Abstract
It is postulated that chickens (Gallus gallus domesticus) became domesticated from wild junglefowls in Southeast Asia nearly 10,000 years ago. Based on 19 individual samples covering various chicken breeds, red junglefowl (G. g. gallus), and green junglefowl (G. varius), we address the origin of domestic chickens, the relative roles of ancestral polymorphisms and introgression, and the effects of artificial selection on the domestic chicken genome. DNA sequences from 30 introns at 25 nuclear loci are determined for both diploid chromosomes from a majority of samples. The phylogenetic analysis shows that the DNA sequences of chickens, red and green junglefowls formed reciprocally monophyletic clusters. The Markov chain Monte Carlo simulation further reveals that domestic chickens diverged from red junglefowl 58,000±16,000 years ago, well before the archeological dating of domestication, and that their common ancestor in turn diverged from green junglefowl 3.6 million years ago. Several shared haplotypes nonetheless found between green junglefowl and chickens are attributed to recent unidirectional introgression of chickens into green junglefowl. Shared haplotypes are more frequently found between red junglefowl and chickens, which are attributed to both introgression and ancestral polymorphisms. Within each chicken breed, there is an excess of homozygosity, but there is no significant reduction in the nucleotide diversity. Phenotypic modifications of chicken breeds as a result of artificial selection appear to stem from ancestral polymorphisms at a limited number of genetic loci.
Introduction
When and how domestication proceeded during the history of modern humans are intriguing questions that have attracted much attention from researchers in natural and cultural sciences. Given the progress in genomic sequencing of various animals, the search for target genes of artificial selection during the domestication process has been a focus of recent studies. Because of the relatively short divergence times of domestic animals from their wild ancestors, genomic differences are likely to be subtle. It has therefore been a great challenge to identify genetic changes that resulted from artificial selection during the domestication process. The discovery of genetic differences among domestic breeds should, however, provide some clues. Some recent studies reveal genetic determinants of changes in body size and coat color in dogs [1], [2]. Also, molecular searches for the origins of domestic animals are conducted on other animals such as pigs, sheep, cows, and chickens [3]–[8].
The origins of domestic chickens (Gallus g. domesticus) have been debated ever since Darwin [9]. Archeological remains of domestic chickens are found in 16 Neolithic sites along the Yellow River in Northeast China as well as in the Indus Valley. Because some of these remains date back to ∼8,000 years ago [10], domestication must have been undertaken at least since that time. It is suggested that domestic chickens originated from junglefowls in Southeast Asia.
Four species of genus Gallus inhabit Southeast Asia: red junglefowl (G. g. gallus), La Fayette's junglefowl (G. lafayettei), gray junglefowl (G. sonnerati), and green junglefowl (G. varius). Red junglefowl has a strong sexual dimorphism with males having red fleshy wattles, and it is most widely distributed over the area. La Fayette's junglefowl morphologically resembles red junglefowl, but it inhabits only in Sri Lanka. Gray junglefowl has body plumage on a gray background color and is distributed from southwest to central India. Morphologically distinct green junglefowl is limited to Java and its immediate vicinity, Bali and Lombok. It has been debated whether any single species of the four, especially red junglefowl, predominantly contributed to the genome of domestic chickens (a single-origin hypothesis) or whether multiple species of the four made a substantial genetic contribution to domestic chickens (a multiple-origin hypothesis).
Darwin [9] proposed a single-origin hypothesis based on the observation that only red junglefowl can produce fertile F1 offspring in a cross with chickens. Subsequently, several hybridization experiments were performed to examine the genetic relationship among the four junglefowls and chickens. Danforth [11] reported complete hybrid fertility between red and gray junglefowls. Steiner [12] stated that although F1 females of red and green junglefowls show reduced fertility, they produce F2 hybrids in backcrosses with red junglefowl. In addition, Morejohn [13] found hybridization of gray junglefowl and chickens in the vicinity of villages. Those observations suggested a possible contribution of the four junglefowls to the origin of domestic chickens and favored the multiple-origin hypothesis.
Recently, molecular approaches have been more commonly used to obtain less ambiguous results than classical approaches. Using the D-loop sequences of mitochondrial DNA (mtDNA) in various gallinaceous birds in family Phasianidae, Fumihito et al. [4] concluded that two red junglefowl subspecies, G. g. gallus and G. g. spadiceus, are the direct ancestors of chickens, but that another subspecies, G. g. bankiva, does not contribute. This conclusion was subsequently supported by other studies using microsatellite DNA [14] and a large number of D-loop sequences [15]. Recently, these molecular data also show that Indian red junglefowl (G. g. murghi) also contributes to the domestication, as well as G. g. gallus and G. g. spadiceus [16]. In contrast, the phylogenetic analysis of the entire mtDNA genome and nuclear DNA (nucDNA) regions for four CR1 (chicken repeat 1) regions and OTC (ornithine carbamoyl transferase) revealed evidence for hybridization of gray junglefowl with chickens, red junglefowl, and La Fayette's junglefowl [17]. In particular, the analysis of the entire mtDNA genome showed the presence of identical haplotypes between gray junglefowl and chickens, and the nucDNA analysis of CR1 and OTC loci demonstrated an intermingling clustering pattern among chickens, red and gray junglefowls. These results raised the possibility that junglefowls other than red junglefowl were also involved in the domestication of chickens. In accordance with this possibility, genes responsible for yellow leg skin could be derived from gray junglefowl during the domestication [18].
In this paper, we determine 30 intron DNA sequences at 25 nuclear loci of four domestic chicken breeds consisting of 10 individuals in total. We also determine the orthologous sequences for the sample of four red junglefowls, four green junglefowls and one quail. Based on these sequence data together with the genome database of red junglefowl (build 1.1) and the GenBank database of turkey, we carry out phylogenetic and population genetic analyses. A particular attention is paid to polymorphisms shared among three Gallus species that can result from either inheritance of ancestral polymorphisms or introgression.
Materials and Methods
Ethics Statement
We adhered to the Guidelines for the Use of Experimental Animals authorized by the Japanese Association for Laboratory Animal Science. All experimental procedures were approved by Institutional Animal Care (National Institute of Agrobiological Sciences) and Use Committee, and all animals were housed and cared for according to guidelines established by the Committee.
Samples
The samples listed in Table 1 were collected from four chicken breeds (Shamo, sample size n = 1; Koshamo, n = 4; Ukokkei, n = 4; and White Leghorn, n = 1), red and green junglefowl species (each with n = 4), and quail (Coturnix japonica, n = 1). Shamo and Koshamo (small Shamo) are breeds of fighting cocks that originated in Thailand and that were imported to Japan by the early Edo period (A.D. 1603–1867). Ukokkei, also known as Silky, uniquely develops one or two extra backward toes. Also given in Table 1 are the locations of our samples and hereafter designated as capitalized abbreviations: SHAMO, KOSHA, UKO, WL, RJF, GJF and QUAIL for samples from each breed and species. CHICKENs mean the whole samples of chicken breeds, namely SHAMO, KOSHA, UKO, and WL, and JFs mean both RJF and GJF.
Table 1. Species and breeds used in this study, sampling locations and years, and sample designations.
| Species | Latin name | Breed | Sample ID | n a | Place of collection (Year of collection) |
| Domestic chickens | Gallus g. domesticus | Shamo | SHAMO | 1 | Okukuji, Japan (2004) |
| White Leghorn | WL | 1 | NILGS b, Tsukuba, Japan (2004) | ||
| Koshamo | KOSHA151, 152, 153, 154 | 4 | Kagoshima, Tokushima, Japan (1994) | ||
| Ukokkei (Silky) | UKO37, 38, 39, 40 | 4 | Yamagata, Niigata, Okinawa, Japan (1994) | ||
| Red junglefowl | G. g. gallus | RJF41, 45, 56, 58 | 4 | Palemberg, Indonesia (1994) | |
| Green junglefowl | G. varius | GJF301, 302, 303, 304 | 4 | Jakarta, Indonesia (1993) | |
| Quail | Coturnix japonica | QUAIL | 1 | NILGS, Tsukuba, Japan (2004) |
n = the number of individuals sampled per species or breed.
NILGS = National Institute of Livestock and Grassland Science.
Whole-blood samples (SHAMO, WL, and QUAIL) or blood blots on filter papers (KOSHA, UKO, RJF, and GJF) were collected from various areas in Japan or Indonesia (Table 1). The blood samples were provided by National Institute of Livestock and Grassland Science (NILGS) in 2004. Animals were maintained on a cycle of 12 h of light and 12 h of darkness. A commercial diet and water were provided. The blood-blotted filter papers of KOSHA and UKO were provided by Dr. Komiyama [19] currently in Tokai University. The filter papers of RJF and GJF were obtained by M. T. All of these were collected in 1993–1994 samples (in 1993, sampled by Dr. Shiraishi in The Research Institute of Evolutionary Biology) and were the same as those used in the previous studies [3], [4]. Sampling in Indonesia was performed with the permission by Indonesian government.
DNA extraction
Genomic DNA was extracted from whole blood using a Blood & Cell Culture DNA kit (QIAGEN). DNA on the filter paper was in limited quantities, so the following extra procedures were undertaken. To extract genomic DNA, the filter paper was cut into small pieces (3 mm in diameter) with a punch. Several of these pieces were immersed in 10 µl of alkaline lysis solution (400 mM KOH, 100 mM DDT, 10 mM EDTA) and mixed gently. After incubation on ice for 10 min, an equal volume of neutralization solution (400 mM HCl, 600 mM Tris HCl) was added. Genomic DNA was thus dissolved from the filter paper into the solution. An aliquot (1 µl) of this solution was used as a template in the whole-genome amplification, which was performed with the GenomiPhi DNA Amplification kit (GE Healthcare).
PCR and sequencing
Thirty introns at 24 autosomal and one Z-linked loci (Table 2) were amplified with Ex Taq (TaKaRa) DNA polymerase using primers previously designed for the comparison between chickens and turkeys [20]. The PCR reaction was performed with an initial denaturation at 95°C for 5 min, followed by 30–40 cycles of denaturation at 95°C for 30 sec, annealing at an appropriate temperature for 40 sec, and extension at 72°C for 1 min. Slightly different annealing temperatures were used for each primer set (available upon request). PCR products were purified with a S.N.A.P. Gel purification kit (Invitrogen) or an ExoSAP-IT kit (Amersham Biosciences). Purified products were directly sequenced or cloned (heterozygous samples) using a TOPO TA cloning kit or a TOPO XL PCR cloning kit (Invitrogen). To obtain reliable sequence data, 8–12 clones were sequenced for each PCR product except for those from SHAMO, WL, and QUAIL. For these three samples, the number of sequenced clones was limited to less than six for technical reasons, and a single sequence was used as a representative of each sample. The BigDye Terminator Cycle Sequencing kit (Applied Biosystems) was used with the corresponding PCR primers. Sequencing was carried out on ABI377 and ABI3100 sequencers (Applied Biosystems). Sequences were read at least twice in both directions. The nucleotide sequences obtained were deposited into DDBJ (accession numbers AB495408–496406).
Table 2. Chromosomal locations and nucleotide lengths of the introns sequenced in this study.
| Intron | Locus (abbreviation) | Length (bp) | Chromosome | GeneID a |
| 1 | Adenylate kinase (AK1int3) | 376 | 17 | D00251 |
| 2 | Annexin V, intron 5 (ANXA5int5) | 821 | 4 | 428767 |
| 3 | Annexin V, intron 7 (ANXA5int7) | 416 | 4 | 428767 |
| 4 | Crystalline, beta A1 (CRYBA1int2) | 801 | 19 | 396499 |
| 5 | Hemoglobin, alpha D (HBADint2) | 315 | 14 | 416651 |
| 6 | Creatine kinase, brain (CKBint3) | 529 | 5 | 396248 |
| 7 | Actin, beta, intron 2 (ACTBint2) | 485 | 2 | X00182 |
| 8 | Actin, beta, intron 3 (ACTBint3) | 312 | 2 | X00182 |
| 9 | Transcriptional repressor-delta EF1 (DEF1int3) | 665 | 2 | 396029 |
| 10 | Fatty acid synthase (FASNint41) | 1335 | 18 | 396061 |
| 11 | Growth hormone 1 (GH1int2) | 449 | 27 | 378781 |
| 12 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDHint2) | 286 | 1 | 374193 |
| 13 | Heat shock 108-kDa protein 1 (HSP108int2) | 698 | 1 | 374163 |
| 14 | Interleukin 8 (IL8int3) | 571 | 4 | 396495 |
| 15 | Ribosomal protein-coding gene L37A (RPL37Aint3) | 1121 | 7 | 769981 |
| 16 | Ribosomal protein-coding gene L5 (RPL5int3) | 613 | 8 | 395269 |
| 17 | Ribosomal protein-coding gene L7A, intron 3 (RPL7Aint3) | 358 | 17 | 417158 |
| 18 | Ribosomal protein-coding gene L7A, intron 4 (RPL7Aint4) | 351 | 17 | 417158 |
| 19 | Luteinizing hormone/choriogonadotropin receptor (LHCGRint7) | 513 | 3 | 395776 |
| 20 | Myosin light chain (MLCint4) | 477 | 7 | 396470 |
| 21 | Nicotinic acetylcholine receptor, gamma-subunit (ACGRint7) | 708 | 9 | 429151 |
| 22 | Opsin intron 1 (OPN1LWint1) | 528 | 19 | 396377 |
| 23 | Opsin intron 3 (OPN1LWint3) | 227 | 19 | 396377 |
| 24 | Opsin intron 4 (OPN1LWint4) | 381 | 19 | 396377 |
| 25 | Rhodopsin visual pigment intron 4 (RHOint4) | 879 | 12 | 396486 |
| 26 | Ribosomal protein-encoding gene L30 (RPL30int2) | 1006 | 2 | 425416 |
| 27 | Transforming growth factor-beta 2 (TGFB2int7) | 561 | 3 | 421352 |
| 28 | Vimentin (VIMint5) | 625 | 2 | 420519 |
| 29 | Spindlin on Z (SPINZint3) | 957 | Z | 395344 |
| 30 | Clathrin heavy chain (CLTCint7) | 676 | 19 | 395272 |
The genome data is build 2.1. For three cases (intron 1, 7, and 8) of which Gene ID is not available, accession numbers are indicated.
Phylogenetic analysis
Orthologous intron sequences were retrieved from the red junglefowl genome database (build 1.1) and those of turkey (Meleagris gallopavo) from GenBank (accession numbers: AY139863, AY139865, AY142943, AY142944, AY144673–AY144682, AY194143, AY298973–AY298989, AY3807788, AY380789). As before, these sequences are designated as RedDB and TURKEY. All the sequences were aligned using CLUSTAL W [21] and then adjusted manually whenever necessary. On the other hand, for the mtDNA D-loop, WL (AP003317), UKO (AB113070, AB086102), KOSHA (AB098676, AB098672, AB098671), RJF (NC_007236, AB007720), GJF (NC_007238, EU329408), TURKEY (NC_010195, AF532414), and QUAIL (NC_003408, NC_004575, X57245) sequences were used.
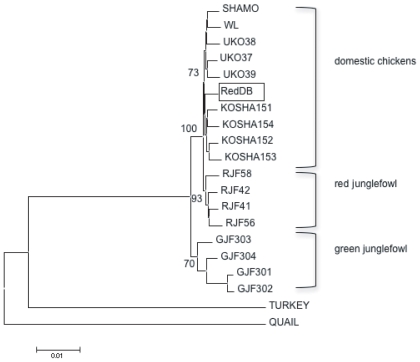
It is to be noted that only one sequence at each intron is available for SHAMO, WL, RedDB, QUAIL, and TURKEY, and thus the sequences from various introns of these individuals were uniquely concatenated. In contrast, the diploid sequences were determined at the 29 autosomal introns for most of KOSHA, UKO, RJF, and GJF (Table S1). Because we do not have any phase information among different introns, one of the diploid sequences at each intron was randomly selected and concatenated to represent an individual sample (a randomly concatenated sequence). For a set of 21 randomly concatenated sequences (four KOSHAs, four UKOs, four RJFs, four GJFs, SHAMO, WL, RedDB, QUAIL, and TURKEY), an individual tree was constructed based on the p-distances or the per-site nucleotide differences in a pair of individuals and the neighbor-joining (NJ) method [22]. The procedure of producing 21 randomly concatenated sequences was repeated to obtain 1,000 individual trees (Figure S1). As a summary of phylogenetic information, the individual p-distances were averaged over the 1,000 repeats (p a distances) and then used to construct the average-difference tree (Figure 1). Assignment of a cluster in the average-difference tree was measured as the proportion of its occurrence among the 1,000 individual trees. The value reflects the robustness of clustering in 1,000 individual trees.
Figure 1. The average-difference tree based on 1,000 concatenated sequences of randomly selected diploid sequences.
The proportion supporting a cluster is shown at each node as the realization of that cluster in the 1,000 individual trees. The TURKEY and QUAIL sequences are used as outgroups. The boxed RedDB indicates that the sequences were taken from the database of red junglefowl. Significant contributions of the domestic chicken genome to this database sequence are evident. The scale shown below the figure is a branch length corresponding to a per-site number of substitutions of 0.01 (1%). Abbreviations for samples are as follows. SHAMO: Shamo, WL: White leghorn, UKO: ukokkei, KOSHA: Koshamo, RJF: red junglefowl, GJF: green junglefowl.
Demographic analyses
Genetic variation within and between species was measured by the nucleotide diversity (π) and the nucleotide divergence (d), respectively. The subscripts of C, R, and G for π stand for domestic chickens, red and green junglefowls, respectively, and those for d stand for two species compared. To estimate the nucleotide divergences of mtDNAs (d mt) and nucDNAs (d nuc) as well as the nucleotide diversity (πmt and πnuc), Jukes and Cantor [23] or Kimura's two-parameter method [24] were used for multiple hits correction.
The “Structure” program by Pritchard et al. [25] was applied to examine the genetic ancestry of 16 samples of KOSHA, UKO, RJF, and GJF with observed genotypes at each intron. In the application, 10,000 iterations were performed for each of the burn-in and thereafter.
The Markov chain Monte Carlo (MCMC) method [26] and the two-species maximum-likelihood (TSL) method [27], [28] were applied to estimate the effective size of the ancestral and extant populations (N e) and the species divergence time (t s). In both methods, the estimates were scaled by the per-year nucleotide substitution rate (μ): θ = 4 N eµg and τ = t sµ, where g is the generation time in units of years. In the MCMC method, all the diploid sequences at each intron were used simultaneously. This method requires a gamma distribution to specify the prior distributions of θ and τ, which were calculated from the actual sequence data. We used two different sets of parameters (Table S2) to confirm the robustness of our estimates. For each parameter set, six runs were applied with a different set of random numbers. For each run, the number of iterations for the burn-in was 10,000, and the number of iterations after the burn-in was 200,000, with a thinning of two. Thus, coalescence-based genealogies were generated 100,000 times, from which the mean, standard errors, median, and 95% confidence limits of estimates were calculated.
The TSL method restricts the number of sequences at a locus to one for each species under study. For this restriction, a sequence was randomly selected from multiple sequences at an intron in each species. This random sampling process was repeated 1,000 times for each pair of species, and 1,000 maximum-likelihood (ML) estimates of θ and τ were obtained, from which the mean, standard error, and median were calculated. Our program for the ML estimation of θ and τ is written in Mathematica (version 4.0, Wolfram Research) and is available upon request.
Results
Phylogenetic analysis of concatenated diploid sequences
We examined one sample of each of SHAMO, WL and RedDB and four samples of each of UKO, KOSHA, RJF and GJF (Table 1). Unfortunately, not all the 30 intron sequences (Table 2) could be obtained from some individuals (see Table S1 for details). Most unsuccessful was UKO40 for which we could not determine the nucleotide sequences at nine introns and therefore excluded from the subsequent analyses. Thus, the total number of Gallus samples in the following analysis was 18.
Using the TURKEY and QUAIL as outgroup sequences, we compared 1,000 individual trees (Figure S1). In most of the trees, RJFs formed a single cluster, whereas RedDB was placed within a cluster of chicken breeds. Several substitutions were specific to a majority of RJFs. While these substitutions were responsible for their tight clustering, they were not shared with RedDB. As noted in the database, RedDB might not be pure red junglefowl, but a hybrid with a chicken breed (possibly White Leghorn). In what follows, we excluded RedDB.
Importantly, species-specific clustering was not seen for GJFs for many individual trees. Two samples of GJF303/304 (Table 1) were often assigned to a cluster of chicken breeds (e.g., individual trees 8, 9, and 1 in Figure S1) so that the introgression occurred from chickens to green junglefowl. The average p a distance (Table S3) of GJF303/304 from CHICKENs (UKO, KOSHA, WL and SHAMO) was 1.2±0.1%, significantly smaller than 1.7±0.1% in the case of GJF301/302 from CHICKENs. Although the average-difference tree (Figure 1) showed that the GJFs were reciprocally monophyletic to the RJFs and the CHICKENs, the branch lengths leading to GJF303/304 were apparently shorter than those leading to GJF301/302. This monophyletic relationships were supported by individual trees as well, but the proportion was 93%, 73% and 70% for the monophyletic cluster of RJFs, CHICKENs and GJFs, respectively (Figure 1).
Within the CHICKEN cluster, four KOSHAs formed a separate clade. Although the proportion of individual trees supporting this clade was low (30%), none of KOSHAs shared any haplotypes with any other CHICKENs at introns 11, 20, 21, and 25 (Table S1). This observation indicated that genetic differentiation in some genomic regions had indeed taken place among different chicken breeds.
NucDNAs vs. mtDNAs
To estimate the substitution rate of nuclear DNA in domestic chickens and junglefowls, we tested whether d nuc accumulated proportionally to elapsed time (see [29] for review). Actually, in the lack of solid information on speciation time, we tested if d nuc is proportional to d mt.
In doing such a test, we first noted that the Phasianidae mtDNA D-loop region was subjected to different degrees of sequence conservation. The region was divided into three domains, each of which contained a unique conserved nucleotide sequences, and because of such conservation, the D-loop region as a whole evolved relatively slowly [30] (see also [3], [31] for other galliforms and anseriforms). However, the sequence alignment clearly showed hypervariability in a certain region (data not shown). It was difficult to align the sequences from position 172 to 382 between chickens and quails/turkeys [30] and even between chickens and green junglefowl. For this reason, we omitted this 210 bp region from the analysis.
The sequence divergence between two related species reflected the nucleotide substitutions that accumulated after the speciation in addition to ancestral polymorphism [27], [32]. Even when the nucleotide substitutions accumulate in a steady fashion, the proportionality between nucDNA and mtDNA divergences does not hold true owing to ancestral polymorphism. A frequently used method for estimating the nucleotide divergences after speciation assumes that the extent of ancestral polymorphism is equal to that of extant populations [32], [33] and subtracts this from the sequence divergence between species. In the present study, however, this assumption was not warranted because domestication had likely influenced the demography of domestic chickens. Instead, we directly compared d nuc and d mt in distantly related pairs in Phasianidae where the effect of ancestral polymorphism on the divergence could be small enough to be ignored without serious errors.
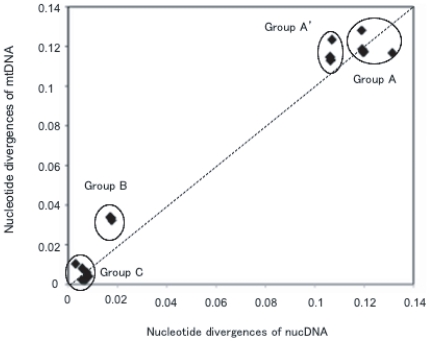
Figure 2 plotted d nuc and d mt for pairs of species, revealing the presence of four distinct groups (Group A, A′, B, and C). Groups A and A′ compared most distantly related species pairs between CHICKENs or JFs (RJFs and GJFs) and QUAIL or TURKEY, respectively. Group B compared GJFs with CHICKENs or RJFs, whereas Group C compared RJFs and CHICKENs as well as samples within CHICKENs or JFs. Thus, Group C included intra-specific comparisons.
Figure 2. Nucleotide divergence among mtDNA D-loop (y axis) and nuclear intron sequences (x axis).
The multiple-hit substitutions were corrected by the method of Jukes and Cantor [23] and Kimura [24]. Group A and A′ comprise comparisons of QUAIL and TURKEY, respectively vs. Gallus samples. Group B comprises comparisons of GJFs vs. RJFs and chickens. Group C represents inter- and intra-group comparisons of RJFs and chickens. The dotted line indicates d mt = d nuc. Each individual data point represents the comparison of a single species pair. Abbreviations for samples are the same as in Figure 1.
In both Groups A′ and B, d mt was slightly larger than d nuc. A greater departure from d mt = d nuc occurred in Group B than in Group A′, suggesting that even the conserved D-loop region underwent multiple-hit substitutions in more distantly related comparisons. Indeed, in the comparison of Group A′, 20 out of 261 variable sites possessed either three different kinds of nucleotides or phylogenetically incompatible substitutions at each site. This trend became more obvious in the most distantly related comparisons of Group A. A point was that d nuc in Group B and C increased almost linearly as d mt increased. We took this as evidence for the presence of molecular clock, or more precisely, if a mitochondrial clock exists in Phasianidae, so does a nuclear clock.
Shared and species-specific haplotypes
The number of haplotypes per intron per breed or species ranged from one to 11. Although this wide range certainly reflected the differences in sample size, the observed number of haplotypes fitted well with the expected number based on the sampling theory [34] (Table 3).
Table 3. Number of haplotypes (H) and the percent nucleotide diversity (π) in the 30 introns.
| Introns | Chickens | RJF | GJF | ||||||||||||
| n a | p | O(F)b | H | E(H)c | n | p | O(F) | H | E(H) | n | p | O(F) | H | E(H) | |
| 1 | 18 | 0.61 | 6 | 7 | 5.5 | 8 | 0.82 | 1 | 4 | 4.5 | 8 | 0.37 | 1 | 5 | 3.2 |
| 2 | 17 | 0.67 | 2 | 8 | 7.9 | 8 | 1.05 | 2 | 3 | 6.4 | 8 | 0.73 | 2 | 3 | 5.7 |
| 3 | 18 | 0.66 | 2 | 8 | 6.1 | 8 | 0.66 | 2 | 3 | 4.4 | 8 | 0.95 | 0 | 5 | 5 |
| 4 | 10 | 0.30 | 3 | 3 | 4.5 | 6 | 0.26 | 0 | 4 | 3.5 | –d | – | – | – | – |
| 5 | 18 | 0.47 | 7 | 2 | 3.9 | 8 | 0 | 4 | 1 | 1 | 8 | 0.33 | 2 | 3 | 2.6 |
| 6 | 18 | 0.25 | 3 | 6 | 4.1 | 8 | 0.23 | 1 | 4 | 3.1 | 8 | 0.13 | 1 | 3 | 2.4 |
| 7 | 18 | 0.33 | 5 | 5 | 4.6 | 8 | 0.27 | 0 | 5 | 3.2 | 8 | 0.56 | 2 | 5 | 4.4 |
| 8 | 16 | 1.14 | 6 | 6 | 6.5 | 8 | 0.85 | 0 | 5 | 4.3 | 8 | 0.90 | 2 | 4 | 4.4 |
| 9 | 16 | 0.71 | 4 | 6 | 7.4 | 8 | 0.82 | 0 | 4 | 5.5 | 8 | 0.79 | 1 | 4 | 5.5 |
| 10 | 18 | 1.26 | 4 | 9 | 12.7 | 8 | 1.06 | 1 | 5 | 7.1 | 8 | 1.37 | 0 | 7 | 7.4 |
| 11 | 18 | 0.65 | 5 | 10 | 6.3 | 8 | 0.69 | 1 | 5 | 4.6 | 8 | 0.40 | 2 | 3 | 3.6 |
| 12 | 18 | 0.50 | 5 | 6 | 4.2 | 8 | 0.83 | 2 | 3 | 4.1 | 8 | 1.65 | 2 | 4 | 5.3 |
| 13 | 16 | 1.26 | 4 | 7 | 9.8 | 8 | 1.63 | 2 | 5 | 6.8 | 8 | 1.79 | 0 | 6 | 7 |
| 14 | 18 | 0.72 | 3 | 8 | 7.5 | 8 | 0.43 | 2 | 3 | 4.2 | 8 | 1.17 | 2 | 4 | 6 |
| 15 | 16 | 0.42 | 5 | 6 | 6.7 | 8 | 0.65 | 3 | 3 | 5.7 | 8 | 1.93 | 2 | 4 | 7.3 |
| 16 | 18 | 0.58 | 4 | 6 | 7 | 8 | 0.40 | 2 | 3 | 4.2 | 8 | 0.67 | 2 | 5 | 5.1 |
| 17 | 18 | 0.49 | 5 | 8 | 4.8 | 8 | 0.65 | 2 | 4 | 4.1 | 8 | 1.93 | 0 | 7 | 6.1 |
| 18 | 18 | 0.67 | 7 | 8 | 5.6 | 8 | 0.28 | 3 | 3 | 2.7 | – | – | – | – | – |
| 19 | 18 | 0.82 | 5 | 5 | 7.5 | 8 | 0.56 | 1 | 4 | 4.4 | 8 | 1.00 | 1 | 5 | 5.5 |
| 20 | 18 | 0.85 | 6 | 8 | 7 | 8 | 0.31 | 2 | 4 | 3.4 | 8 | 1.23 | 1 | 6 | 5.7 |
| 21 | 16 | 0.40 | 2 | 9 | 5.9 | 8 | 0.24 | 4 | 2 | 3.6 | 8 | 0.71 | 1 | 5 | 5.5 |
| 22 | 16 | 0.72 | 7 | 6 | 6.8 | 8 | 0.89 | 1 | 3 | 5.3 | 8 | 1.17 | 2 | 3 | 5.8 |
| 23 | 18 | 0.86 | 5 | 6 | 5.1 | 8 | 0.87 | 0 | 3 | 3.8 | 8 | 0.57 | 2 | 3 | 3.1 |
| 24 | 18 | 0.49 | 6 | 6 | 5 | 8 | 1.90 | 3 | 4 | 6.1 | 8 | 1.90 | 1 | 4 | 6.1 |
| 25 | 18 | 0.57 | 4 | 9 | 8.2 | 8 | 0.48 | 1 | 6 | 5.2 | 8 | 0.60 | 2 | 6 | 5.6 |
| 26 | 16 | 1.09 | 2 | 10 | 9.5 | 8 | 1.08 | 1 | 4 | 6.3 | 8 | 1.77 | 0 | 8 | 7.1 |
| 27 | 18 | 0.12 | 8 | 4 | 2.9 | 8 | 0.33 | 2 | 3 | 3.7 | 8 | 0.71 | 0 | 4 | 5.1 |
| 28 | 18 | 1.22 | 4 | 6 | 9.8 | 8 | 0.27 | 2 | 2 | 3.5 | 8 | 0.63 | 0 | 5 | 5 |
| 29 | 13 | 0.08 | – | 3 | 2.7 | 7 | 0 | 0 | 1 | 1 | 8 | 0.15 | – | 4 | 2.8 |
| 30 | 18 | 0.47 | 7 | 5 | 6.6 | 8 | 0.36 | 1 | 5 | 4.2 | 8 | 0.86 | 2 | 4 | 5.7 |
n = the number of sampled chromosomes.
O(F) = the observed number of homozygotes.
E(H) = the expected number of haplotypes or alleles based on πL and Ewens's sampling theory [34], where L is the length of an intron, which is given in Table 2.
A dash indicates that the sequences were not obtained in this study.
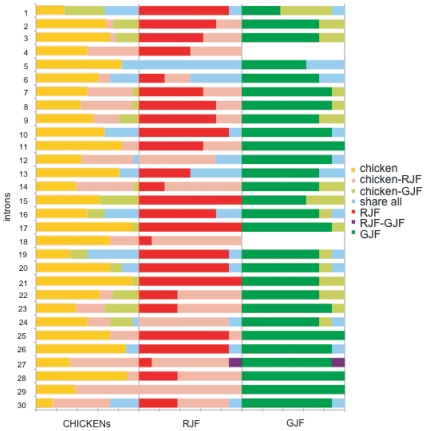
To quantify the extent of genetic information shared between CHICKENs and JFs, we examined the proportion of shared haplotypes at each intron (Figure 3). The average proportion of shared haplotypes per intron was about 21% between CHICKENs and RJFs, and it was about 11% between CHICKENs and GJFs. Thirteen haplotypes at 12 loci were shared among the three species groups, 24 at 19 loci shared between RJFs and CHICKENs, and 18 at 16 loci shared between GJFs and CHICKENs. In the case of GJFs, all of the shared haplotypes were represented in either GJF303 or GJF304, or both. In one case, a haplotype at intron 1 (AKI) shared with CHICKENs was found not only in both GJF303 and 304 but also in GJF301. In contrast to a relatively high proportion of haplotypes shared between CHICKENs and JFs, haplotypes shared between RJFs and GJFs were so rare that there was only one at intron 27 (TGFB2).
Figure 3. The proportion of shared haplotypes at 30 introns.
The 30 intron numbers in Table 2 are shown on the left. Different colors indicate different patterns of shared haplotypes as specified by the key on the right margin. CHICKENs in this Figure include only UKO and KOSHA samples. Abbreviations for samples are the same as in Figure 1.
Of course, there existed species- or breed-specific haplotypes as well. The proportion per intron ranged from 20% to 90% in CHICKENs, but for a particular breed at least one haplotype at each intron was shared with another breed or JFs. In contrast, there were species-specific haplotypes only at three introns (RPL37A, RPL7Aint3, ACGR) in RJFs and four introns (GH1, RHOint4, VIM, and SPINZ) in GJFs. As expected, the proportion of species-specific haplotypes was higher in GJFs than in RJFs.
Genetic variation within and between species
We estimated the nucleotide diversity (π) and divergence (d) at 30 introns within and between domestic chickens and junglefowls (Table 3, Table S4). The average π value was πC = 0.65±0.32% for CHICKENs and πR = 0.58±0.37% for RJFs. On the other hand, πG = 0.97±0.56% for GJFs and six introns (GAPD, HSP108, RPL37A, RPL7Aint3, OPSINint4, RPL30) individually exhibited high π values (>1.7%). However, if GJF303/304 were excluded, the π values at GAPD, RPL37A, OPSINint4, and RPL30 decreased substantially (Figure S2). The πG values at HSP108 and RPL7Aint3 introns were 1.8±1.1% and 1.9±1.4%, but they remained high at 1.7±1.0% and 1.3±0.9% even if GJF303/304 were removed. The cause of this large πG was probably due to a presence of haplotypes which were more closely related to CHICKENs or RJFs than to other haplotypes in GJFs.
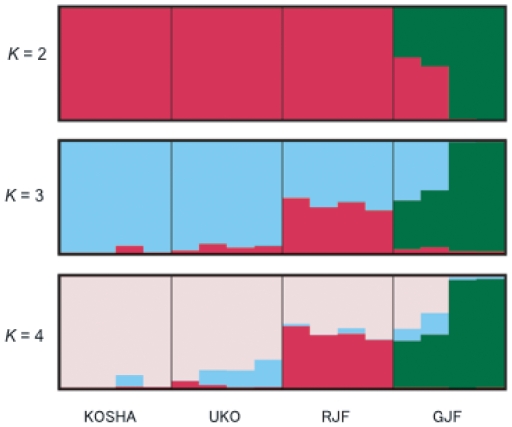
Population structure
To obtain an overview of the genetic components of CHICKENs, RJFs, and GJFs, we used the program Structure [25]. We tested several different models, but the results did not differ greatly. Figure 4 shows the result of the ‘admixture model’ with ‘allele frequency correlations.’ The most likely value of K was estimated as 2, suggesting that genetic components in these samples were likely to be classified into two groups. The first component was found mainly in RJFs and CHICKENs, and partially in GJFs, whereas the second component was exclusively found in GJFs. In this regard, RJFs and CHICKENs did not differ much genetically. In contrast, for K = 3 or 4, RJFs and CHICKENs became distinguishable. On one hand, approximately a half of the RJF genome was characteristic of the species, and a substantial portion of CHICKENs penetrated into RJFs and GJFs. On the other hand, GJFs and RJFs shared few genetic components, which was consistent with the low proportion of shared haplotypes (Figure 3). It appeared that unlike chickens, red and green junglefowls had been strongly isolated from each other since their speciation.
Figure 4. ‘Structure’ graph for individual proportions of shared ancestry.
Ancestry was estimated from diploid intron sequences in the samples from four KOSHAs, four UKOs, four RJFs, and four GJFs. Predefined populations are shown on the abscissa. Different colors within each block indicate different ancestries. Abbreviations for samples are the same as in Figure 1
MCMC and TSL analyses
We estimated the species divergence time (t s) and the effective size (N e) of the ancestral and extant populations from the present data. There are several such estimation methods including the TSL [27], [28], the tree mismatch [32], and the MCMC [26] methods. Because multiple sequences were available from each species, we preferred the MCMC method that does not give any limitation to the number of sequences from a species. Nevertheless, we were interested in comparing the TSL estimates with the MCMC estimates.
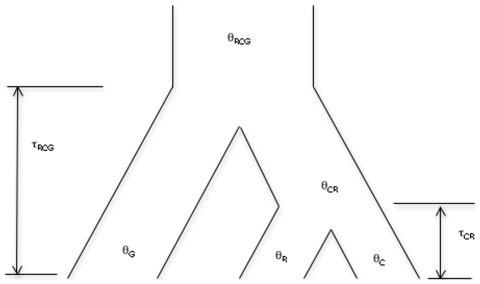
In the case of three species (Figure 5), θ = 4 N eµg and τ = t sµ were concerned with two species-divergence times (τCR and τRCG) and five effective sizes of ancestral and extant populations (θC, θR, θG, θCR, and θRCG). We conducted several independent runs of the MCMC method with different parameter sets in the prior distribution. The estimates were not much different for different parameter sets (data not shown).
Figure 5. The three species and demographic parameters as estimated by MCMC and TSL methods.
With the TSL method, only pairs of species are taken into account at once, such that one of the three consecutive subscripts of a demographic parameter is shown in parentheses in the text. The symbols of θ and τ stand for the nucleotide divergence due to the extent of polymorphism and the species divergence, respectively. The subscript G, R, C means the three species, green junglefowl, red junglefowl and domestic chickens, respectively.
We compared the MCMC results with and without GJF303/304. In their exclusion, both θG and θRCG decreased substantially and the decrease of θRCG was compensated by an increase in τRCG. We excluded GJF303/304 for further analyses. In the TSL method, we used 1,000 sets of randomly selected diploid sequences at each intron for each species (Table 4). For distantly related species, the MCMC method mostly resulted in d<θ+2τ, but the TSL method almost always resulted in d = θ+2τ [27], [28]. For closely related species, the two methods also resulted in different estimates of θ and τ, although θ+2τ did not differ much from the average nucleotide divergences.
Table 4. Comparison of means ± standard deviations between MCMC (Markov chain and Monte Carlo) and TSL (two species likelihood) estimates (%) based on 26 intron sequences with and without GJF303 and 304.
| Estimatesa | MCMCb [95% confidence limits] | TSL [median]c | |
| With GJF303 and 304 | |||
| θC | 0.24±0.06 [0.13, 0.37] | – | |
| θR | 0.10±0.03 [0.05, 0.16] | – | |
| θG | 0.58±0.08 [0.43, 0.75] | – | |
| θCR | 1.60±0.14 [1.34, 1.90] | 0.58±0.16 [0.58] | |
| θRCG | 1.94±0.30 [1.43, 2.58] | θCG = 1.10±0.25 [1.07],θRG = 1.06±0.22 [1.04] | |
| τCR | 0.01±0.003 [0.007, 0.02] | 0.13±0.08 [0.12] | |
| τRCG | 0.12±0.01 [0.10, 0.14] | τCG = 0.31±0.15 [0.32],τRG = 0.31±0.13 [0.33] | |
| Without GJF303 and 304 | |||
| θC | 0.26±0.06 [0.15, 0.39] | – | |
| θR | 0.11±0.03 [0.06, 0.18] | – | |
| θG | 0.34±0.06 [0.25, 0.46] | – | |
| θCR | 0.71±0.14 [0.47, 1.00] | 0.57±0.16 [0.58] | |
| θRCG | 1.61±0.15 [1.34, 1.92] | θCG = 0.80±0.10 [0.80],θRG = 0.86±0.08 [0.86] | |
| τCR | 0.01±0.003 [0.008, 0.02] | 0.13±0.08 [0.12] | |
| τRCG | 0.63±0.06 [0.52, 0.74] | τCG = 0.59±0.03 [0.58],τRG = 0.55±0.02 [0.55] | |
The symbols of θ and τ stand for the nucleotide divergence due to the extent of polymorphism and the species divergence, respectively. θ = 4 N eµg and τ = t sµ, where g is the generation time in units of years, N e is the effective population size, t s is the species divergence time and µ is the per-year nucleotide substitution rate.
Scale and shape parameters for gamma distributions of prior MCMC analyses are given in Table S2, and the result for set 1 is shown here.
Two different pairs of species were used for the TSL method. The estimates for the pair of chickens and green junglefowl are shown as θCG and τCG, and those for the pair of red and green junglefowls are indicated as θRG and τRG.
Disucussion
Inferring a species tree from gene trees
There are several methods for inferring a species tree from multiple gene trees. Generally, these methods can be classified into one of two approaches: the combined or separate approach. In the combined approach, sequences from multiple loci are concatenated, and the resulting data set is analyzed using phylogenetic methods. In the separate approach, the sequence data from each locus is first analyzed individually, and then a reconciliation of different gene trees is obtained.
In this study, we used a slightly modified version of the combined approach. Because of diploid sequences available at 26 loci and because of numerous phase combinations per individual, a consensus tree was constructed based on the average distances of 1,000 sets of random sampling of concatenated sequences (the average-difference tree; Figure 1). Basically, each topology of 1,000 individual trees based on individual sampling is similar to that of the average-difference tree. In contrast, when a tree at each intron is constructed separately, the resulting trees shows that the individual samples are intermingled with each other and do not have a sufficient power to distinguish among CHICKENs, RJFs, and GJFs. In the lack of phase information, combining data at different loci is advantageous to minimize noise [35], [36]. The average-difference tree clearly shows that there is a monophyletic relationship among chickens, red and green junglefowls and that chickens are more closely related to red junglefowl than green junglefowl.
Estimation of species divergence times from d mt and d nuc
The relationship between d mt and d nuc shows a rough linearity, although there is some indication of saturations in d mt when distantly related species are compared (Groups A and A′ in Figure 2). To avoid the saturation effect as much as possible, we used the divergences in Group B with d nuc = 1.9±0.9% and d mt = 3.3±0.1% (Figure 2). The ratio of d mt to d nuc is 1.7, indicating that the conserved mtDNA D-loop region of Gallus evolves only 1.7 times faster than nucDNA. This is in sharp contrast to mammals where the D-loop region may have evolved approximately 100 times faster than nucDNA [37]. Thus, even for such deep phylogenies as those in family Phasianidae, mtDNA together with nucDNA can give reliable evolutionary signals.
We used the divergence time between quails and chickens to calibrate the absolute nucDNA substitution rate. Three independent but similar estimates of the divergence time were available: The fossil record dates the species divergence to 32–38 million years (myr) ago [38], mtDNA sequences to 33–36 myr ago [39], and micro-complement fixation analyses to 38 myr ago [40]. We assume the divergence time to be 35 myr ago. Using the average d nuc = 12±0.5% in the comparison of chickens with quails and ignoring ancestral polymorphisms, we estimate the rate as 1.7±0.07×10−9 per site per year. This rate is almost the same as that in the previous report [20] and slightly greater than the rate in primates [28]. Likewise, the mtDNA substitution rate is simply 1.7 times higher and estimated as 2.9×10−9 per site per year.
The above calibrated rate and d nuc = 11±0.03% between turkeys and junglefowls lead to their divergence time of 31±0.7 myr. Likewise, d nuc = 1.9±0.9% between green junglefowl and chickens indicates the divergence time of 5.5±2.7 myr, which is more recent than the previous estimates [39] (see also [41], [42]). In either case, the actual divergence time may be even shorter than what the d value indicates, because d for closely related species might be substantially increased by ancestral polymorphism.
Demographic history of green junglefowl and introgression of the chicken genome
The MCMC and TSL methods attempt to remove the effect of ancestral polymorphisms on estimates of the species divergence time. The MCMC estimate of τRCG is 0.63±0.06% (Table 4) and this is indeed smaller than d nuc/2 = 0.95%. The net species divergence time (t RCG) of green junglefowl becomes 3.6 myr (95% confidence interval is 3.0–4.3 myr). Similarly, the TSL estimate of τ CG or τRG is 0.59±0.03% or 0.55±0.02%, which indicates that t CG = 3.4±0.2 myr or t RG = 3.2±0.1 myr. Both the MCMC and TSL estimates of t RCG are thus in agreement with each other. These estimates are smaller than not only 4–5 myr based on the fossil record (see [37] and references therein), but also the d nuc-based estimate of 5.5 myr. On the other hand, they are consistent with a volcanic origin of Java in the Plio-Pleistocene era (3–4 myr). Because green junglefowl is limited to Java and its immediate vicinity, the speciation may have coincided with this geological event [41].
The presence of haplotypes shared with chickens can be explained by ancestral polymorphisms and/or introgression. To distinguish these two causes, we calculate the probability (P C) that two shared haplotypes are identical by descent for a given time duration. Using μ = 1.7×10−9 per site per year and the MCMC estimate of t RCG of 3.6 myr, P C becomes <0.005 if the sequence length is >250 bp. This indicates that a pair of orthologous sequences can be identical with the probability of less than 0.005. Among the 26 intron sequences analyzed, 25 introns are >250 bp long, and 22 or 17 haplotypes at the 25 introns are identical between chickens and GJF303 or 304. We therefore conclude that these identical sequences found between chickens and green junglefowl are caused by recent introgression of the chicken genome into green junglefowl.
The multiple-origin hypothesis of domestic chickens postulates different junglefowl species that were involved in the origin of domestic chickens. Although red and green junglefowls can still hybridize with chickens and the genome of one species is actually transferred over the species boundary, these observations are not direct evidence for the multiple-origin hypothesis of domestic chickens. Rather, green junglefowl has been genetically distinct from chickens since green junglefowl speciated around the time of the uplift of the Java islands.
The possibility that other species such as La Fayette's and gray junglefowls were involved in the domestication remains to be elucidated. Unfortunately, our samples include neither of these junglefowls nor any other subspecies of red junglefowl. We do, however, hope that we will be able to depict a clear picture once such samples are used in the present framework.
Ancient divergence and a large ancestral population of red junglefowl
At 27 out of 30 introns in red junglefowl, shared haplotypes with chickens are observed. The proportion of shared haplotypes at each intron is much higher in red junglefowl than in green junglefowl. Importantly, these shared haplotypes are evenly distributed over all samples of red junglefowl, whereas this is not the case for those of green junglefowl.
According to archeological findings, the divergence time of domestic chickens from junglefowls is estimated to be on the order of 10,000 years. The MCMC method reveals, however, that the extent of nucleotide divergence after the split of red junglefowl from the chicken ancestor is as small as τCR = 0.01% (Table 4). With the calibrated μ, the divergence time t CR becomes 58,000±16,000 years ago. This dating is nearly six times older than what the archeological remains suggest. Because the MCMC model assumes the complete separation of species, it may still underestimate t CR if subsequent introgression occurred. The relatively small 95% confidence limit of the MCMC estimate (Table 4) does not reconcile the discrepancy. In any case, more intron sequences from appropriate samples from red junglefowl will be needed to prove or disprove the rather old species divergence of red junglefowl.
During this period of divergence time, the expected number of substitutions is less than one even in the longest intron in our sample. It is thus most likely that shared haplotypes are identical by descent. Nonetheless we cannot exclude the possibility of introgression. Given that introgression takes place between green junglefowl and chickens, it is even more likely to occur between a more closely related pair of species, red junglefowl and chickens.
The genetic variation within chicken breeds
The MCMC estimate of θC = 0.26% for the entire CHICKENs is twice as large as θR = 0.11% for RJFs (Table 4). An important implication of θC is that distinct ancestral lineages of red junglefowl could have been used for domestication. Again, with the calibrated μ and the generation time (g) of one year, the effective size of domestic chicken population becomes as large as 4×105. This estimate is 2.5 times larger than the estimate of 1.6×105 for the extant red junglefowl population, but it is slightly smaller than the estimate of 5×105 for the extant green junglefowl population. On the other hand, the effective size of the ancestral population of red junglefowl and chickens is estimated as 106 from θCR = 0.71% (with a 95% confidence interval of 0.7–1.4×106) and the ancestral size of three species is estimated as 2.3×106. Compared with these values, the effective population size has been reduced to less than half in all the three extant species.
The genetic variation maintained within chicken breeds is also of particular interest. Koshamo and Ukokkei clearly show an excess of homozygotes as compared with the Hardy-Weinberg equilibrium (Table S5). Out of eight samples, the number of expected and observed homozygotes averaged over 29 autosomal introns is 2.2 and 4.7, respectively. This excess most likely reflects intensive inbreeding within each breed. However, the π value within Koshamo (0.52±0.36%) and Ukokkei (0.63±0.35%) is actually similar to the value for chickens as a whole, 0.65±0.32%. These values are also similar to the π values estimated for other breeds using SNP data across the entire genome [43]: 0.43% for Broiler, 0.37% for Layer, and 0.55% for Ukokkei. Clearly, each individual tends to be genetically homogeneous but each chicken breed may not. Only when we examine commercially pure lines of Broilers and Layers, significant reduction in genetic variation is observed [44].
To examine this feature in more detail, we applied the MCMC method to two breeds; Koshamo and Ukokkei with an extremely short divergence time. The estimated θ is 0.05% for Koshamo and 0.16% for Ukokkei with τ = 0.005% and ancestral θ = 0.68%. The extent of polymorphism significantly differs from one breed to another, but most of the genetic variation can be attributed to a common ancestral population, with only a minor proportion associated with introgression. More interestingly, although the small τ value indicates 29,000 years, the two ancestral lineages leading to Koshamo and Ukokkei appear to have been distinct well before their domestication. Many distinct lineages of the ancestor may have been involved in chicken domestication, but they are not represented in the extant populations of junglefowls. If this is the case, we would not expect the MCMC estimates of species divergence times to coincide with the actual time of chicken domestication.
Conclusion
In conclusion, domestic chickens are closely related to red junglefowl, although genetic contribution from other junglefowls remains to be answered. The ancestral population size for domestic chickens is large and each breed still maintains a substantial portion of ancestral polymorphisms at the genomic level. Despite this, domestic chickens have undergone substantial changes in morphology and physiology. In this paper, we have demonstrated that past artificial selection does not substantially reduce the genetic variation of the domestic chicken genome. If this is the case, it is inevitable to conclude that the number of genes involved in the domestication process is likely to be small.
Supporting Information
Examples of individual trees. Each tree is constructed from randomly concatenated sequences (see text). Scale of each tree is indicated at the bottom of each tree. Red rectangle indicates RedDB, and green rectangle means either GJF303 or GJF304, which is not in a green junglefowl cluster. Brackets at the right side indicate a cluster of C (chickens), R (red junglefowl), and G (green junglefowl), respectively.
(0.33 MB PDF)
Effect of introgressed sequences from chickens to GJF on the nucleotide diversity (pai) of GJF. Pai values at each intron with and without GJF303/304 were located on both sides of the graph. Among 30 introns, intron 13 and 17 were only the exception where removal of introgressed sequences did not reduce the pai value.
(0.06 MB PDF)
Haplotypes (numbers) of each individual.
(0.34 MB PDF)
Two sets of scale and shape parameters (a, b) of the gamma distribution in MCMC analyses and prior distribution (%).
(0.03 MB PDF)
The average distances of 26 concatenated intron sequences.
(0.05 MB PDF)
Per-site nucleotide divergence (d) and standard deviation (s.d.) among chickens, RJF and GJF.
(0.33 MB PDF)
Hardy-Weinberg equilibrium test in chicken breeds.
(0.44 MB PDF)
Acknowledgments
We are thankful to Dr. Moriwaki for his help for the discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by a grant (no. 16107001) from the Japan Society for Promotion of Science (http://www.jsps.go.jp/j-grantsinaid/index.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. DOI: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, et al. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. DOI: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fumihito A, Miyake T, Takada M, Ohno S, Kondo N. The genetic link between the Chinese bamboo partridge (Bambusicola thoracica) and the chicken and junglefowls of the genus Gallus. Proc Natl Acad Sci U S A. 1995;92:11053–11056. doi: 10.1073/pnas.92.24.11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fumihito A, Miyake T, Takada M, Shingu R, Endo T, et al. Monophyletic origin and unique dispersal patterns of domestic fowls. Proc Natl Acad Sci U S A. 1996;93:6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giuffra E, Kijas JMH, Amarger V, Carlborg Ö, Jeon J-T, et al. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kijas JMH, Andersson L. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J Mol Evol. 2001;52:302–308. doi: 10.1007/s002390010158. DOI: 10.1007/s002390010158. [DOI] [PubMed] [Google Scholar]
- 7.Götherström A, Anderung C, Hellborg L, Elburg R, Smith C, et al. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proc R Soc Lond B. 2005;272:2345–2350. doi: 10.1098/rspb.2005.3243. DOI: 10.1098/rspb.2005.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE. 2009;4:e4668. doi: 10.1371/journal.pone.0004668. DOI: 10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin CR. London: John Murray; 1868. The variation of animals and plants under domestication. [Google Scholar]
- 10.West B, Zhou B-X. Did chickens go north? New evidence for domestication. J Archaeol Sci. 1988;15:515–533. [Google Scholar]
- 11.Danforth CH. Gallus sonnerati and the domestic fowl. J Hered. 1958;49:167–169. [Google Scholar]
- 12.Steiner H. Ueber letal Fehentwicklung der Zeiten Zackommenschafts-Generation bei tieischen Arbastarden Arcb. Julius Klaus-Stift. Verebungforscb. Socialanthropol Rossenbg. 1945;20:236–251. [Google Scholar]
- 13.Morejohn GV. Breakdown of isolation mechanisms in two species of captive junglefowl. Evolution. 1968;22:576–582. doi: 10.1111/j.1558-5646.1968.tb03993.x. [DOI] [PubMed] [Google Scholar]
- 14.Hillel J, Groenen MAM, Tixier-Boichard M, Korol AB, David L, et al. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet Sel Evol. 2003;35:533–557. doi: 10.1186/1297-9686-35-6-533. DOI: 10.1051/gse:2003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, et al. Multiple maternal origins of chickens: Out of the Asian jungles. Mol Phylogenet Evol. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. DOI: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Kanginakudru S, Metta M, Jakati RD, Nagaraju J. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol Biol. 2008;8:174. doi: 10.1186/1471-2148-8-174. DOI: 10.1186/1471-2148-8-174.10.1016/j.ympev.2005.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishibori M, Shimogiri T, Hayashi T, Yasue H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim Genet. 2005;36:367–375. doi: 10.1111/j.1365-2052.2005.01318.x. DOI: 10.1111/j.1365-2052.2005.01318.x. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4:e1000010. doi: 10.1371/journal.pgen.1000010. DOI: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komiyama T, Ikeo, K, Gojobori T. The evolutionary origin of long-crowing chicken: its evolutionary relationship with fighting cocks disclosed by the mtDNA sequence analysis. Gene. 2004;333:91–99. doi: 10.1016/j.gene.2004.02.035. DOI: 10.1016/j.gene.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 20.Axelsson E, Webster MT, Smith NGC, Burt DW, Ellegren H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. DOI: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HM, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahata N, Satta Y, Klein J. Divergence time and population size in the lineage leading to modern humans. Theor Pop Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- 28.Takahata N, Satta Y. Evolution of the primate lineage leading to modern humans: Phylogenetic and demographic inferences from DNA sequences. Proc Natl Acad Sci U S A. 1997;94:4811–4815. doi: 10.1073/pnas.94.9.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahata N. Molecular clock: An anti-neo-Darwinian legacy. Genetics. 2007;176:1–6. doi: 10.1534/genetics.104.75135. DOI: 10.1534/genetics.104.75135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randi E, Lucchini V. Organization and evolution of the mitochondrial DNA control region in the Avian Genus Alectoris. J Mol Evol. 1998;47:449–462. doi: 10.1007/pl00006402. [DOI] [PubMed] [Google Scholar]
- 31.Quinn TW, Wilson AC. Sequence evolution in and around the mitochondrial control region in birds. J Mol Evol. 1993;37:417–425. doi: 10.1007/BF00178871. [DOI] [PubMed] [Google Scholar]
- 32.Nei M. New York: Columbia University Press; 1987. Molecular Evolutionary Genetics. [Google Scholar]
- 33.Nei M, Kumar S. New York: Oxford University Press; 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- 34.Ewens WJ. The sampling theory of selectively neutral alleles. Theor Pop Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. DOI: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 36.Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. DOI: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 37.Avise JC. New York: Chapman & Hall; 1993. Molecular Markers, Natural History and Evolution. [Google Scholar]
- 38.Brodkorb P. Catalogue of fossil birds, part 2 (Anseriformes through Galliformes). Bull Florida State Mus Biol Sci. 1964;8:195–335. [Google Scholar]
- 39.van Tuinen M, Dyke GJ. Calibration of galliform molecular clocks using multiple fossils and genetic partitions. Mol Phylogenet Evol. 2004;30:74–86. doi: 10.1016/s1055-7903(03)00164-7. DOI: 10.1016/S1055-7903(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 40.Prager EM, Brush AH, Nolan RA, Nakanishi M, Wilson AC. Slow evolution of transferrin and albumin in birds according to micro-complement fixation analysis. J Mol Evol. 1974;3:243–262. doi: 10.1007/BF01796041. [DOI] [PubMed] [Google Scholar]
- 41.Helm-Bychowski KM, Wilson AC. Rates of nuclear DNA evolution in pheasant-like birds: Evidence from restriction maps. Proc Natl Acad Sci U S A. 1986;83:688–692. doi: 10.1073/pnas.83.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimcheff DE, Drovetski SV, Mindell DP. Phylogeny of Tetraoninae and other galliform birds using mitochondrial 12S and ND2 genes. Mol Phylogenet Evol. 2002;24:203–215. doi: 10.1016/s1055-7903(02)00230-0. DOI: 10.1016/S1055-7903(02)00230-0. [DOI] [PubMed] [Google Scholar]
- 43.International Chicken Polymorphism Map Consortium. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. DOI: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muir WM, Wong GKS, Zhang Y, Wang J, Groenen MAM, et al. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc Natl Acad Sci U S A. 2008;105:17312–17317. doi: 10.1073/pnas.0806569105. DOI: 10.1073/pnas.0806569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of individual trees. Each tree is constructed from randomly concatenated sequences (see text). Scale of each tree is indicated at the bottom of each tree. Red rectangle indicates RedDB, and green rectangle means either GJF303 or GJF304, which is not in a green junglefowl cluster. Brackets at the right side indicate a cluster of C (chickens), R (red junglefowl), and G (green junglefowl), respectively.
(0.33 MB PDF)
Effect of introgressed sequences from chickens to GJF on the nucleotide diversity (pai) of GJF. Pai values at each intron with and without GJF303/304 were located on both sides of the graph. Among 30 introns, intron 13 and 17 were only the exception where removal of introgressed sequences did not reduce the pai value.
(0.06 MB PDF)
Haplotypes (numbers) of each individual.
(0.34 MB PDF)
Two sets of scale and shape parameters (a, b) of the gamma distribution in MCMC analyses and prior distribution (%).
(0.03 MB PDF)
The average distances of 26 concatenated intron sequences.
(0.05 MB PDF)
Per-site nucleotide divergence (d) and standard deviation (s.d.) among chickens, RJF and GJF.
(0.33 MB PDF)
Hardy-Weinberg equilibrium test in chicken breeds.
(0.44 MB PDF)