comparison by Bonferroni-Dunn post-hoc test. Data are mean ± SEM from n=4 (α-CREB), n=3 (α-MEF2D, α-CBP), or n=2 (α-pol II, α-H3) independent experiments in which each sample was measured in triplicate.
CREmKI mice are viable, fertile, born in the expected Mendelian ratios, and visually indistinguishable from loxP control littermates. In addition, CREmKI animals have comparably sized brains to loxP control animals, show no gross anatomical alterations in cortical, cerebellar, or hippocampal structure, no obvious changes in gait, and no obvious defects in cortical layering (see Methods, Supplemental Figure 4, and data not shown). These observations suggest that, in contrast to the complete loss of Bdnf in Bdnf−/− mutants, the introduction of a subtle knock-in mutation into Bdnf promoter IV to specifically impair activity-dependent Bdnf expression does not have a strong effect on neuronal survival.
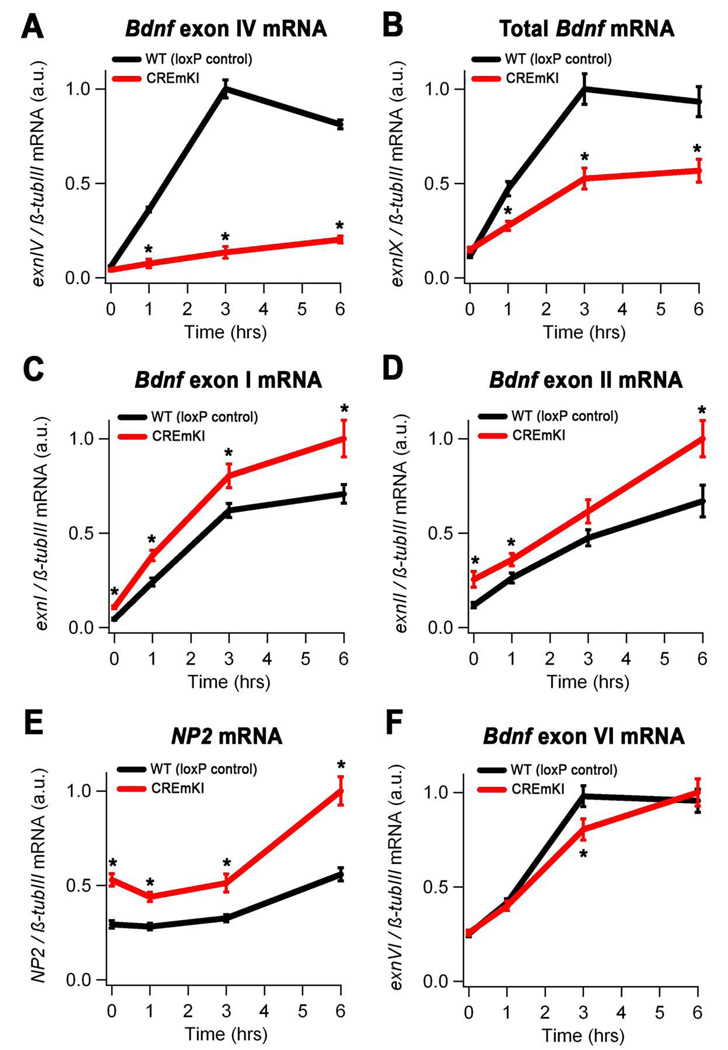
To investigate how the CREmKI mutation affects activity-regulated Bdnf expression in response to glutamate receptor activation, we treated 12 DIV cortical neurons, which have developed many more synaptic contacts than 5DIV neurons, with NMDA to stimulate NMDA-type glutamate receptors, a major mediator of excitatory neurotransmission. Similar to our findings under membrane-depolarizing conditions, we observed significant impairments in the activation of Bdnf promoter IV and total Bdnf expression in CREmKI neurons as compared to loxP control neurons (Figure 3A and 3B) in response to NMDA stimulation. Interestingly, when we examined the expression of the remaining major Bdnf transcripts (Figure 1A) as well as other activity-regulated genes in 12 DIV CREmKI neurons, we were surprised to find that the induction of most immediate-early genes, including neuronal pentraxin 2 (NP2), Arc, c-fos, as well as Bdnf exons I and II, is significantly upregulated in response to stimulation with NMDA (Figure 3C–F and data not shown). These observations raise the interesting possibility that the general increase in neuronal activity-dependent transcription in 12 DIV CREmKI neurons may be secondary to an increase in their overall level of excitability. These differences may not be obvious after only five days in culture when neurons are not extensively synaptically connected (Figure 2G–I).
Figure 3.
Neuronal activity-dependent Bdnf transcription in CREmKI cortical neurons is impaired in response to glutamate receptor activation. Levels of Bdnf exon IV (A), total Bdnf (exon IX) (B), Bdnf exon I (C), Bdnf exon II (D), NP2 (E), and Bdnf exon VI (F) mRNA in 12DIV cortical neurons prepared from CREmKI or loxP control littermates that were either mock-stimulated or treated with 20µM NMDA for the indicated amounts of time. Asterisk denotes P<0.01, repeated-measures ANOVA, pairwise comparisons at indicated timepoints, Bonferroni-Dunn post-hoc correction. Data are mean ± SEM from n=2 independent experiments in which each sample was measured in triplicate.
Mutation of the CREB-binding site in Bdnf promoter IV impairs sensory experience-driven Bdnf transcription in the intact brain
We next examined whether the activity-dependent expression of Bdnf in the intact CREmKI brain is affected in response to a physiologically relevant environmental stimulus. Adult mice were maintained in total darkness for fourteen days to reduce levels of sensory activity-driven transcripts in the visual cortex. RNA was then harvested from the primary visual cortex of mice that were either maintained in darkness or exposed to light for 90 minutes, and the levels of Bdnf transcripts were measured by qPCR. We found that the nearly 20-fold increase in the levels of exon IV-containing transcripts measured in loxP control visual cortex in response to light exposure is reduced by approximately 75% in CREmKI visual cortex (Figure 4A). The activity-dependent induction of gene expression measured in the visual cortex is specific to visual experience because it is not observed in the somatosensory cortex of either loxP control or CREmKI brains (Supplemental Figure 5). Interestingly, we also observed that many other immediate-early genes, including c-fos, Arc, and NP2, are more highly induced in the visual cortex of CREmKI animals in response to light stimulation when compared to loxP controls (Figure 4B and data not shown). Moreover, measurements of activity-dependent gene expression in the cortex of CREmKI and loxP control animals given seizures by administration of the chemoconvulsant kainic acid resulted in similar findings (Figure 4C–E and Supplemental Figure 6). The heightened induction of many activity-regulated genes in response to neuronal activity in CREmKI animals compared to controls again suggests the possibility of changes in the excitability of neuronal circuits in the CREmKI brain.
The impairment in activity-dependent Bdnf promoter IV transcription in response to light exposure in CREmKI visual cortex results in an approximately 50% decrease in the induction of total Bdnf transcripts driven by sensory stimulation (Figure 4A), as compared to loxP control animals. This reduction in Bdnf mRNA is reflected in an approximately 60% decrease in the induction of BDNF protein levels in visual cortex in response to visual experience (Figure 4F). Importantly, the CREmKI mutation does not appear to affect Bdnf expression under conditions of significantly reduced neuronal activity, since levels of both promoter IV-derived and total Bdnf transcripts are similar in sensory-deprived loxP control and CREmKI visual cortex (Figure 4G, P>0.05). Taken together, these results provide strong evidence that the neuronal activity-dependent component, but not the basal component, of Bdnf transcription is selectively impaired in the intact brains of CREmKI animals in response to a physiologically relevant sensory experience and demonstrate the utility of the CREmKI animal model for examining the functional significance of neuronal activity-regulated gene expression for nervous system development and function.
Neuronal activity-dependent Bdnf expression regulates the development of inhibition in the nervous system
Our observations that the neuronal activity-dependent induction of many immediate early genes, including non-promoter IV-derived Bdnf transcripts, c-fos, NP2, or Arc, is enhanced in mature CREmKI neuronal cultures or in the intact brain of CREmKI mutants suggested that neuronal connectivity may be altered in CREmKI brains. In addition, a large body of work has independently established both BDNF and neuronal activity as positive regulators of inhibitory synapse development. For example, application of recombinant BDNF or BDNF overexpression promotes the development of inhibition (Huang et al., 1999; Ohba et al., 2005), whereas reduction of neuronal activity using pharmacological blockers or by sensory deprivation retards the maturation of inhibition (Benevento et al., 1995; Hendry and Jones, 1986; Rutherford et al., 1997). A compelling hypothesis that arises from this work suggests that neuronal activity regulates the development of inhibitory synapses through its effects on Bdnf gene transcription. The CREmKI mutant allows us to directly test this hypothesis by asking whether the impairment in activity-dependent Bdnf expression affects the development of inhibition in the nervous system.
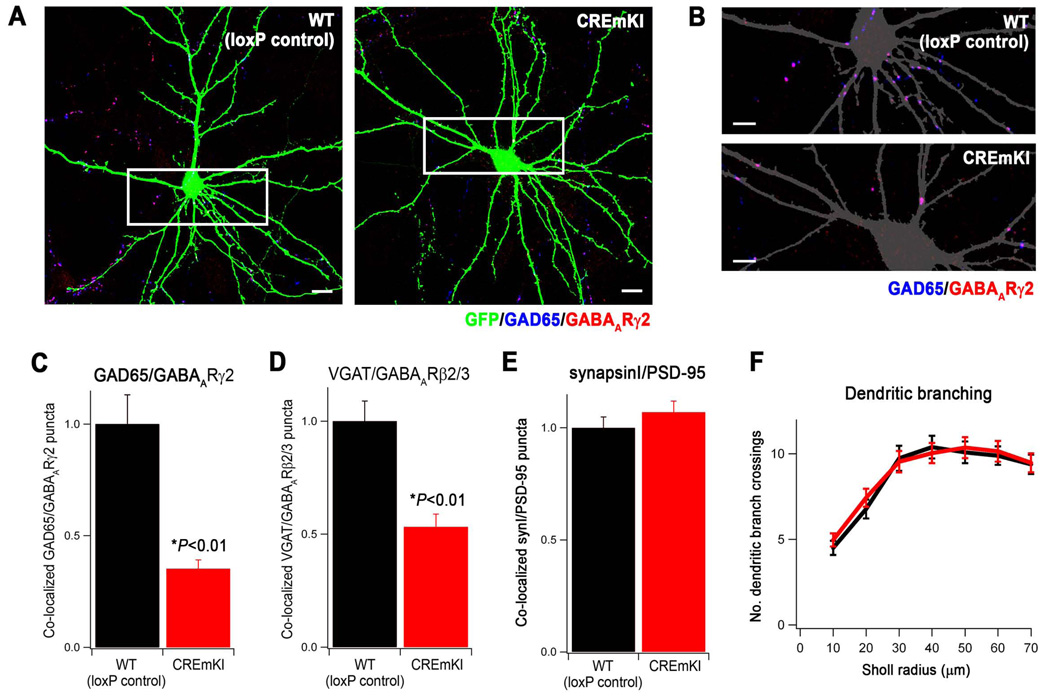
To specifically investigate the effect of the CREmKI mutation on the development of inhibitory synapses, we first prepared low-density cortical cultures from CREmKI and loxP control littermates that allowed us to quantify synapse density by immunostaining. Neurons were transfected with a plasmid encoding green fluorescent protein (GFP) at 5DIV, and fixed and immunostained at 18 DIV with antibodies that recognize the presynaptic inhibitory marker GAD65 and the postsynaptic inhibitory marker GABAA receptor γ2 subunit (GABAARγ2). The number of inhibitory synapses that form on a neuron was quantified as the number of co-localized GAD65 and GABAARγ2 puncta that overlap with GFP signal (Figure 5A, 5B).
Figure 5.
Impaired activity-dependent Bdnf expression reduces the number of inhibitory synapses formed on CREmKI neurons in culture. A) Representative images of GFP-transfected, E17.5 + 18DIV CREmKI and loxP control littermate cortical neurons, immunostained with the indicated antibodies to label pre- and post-synaptic inhibitory synapse terminals. Scale bar, 10µm. B) Enlargement of the boxed area in A shows details of dendrites. The GFP signal is converted to gray to allow better visualization of the synaptic puncta. Scale bar, 5µm. C–E) Quantification of the average density of GAD65/GABAARγ2 (C, 78–81 cells/condition), VGAT/GABAARβ2/3 (D, 65–66 cells/condition), and synapsinI/PSD-95 (E, 63–65 cells/condition) co-clusters along the dendrites of GFP-transfected CREmKI and loxP control neurons. Asterisk denotes P<0.01, two-way ANOVA with pairwise comparison by Bonferroni-Dunn post-hoc test. Data are mean±SEM from 3–4 independent experiments. F) Quantification of dendritic branch complexity by Sholl analysis plots the number of dendritic branches intersecting concentric circles of increasing radii centered on the cell body. P>0.05 by repeated-measures ANOVA; data are mean±SEM from 26–28 cells/condition from 2 independent experiments.
Using these criteria, we observed that significantly fewer inhibitory synapses form on CREmKI cortical neurons than on loxP control littermate neurons in culture (Figure 5A–C). A similar result was also seen when inhibitory synapses were quantified using an independent set of markers, the inhibitory presynaptic marker VGAT and the inhibitory postsynaptic marker GABAA receptor β2/3 subunits (Figure 5D). The decrease in synapse number in CREmKI neurons appears specific to inhibitory synapses because no difference in the number of excitatory synapses was detected when CREmKI neurons were compared to loxP control neurons using the presynatic marker synapsin I and the excitatory postsynaptic marker PSD95 (Figure 5E). In addition, the reduction in inhibitory synapse number in CREmKI neurons does not appear to be secondary to poor cell health since no significant differences between CREmKI and loxP control littermate neuronal cultures were observed in the number of pyknotic nuclei present (data not shown) or in the extent of dendritic arborization, as quantified by Sholl analysis (Figure 5F). These results demonstrate that the activity-dependent expression of Bdnf is important for the development of inhibitory, but not excitatory, synapses in neurons maintained in culture.
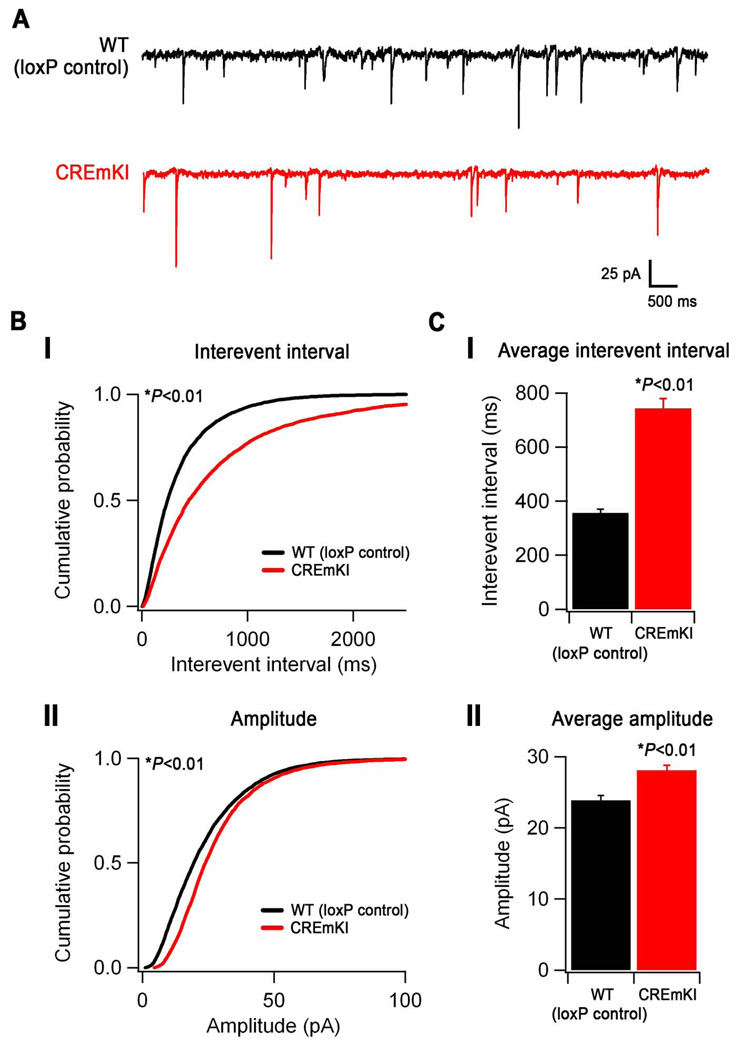
Although many of the key steps of synaptogenesis are recapitulated by dissociated neurons as they re-establish their connections in culture (Ziv and Garner, 2004), we also wanted to examine the role of activity-dependent Bdnf expression in regulating the development of inhibition in the context of an endogenous neural circuit. Since our previous data indicate that visual experience-dependent Bdnf transcription is impaired in CREmKI brains, and because previous studies have implicated Bdnf in the regulation of cortical inhibition and plasticity in the visual cortex, we chose to measure the number of functional inhibitory synapses that form on neurons in an intact circuit in the primary visual cortex of CREmKI animals. We prepared acute slices containing primary visual cortex (V1) from the brains of P16-P18 CREmKI or loxP control littermates and obtained whole-cell patch clamp recordings from layer II/III pyramidal neurons in V1 to measure spontaneous miniature inhibitory postsynaptic currents (mIPSCs) (Figure 6A). Pharmacologically isolated mIPSCs were demonstrated to be GABAAR-mediated, since they were abolished by treatment with picrotoxin (data not shown). We observed a significant decrease in the frequency of mIPSC events in CREmKI neurons (interevent interval, 742.4±36.8 vs. 355.4±14.7 ms, CREmKI and loxP control, respectively; Figure 6B–C(i)), suggesting that impaired activity-dependent Bdnf transcription in CREmKI cortex results in fewer functional inhibitory synapses as compared to loxP control. Interestingly, we also found a significant increase in the amplitude of mIPSCs in CREmKI neurons (28.1±0.7 vs. 23.9±0.7 pA, CREmKI and loxP control, respectively; Figure 6B–C(ii)), suggesting that activity-dependent Bdnf expression can regulate inhibitory synapse strength. Whereas the change in mIPSC amplitude in CREmKI neurons might reflect the direct action of activity-dependent Bdnf on vesicle neurotransmitter content or postsynaptic GABAA receptor number, it could also occur secondarily as a homeostatic adjustment to the decrease in inhibitory input number. Nevertheless, these findings are consistent with our measurements by immunofluorescence staining in cultured cortical neurons indicating that fewer inhibitory synapses form on CREmKI neurons and strongly support the hypothesis that activity-dependent Bdnf transcription plays a critical role in GABAergic synapse development in the nervous system.
Figure 6.
Neuronal activity-dependent Bdnf expression controls the development of cortical inhibition. A) Representative traces of mIPSCs recorded from layer II/III V1 pyramidal neurons in loxP control and CREmKI acute cortical slices. B) Cumulative probability distributions of mIPSC interevent intervals (I) and amplitudes (II) recorded from loxP control and CREmKI neurons. P<0.01 by either Kolmogorov-Smirnov test or Monte Carlo simulation (see Supplemental Methods). C) Average interevent interval (I) and amplitude (II) of mIPSCs recorded from loxP control and CREmKI neurons. Data are mean ± SEM, P<0.01 by Student’s t-test. Data are from 18–20 cells/genotype recorded from 12 pairs of littermates.
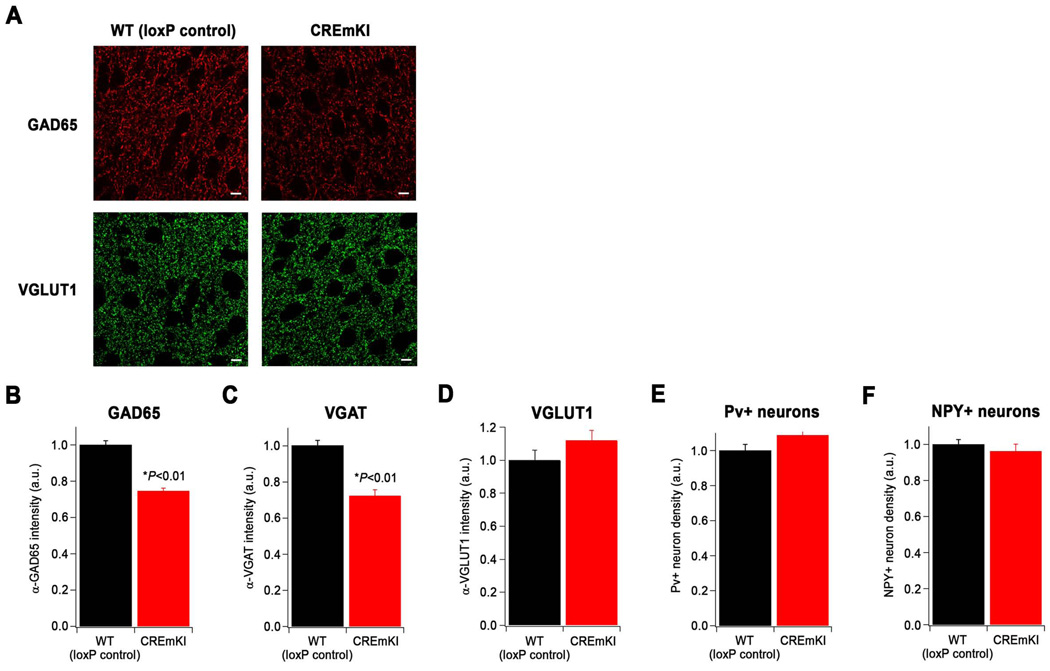
Because mIPSCs reflect the spontaneous release of individual neurotransmitter vesicles at presynaptic sites of contact, changes in mIPSC frequency correlate with changes in inhibitory synapse number, but they can also be affected by changes in presynaptic properties such as the probability of neurotransmitter vesicle release. As an independent measurement of inhibitory synapse number in CREmKI neurons connected together in an endogenous circuit, we immunostained histological sections of visual cortex prepared from P20-P24 CREmKI or loxP control littermates for inhibitory synaptic markers. We observed modest, but significant decreases in the intensity of staining for the inhibitory presynaptic markers GAD65 and VGAT in layer II/III of CREmKI visual cortex as compared to loxP control visual cortex (Figure 7A–C), consistent with a reduction in inhibitory synapse number in CREmKI neurons. However, immunostaining for the presynaptic marker synapsin I (data not shown) or the excitatory presynaptic marker VGLUT1 (Figure 7A, 7D) is similar in CREmKI and loxP control visual cortex, consistent with our previous observation that excitatory synapse development is unaffected in CREmKI neurons in culture. Taken together, our data demonstrate a role for neuronal activity-dependent Bdnf expression in the development of inhibition in the cortex, and suggest that impairments in activity-dependent Bdnf transcription lead to a shift in the ratio of excitation to inhibition in the brain that may increase the excitability of the CREmKI brain.
Figure 7.
Reduced immunoreactivity for inhibitory synaptic markers in CREmKI cortex. A) Representative images of immunostaining with antibodies recognizing GAD65 (red), which is enriched in inhibitory presynaptic terminals, and VGLUT1 (green), which is enriched at excitatory presynaptic terminals, in layer II/III of CREmKI and loxP control primary visual cortex. Scale bar, 5µm. B–D) Quantification of the average intensity of α-GAD65 (B, 74–77 fields/genotype), α-VGAT (C, 48 fields/genotype), and α-VGLUT1 (D, 48 fields/genotype) immunostaining in CREmKI and loxP control littermates. Asterisk indicates P<0.01, two-way ANOVA with pairwise comparison by Bonferroni-Dunn post-hoc test. Data are mean ± SEM from 3–4 pairs of littermates. E–F) Quantification of the number of paralbumin- (E) and NPY- (F) positive inhibitory neurons in the primary visual cortex of CREmKI and loxP control littermates, normalized to the number of total nuclei. P>0.05 for either parvalbumin-positive or NPY-positive neurons, two-way ANOVA. Data are mean ± SEM collected from n=24 hemispheres from 3 pairs of littermates.
To begin to gain insight into the cellular mechanisms by which activity-dependent Bdnf expression regulates inhibitory synapse number, we next asked whether the decrease in inhibition observed in CREmKI brains might be a simple consequence of changes in the number of cortical inhibitory neurons, since total Bdnf null mice exhibit a dramatic reduction in the density of parvalbumin-positive and NPY-positive inhibitory neurons in the cortex (Jones et al., 1994). We focused our analysis on these populations of interneurons and employed stereological methods to quantify the number of parvalbumin-or NPY-positive neurons in the visual cortex of CREmKI or loxP littermate control mice. To control for any possible changes in cell density, interneuron counts were normalized to the total number of cell nuclei as quantified by the DNA-binding dye Hoechst 33342. We observed no significant difference in the number of either parvalbumin- or NPY-positive interneurons in CREmKI visual cortex (Figure 7E, 7F), suggesting that reductions in inhibitory synapse number in CREmKI neurons are not secondary to changes in inhibitory neuron number. Since no molecular marker exists that can easily label all known GABAergic cell types, we cannot exclude the possibility that the differentiation of other classes of interneurons is affected in CREmKI mutants. However, since parvalbumin- and NPY-positive interneurons together comprise more than 50% of the inhibitory neurons in the cortex, and since these specific populations of interneurons have been previously shown to be Bdnf-dependent in the context of the complete loss of Bdnf, our data suggest that the activity-dependent component of Bdnf expression is important for the formation or maintenance of inhibitory synapses by GABAergic neurons, but not for the survival or differentiation of GABAergic neurons.
Loss of CREB binding leads to disassembly of the transcriptional activating complex at Bdnf promoter IV
As the loss of activity-dependent Bdnf expression in CREmKI animals is due to the specific mutation of the CaRE3/CRE in Bdnf promoter IV, a sequence that is bound by the transcription factor CREB, our results also potentially point to a role for CREB in inhibitory synapse development that has not been previously described. CREB is a well-established regulator of calcium-dependent gene expression that has important functions in nervous system development and function, including survival, synaptic plasticity, and learning and memory (Flavell and Greenberg, 2008; West et al., 2002). The central role of CREB in neuronal survival and maintenance has hampered the study of CREB function in mature neurons; for instance, postnatal disruption of CREB in the forebrain leads to widespread neurodegeneration (Mantamadiotis et al., 2002), preventing the study of other CREB functions in these neurons. However, given that CREB regulates a transcriptional program comprising hundreds, if not thousands of genes (Impey et al., 2004), only a portion of which are involved in survival, many additional functions of CREB in the nervous system undoubtedly remain to be discovered. Our findings with CREmKI mice suggest that one of these additional functions of CREB is the control of inhibitory synapse development via the calcium-dependent regulation of Bdnf expression.
To explore this idea, we used chromatin immunoprecipitation (ChIP) to directly assess the effect of the CREmKI mutation on the binding of CREB to endogenous Bdnf promoter IV in the brain. Forebrains from CREmKI and loxP littermate control mice were treated with formaldehyde to covalently crosslink endogenous protein-DNA complexes, which were then mechanically sheared and immunoprecipitated with an antibody against CREB. The specificity of the anti-CREB antibody for use in ChIP experiments has been previously validated in neurons that lack CREB protein (Riccio et al., 2006). qPCR measurements indicated that whereas Bdnf promoter IV-containing DNA fragments are enriched in CREB immunoprecipitates prepared from loxP control brains as compared to control immunoprecipitates from loxP control brains, CREB immunoprecipitates from littermate CREmKI brains do not contain detectable levels of Bdnf promoter IV gDNA (Figure 8B). The failure to detect CREB binding at promoter IV in CREmKI brains is not due to differences in the level of CREB expression (Figure 8A) or the ability to immunoprecipitate CREB under crosslinking conditions between CREmKI and loxP control brains (Figure 8A), and we detected robust CREB binding to other known CRE-containing CREB target promoters, such as c-fos or Nr4a1, in CREmKI brains (Figure 8B and data not shown). In contrast, we detected no binding of CREB in either loxP control or CREmKI brains to negative control regions in Bdnf intron VII, in the Bdnf 3’ untranslated region (UTR), or within the Nr4a1 gene (Figure 8B and data not shown). Taken together, these data provide the first direct demonstration that Bdnf promoter IV is an endogenous target gene of CREB in the brain and demonstrate that mutation of the promoter IV CaRE3/CRE element abrogates CREB binding to Bdnf promoter IV in the intact brain. Together with our earlier experiments demonstrating that Bdnf promoter IV transcription is induced upon activity-dependent CREB phosphorylation (Tao et al., 1998), these findings strongly suggest that the impairment in neuronal-activity dependent induction of Bdnf expression in CREmKI mice is due to a loss of CREB-binding and regulation at promoter IV.
Since our initial characterization of CaRF (Tao et al., 2002), USF1/2 (Chen et al., 2003b), and CREB (Tao et al., 1998) as important regulators of Bdnf promoter IV activation, ongoing work in our lab and others has described additional components of the transcriptional regulatory complex at promoter IV, including the transcription factors BHLHB2 (Jiang et al., 2008), NPAS4 (Lin et al., 2008), and MEF2 (S.W.F., T.K.K., and M.E.G., manuscript in preparation); the methyl-DNA binding protein MeCP2 (Chen et al., 2003a; Martinowich et al., 2003); and the transcriptional co-activator CBP (West et al., 2001). These factors are believed to function together with CREB to confer calcium-and neural cell type-specific regulation of Bdnf transcription, but little is understood about how they cooperate with one another to regulate transcription or how they affect the recruitment of one another to Bdnf promoter IV. We took advantage of the CREmKI mutant to assess the contribution of CREB to the assembly of various components of the transcriptional activating complex at Bdnf promoter IV.
We first examined whether loss of CREB binding affects the recruitment of the transcriptional co-activator CBP to Bdnf promoter IV. Phosphorylation of CREB at Ser133 in response to rises in intracellular calcium recruits the binding of CBP, which functions as a transcriptional co-activator both through its intrinsic histone acetyltransferase activity and through its stabilization of components of the pre-initiation complex at promoters (Hong et al., 2005; Lonze and Ginty, 2002). Although CBP has been best studied in the context of co-regulation with CREB, it is a general transcriptional co-factor that cooperates with a wide variety of other transcription factors, including MEF2 (Sartorelli et al., 1997). Performing ChIP with an antibody directed against CBP, we observed significant binding of CBP to Bdnf promoter IV in loxP control brains that was almost completely absent in CREmKI littermate brains (Figure 8C). By contrast, in both CREmKI and loxP control brains, we detected strong binding of CBP to the c-fos promoter and relatively little binding of CBP in the Bdnf 3’ UTR negative control region (Figure 8C). These data indicate that loss of CREB binding to Bdnf promoter IV disrupts the binding of the general transcriptional co-activator CBP to the promoter as well. To determine how loss of CBP binding to Bdnf promoter IV affects the recruitment of the basal transcriptional machinery, we also performed ChIP with an antibody (monoclonal 8WG16) directed against the hypophosphorylated state of the largest subunit of RNA polymerase II, which is specifically enriched at transcriptional start sites (Brodsky et al., 2005). In the absence of CREB binding to promoter IV in CREmKI brains, we also observed a significant reduction in the level of Pol II docked at the promoter (Figure 8D), consistent with the impairment in activity-dependent transcription from Bdnf promoter IV (Figure 2C, Figure 3A, Figure 4A). Pol II binding at the c-fos promoter was not affected in CREmKI brains (Figure 8D), indicating that the loss of polymerase binding in CREmKI brains is specific to promoter IV. Together, these results indicate that the loss of binding of a single sequence-specific factor, CREB, to Bdnf promoter IV is sufficient to disrupt the assembly of the general transcriptional activation machinery used by many transcription factors at Bdnf promoter IV.
Finally, we asked whether loss of CREB binding to Bdnf promoter IV affects the recruitment of other sequence-specific transcription factors that are believed to cooperate with CREB to mediate calcium-specific gene expression. We focused on the calcium-regulated factor MEF2 because ongoing studies in our lab have extensively validated the antibody directed against MEF2D for use in ChIP (S.W.F, T.K.K. and M.E.G., manuscript in preparation). Using this antibody, we observed strong binding of MEF2D to Bdnf promoter IV in loxP control brains that is significantly reduced in CREmKI littermate brains (Figure 8E). In contrast, we detected enrichment of the Nr4a1 promoter, a well-characterized target of MEF2 transcriptional regulation, in anti-MEF2D immunoprecipitates from both CREmKI and loxP control brains (Figure 8E), and little MEF2D binding to negative control regions in the Bdnf and Nr4a1 genes in either CREmKI or loxP control brains (Figure 8E and data not shown). Thus, the loss of CREB binding to Bdnf promoter IV disrupts the binding of MEF2D to Bdnf promoter IV as well. Our failure to observe the binding of many components of the transcriptional activating complex — CBP, polII, and MEF2D — to Bdnf promoter IV in CREmKI brains does not reflect a general inability to immunoprecipitate promoter IV-containing gDNA fragments from CREmKI brains because promoter IV fragments are found to be equally enriched in CREmKI and loxP control chromatin immunoprecipitates collected with an antibody that recognizes histone H3, a core structural component of chromatin (Figure 8F). Taken together, our findings indicate that loss of CREB binding to Bdnf promoter IV disrupts the multi-factor transcriptional activating complex at promoter IV and suggest a new function for CREB in nucleating the assembly of transcriptional complexes at its target promoters.
DISCUSSION
Despite more than two decades of investigation into the program of gene expression that is regulated by neuronal activity in the brain, a significant gap persists in our understanding of the biological significance of the activity-dependent component of gene transcription. In this study, the introduction of a subtle mutation into the endogenous Bdnf gene in mice to disrupt the ability of CREB to bind Bdnf promoter IV represents a powerful approach for selectively evaluating the functional significance of activity-dependent Bdnf transcription. We show that disruption of the ability of CREB to regulate Bdnf promoter IV results in an animal in which the activity-dependent transcription of Bdnf is impaired in response to sensory experience-driven synaptic activation in the intact brain. Using this animal, we find that activity-dependent Bdnf expression is required for the appropriate development of inhibition in the cortex, but does not appear to affect the survival or differentiation of GABAergic neurons. These results demonstrate the biological importance of activity-regulated gene transcription for the establishment of appropriate connectivity in the nervous system. In addition, our findings also indicate a requirement for promoter IV-derived Bdnf in the development of inhibition in the cortex and thus provide insight into the functional significance of the complex, multi-promoter organization of the Bdnf gene.
Most previous studies examining the role of activity-regulated gene transcription in the nervous system have taken the approach of disrupting either genes whose transcription, or transcription factors whose function, is regulated by neuronal activity. For example, the observation of defects in synaptic plasticity and learning in hypomorphic mutants of the transcription factor CREB (Balschun et al., 2003; Bourtchuladze et al., 1994; Gass et al., 1998) has been taken as evidence of the importance of neuronal activity-dependent transcription in these adaptive processes, since CREB is a well-established regulator of calcium-dependent gene expression. Experiments with this design, however, cannot easily dissociate the functions of a gene that are due specifically to its regulation by neuronal activity from the many other influences on its action. In the case of CREB, for instance, interpretation of its role in synaptic and behavioral plasticity is complicated by its essential role in neuronal survival and maintenance (Mantamadiotis et al., 2002; Rudolph et al., 1998). The current study is distinguished by our ability to make use of the substantial existing mechanistic understanding of Bdnf transcription to specifically disrupt neuronal activity-dependent Bdnf transcription in vivo, via the genetic mutation of the CREB-binding site in Bdnf promoter IV. This objective can be achieved because, although CREB can be activated by multiple signaling pathways including those mediated by rises in cyclic AMP, exon IV-containing Bdnf transcripts are induced selectively in response to the influx of extracellular calcium. Furthermore, since promoter IV-derived Bdnf transcripts comprise the majority of activity-induced Bdnf transcripts expressed in the cortex, the disruption of activity-regulated promoter IV activity results in a substantial impairment in overall neuronal activity-dependent Bdnf transcription in the cortex.
Several phenotypic aspects of the CREmKI mutant animal also suggest that the changes in the development and function of its nervous system specifically reflect defects in the neuronal activity-regulated expression of Bdnf. For instance, in contrast to total Bdnf−/− knockout mice which typically die in the first few days of life (Ernfors et al., 1994; Jones et al., 1994), CREmKI mutants are viable, fertile, and appear indistinguishable from control littermates. In addition, at a gross anatomical level CREmKI brains do not appear significantly different from wildtype or loxP control littermate brains (Supplemental Figure 4A), and cortical layering in CREmKI brains appears normal (Supplemental Figure 4B), suggesting that impairment of activity-dependent promoter IV expression does not have strong effects on neuronal survival. Furthermore, whereas total Bdnf−/− null animals have a dramatic reduction in the number of inhibitory neurons in the cortex (Jones et al., 1994), we do not observe a change in inhibitory neuron number in CREmKI cortex (Figure 7E–F), again indicating that not all Bdnf-dependent phenotypes are affected by deficits in activity-dependent promoter IV activation. Like in Bdnf+/− heterozygous mutants, the level of cortical BDNF protein in CREmKI animals reared in standard housing conditions is approximately 50% of controls (Supplemental Figure 3), and, like CREmKI animals, Bdnf+/− heterozygotes also exhibit deficits in the development of inhibition in the visual cortex (Abidin et al., 2008). However, a simple 50% reduction in BDNF protein level cannot account for the phenotype of the CREmKI animal because, unlike Bdnf+/− heterozygous mutants that develop obesity by three months of age (Kernie et al., 2000; Lyons et al., 1999), preliminary evaluation suggests that CREmKI mutants do not exhibit changes in body weight regulation (Supplemental Figure 7). Taken together, these observations are consistent with the idea that the defects in cortical inhibition seen in the CREmKI mutant reflect a requirement for Bdnf function that depends specifically on the activity-dependent expression of the gene. It is important to note that, since activity-dependent Bdnf expression is significantly attenuated, but not completely eliminated in the CREmKI animal, we cannot exclude the possibility that many of the well-characterized phenotypes of Bdnf−/− and Bdnf+/− mice that are not apparent in the CREmKI animal may ultimately prove to also be dependent on activity-dependent Bdnf transcription.
The finding that activity-dependent Bdnf expression is important for the development of GABAergic synapses in the visual cortex indicates an important role for activity-regulated gene transcription in the control of cortical excitability. The establishment and maintenance of appropriate excitatory-inhibitory balance is critical to normal brain physiology and function; defects in excitatory-inhibitory balance can lead to the emergence of seizures and aberrant critical periods for cortical plasticity (Hensch and Fagiolini, 2005; Sun, 2007). Previous studies have demonstrated that a reduction in excitatory drive caused by sensory deprivation leads to a decrease in GABA immunoreactivity in the visual cortex (Benevento et al., 1995; Hendry and Jones, 1986), suggesting the hypothesis that the level of neuronal activity can adjust the strength of cortical inhibition to maintain appropriate firing rates in cortical circuits. Since BDNF levels can serve as a molecular “sensor” of global levels of neuronal activity, it has been suggested that the induction of Bdnf expression in response to increases in the level of neuronal activity may act to dampen cortical excitability by promoting the development and/or strengthening of inhibitory synapses in local circuits (Genoud et al., 2004; Rutherford et al., 1997). However, direct evidence that the neuronal activity-dependent regulation of Bdnf expression controls the level of excitability of cortical circuits has been lacking. Our finding that the disruption of neuronal activity-regulated Bdnf expression impairs the development of inhibitory synapses provides strong evidence in support of this hypothesis. Since levels of Bdnf no longer function as an accurate sensor of neuronal activity in CREmKI brains, cortical circuits appear to form as if under conditions of chronic sensory activity deprivation and fail to develop the appropriate level of inhibition. Important future experiments include pinpointing the source(s) of activity-dependent BDNF expression and release, the cellular target(s) on which it acts, and understanding whether activity-regulated BDNF functions equivalently at all inhibitory inputs or may have local effects near specific or active synapses.
In addition to demonstrating the biological significance of activity-regulated Bdnf transcription for brain development and function, our findings provide new insight into the action of CREB in the nervous system. We show that loss of the ability of CREB to bind and regulate Bdnf promoter IV in CREmKI brains results in defects in the development of cortical inhibition, implicating CREB, or possibly its closely related family members CREM and ATF-1, as having a previously unappreciated function in the development of inhibition in the nervous system. Our results with the CREmKI animal demonstrate that assessing the function of transcriptional regulators via the mutation of their binding sites in their target promoters is a powerful approach that can deconvolve the many functions of the factor (Xiang et al., 2006). Additionally, in the case of a transcriptional regulator like CREB that belongs to a multi-factor family, the approach of mutating the factor binding site in target promoters of interest avoids a potential complication of gene knockout studies, in which compensation of one family member for another’s function is common.
Our results also provide insight into the role of CREB in the formation of transcriptional regulatory complexes at gene promoters. We find that loss of CREB binding in CREmKI brains results in disruption of core components of the transcriptional activating complex at Bdnf promoter IV, including CBP and PolII, as well as loss of another sequence-specific transcription factor MEF2 that cooperates with CREB to regulate promoter IV activity (S.W.F., T.K.K., and M.E.G, manuscript in preparation). These results suggest a central function for CREB in nucleating the assembly of the multi-factor transcriptional regulatory complex that mediates calcium-dependent exon IV transcription. This idea is supported by our observation that additional mutation of the CaRE1 and CaRE2 sites in addition to the CaRE3/CRE site does not impair activity-dependent promoter IV induction significantly more than mutation of the CREB-binding site alone. Although there is evidence that CREB, both directly and through its association with CBP, can bind and stabilize components of the pre-initiation complex (Lonze and Ginty, 2002), whether CREB recruits other sequence-specific transcriptional activators to its target promoters was less clear. One possibility is that the co-activator CBP has additional functions as a molecular scaffold to bridge the interactions among the many factors in a transcriptional complex, such that recruitment of CBP by activated CREB leads to the stabilization of additional sequence-specific factors at the promoter. Alternatively, CREB may interact directly with other sequence-specific factors to recruit them to promoter IV, as has recently been shown for the transcriptional regulator MeCP2 (Chahrour et al., 2008). Further characterization of the multi-factor transcriptional complex, as well as putative changes in chromatin state, at Bdnf promoter IV in CREmKI neurons may yield new insight into the general mechanisms by which CREB coordinates the stimulus-specific activation of its target promoters.
Taken together, our results demonstrate that the neuronal activity-dependent component of gene expression is an important functional mediator of the effects of neuronal activity in reorganizing synaptic connectivity in the nervous system. It is remarkable that the impairment in the ability of neuronal activity to trigger the expression of just one of eight promoters of a single gene, Bdnf, results in significant defects in the development of cortical inhibition, highlighting the importance of the exquisite level of transcriptional control over Bdnf expression in the nervous system. Our finding that activity-regulated Bdnf expression is an important mediator of the effect of neuronal activity on the development of cortical inhibition suggests that, more generally, a major function of the program of neuronal activity-regulated gene expression may be to regulate the level of excitability of cortical circuits. Aberrant excitatory-inhibitory balance in the cortex can have significant consequences for the nervous system, including the development of seizures and abnormal critical periods for cortical plasticity, two common features of mental retardation, neurodevelopmental and autism spectrum disorders (Hensch and Fagiolini, 2005; Rubenstein and Merzenich, 2003). These observations highlight the importance of continued investigation into the full program of neuronal activity-regulated genes, and the synapse-to-nucleus signaling pathways that regulate their expression, for understanding how experience shapes the nervous system and how defects in this process may lead to disorders of cognition.
METHODS
See Supplemental Methods for details of animal husbandry and colony management, neuronal cell culture, immunocytochemistry, immunohistochemistry, image analysis, and electrophysiology
Generation of Bdnf pIV−/−, TMKI, CREmKI, and loxP control mice
See Supplemental Methods for description of gene targeting. The Bdnf pIV deletion spans basepairs −160 to +281 relative to the TSS. All experiments were performed with animals in a sv129/C57BL/6 mixed genetic background, with the mutation backcrossed into the C57BL/6 background between four and six generations.
Visual stimulation and seizure induction
Eight- to twelve-week old adult mice reared in a 12-hour:12-hour light:dark cycle were transferred into constant darkness for 14 days. Animals in the stimulated (+) condition were exposed to light for 90 minutes. Seizures were induced in 8–12 week old adult mice by intraperitoneal injection of kainic acid (20 mg/kg). At the indicated times post-injection, mice were killed and the cortex isolated. See Supplemental Methods for details.
Gene expression analysis
cDNA was synthesized by reverse transcription of total RNA using oligo-dT priming, and transcript levels were measured by quantitative real-time PCR using SYBR Green detection. BDNF protein levels were quantified by two-site ELISA. See Supplemental Methods for details.
Chromatin immunoprecipitation
For anti-CREB or anti-MEF2D ChIP, the antibody was pre-incubated with either the antigen peptide against which the antibody was raised (negative control IP) or an irrelevant peptide (experimental IP) prior to use in immunoprecipitation. For all other factors, normal mouse or rabbit IgG was used for the negative control IP. See Supplemental Methods for details.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. E. West for stimulating discussions and guidance in the conceptual design of this study; P. L. Greer, A. E. West, J. Zieg, and E. C. Griffith for critical readings of the manuscript; M. Gee for assistance with mouse colony management; B. L. Bloodgood and C. Chen for technical consultations about electrophysiological recordings and analysis; C. Chen and M. Fagiolini for the use of their dark-rearing facilities; the MRDDRC Gene Manipulation Core (M. Thompson, Y. Zhou, and H. Ye); the MRDDRC Imaging Core (L. Bu); and the MRDDRC Histology Core (M. Liana).
M.E.G. acknowledges the generous support of the F. M. Kirby Foundation to the Neurobiology Program at Children's Hospital. This work was supported by a John and Fannie Hertz Foundation fellowship to E.J.H., NIH grant R01-NS-048276 to M.E.G., and MRDDRC grant NIH-P30-HD-18655.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abidin I, Eysel UT, Lessmann V, Mittmann T. Impaired GABAergic inhibition in the visual cortex of brain-derived neurotrophic factor heterozygous knockout mice. J Physiol. 2008;586:1885–1901. doi: 10.1113/jphysiol.2007.148627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abidin I, Kohler T, Weiler E, Zoidl G, Eysel UT, Lessmann V, Mittmann T. Reduced presynaptic efficiency of excitatory synaptic transmission impairs LTP in the visual cortex of BDNF-heterozygous mice. Eur J Neurosci. 2006;24:3519–3531. doi: 10.1111/j.1460-9568.2006.05242.x. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balschun D, Wolfer DP, Gass P, Mantamadiotis T, Welzl H, Schutz G, Frey JU, Lipp HP. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- Benevento LA, Bakkum BW, Cohen RS. gamma-Aminobutyric acid and somatostatin immunoreactivity in the visual cortex of normal and dark-reared rats. Brain Res. 1995;689:172–182. doi: 10.1016/0006-8993(95)00553-3. [DOI] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AR, Chen C, Schwartz PM, Segal RA. Brain-derived neurotrophic factor modulates cerebellar plasticity and synaptic ultrastructure. J Neurosci. 2002;22:1316–1327. doi: 10.1523/JNEUROSCI.22-04-01316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003a;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 2003b;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System. Annual Review of Neuroscience. 2008;31 doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K, Wong RO. A comparison of experience-dependent plasticity in the visual and somatosensory systems. Neuron. 2005;48:465–477. doi: 10.1016/j.neuron.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Gass P, Wolfer DP, Balschun D, Rudolph D, Frey U, Lipp HP, Schutz G. Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn Mem. 1998;5:274–288. [PMC free article] [PubMed] [Google Scholar]
- Genoud C, Knott GW, Sakata K, Lu B, Welker E. Altered synapse formation in the adult somatosensory cortex of brain-derived neurotrophic factor heterozygote mice. J Neurosci. 2004;24:2394–2400. doi: 10.1523/JNEUROSCI.4040-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Jones EG. Reduction in number of immunostained GABAergic neurones in deprived-eye dominance columns of monkey area 17. Nature. 1986;320:750–753. doi: 10.1038/320750a0. [DOI] [PubMed] [Google Scholar]
- Hensch TK, Fagiolini M. Excitatory-inhibitory balance and critical period plasticity in developing visual cortex. Prog Brain Res. 2005;147:115–124. doi: 10.1016/S0079-6123(04)47009-5. [DOI] [PubMed] [Google Scholar]
- Hong EJ, West AE, Greenberg ME. Transcriptional control of cognitive development. Curr Opin Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu XZ, Xu K, et al. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci. 2008;28:1118–1130. doi: 10.1523/JNEUROSCI.2262-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KR, Farinas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kernie SG, Liebl DJ, Parada LF. BDNF regulates eating behavior and locomotor activity in mice. Embo J. 2000;19:1290–1300. doi: 10.1093/emboj/19.6.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-w. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AK, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of GABAergic synapse development by Npas4. Nature. 2008 doi: 10.1038/nature07319. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarollo L. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci U S A. 1999;96:15239–15244. doi: 10.1073/pnas.96.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Ohba S, Ikeda T, Ikegaya Y, Nishiyama N, Matsuki N, Yamada MK. BDNF locally potentiates GABAergic presynaptic machineries: target-selective circuit inhibition. Cereb Cortex. 2005;15:291–298. doi: 10.1093/cercor/bhh130. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Riccio A, Alvania RS, Lonze BE, Ramanan N, Kim T, Huang Y, Dawson TM, Snyder SH, Ginty DD. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Robertson LM, Kerppola TK, Vendrell M, Luk D, Smeyne RJ, Bocchiaro C, Morgan JI, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D, Tafuri A, Gass P, Hammerling GJ, Arnold B, Schutz G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc Natl Acad Sci U S A. 1998;95:4481–4486. doi: 10.1073/pnas.95.8.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford LC, DeWan A, Lauer HM, Turrigiano GG. Brain-derived neurotrophic factor mediates the activity-dependent regulation of inhibition in neocortical cultures. J Neurosci. 1997;17:4527–4535. doi: 10.1523/JNEUROSCI.17-12-04527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz PM, Borghesani PR, Levy RL, Pomeroy SL, Segal RA. Abnormal cerebellar development and foliation in BDNF−/− mice reveals a role for neurotrophins in CNS patterning. Neuron. 1997;19:269–281. doi: 10.1016/s0896-6273(00)80938-1. [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Sun QQ. The missing piece in the 'use it or lose it' puzzle: is inhibition regulated by activity or does it act on its own accord? Rev Neurosci. 2007;18:295–310. doi: 10.1515/revneuro.2007.18.3-4.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Belluardo N, Persson H, Metsis M. Developmental regulation of brain-derived neurotrophic factor messenger RNAs transcribed from different promoters in the rat brain. Neuroscience. 1994;60:287–291. doi: 10.1016/0306-4522(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Xiang H, Wang J, Boxer LM. Role of the cyclic AMP response element in the bcl-2 promoter in the regulation of endogenous Bcl-2 expression and apoptosis in murine B cells. Mol Cell Biol. 2006;26:8599–8606. doi: 10.1128/MCB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci. 2004;5:385–399. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.