Abstract
Background
Various evolutionary models have been proposed to interpret the fate of paralogous duplicates, which provides substrates on which evolution selection could act. In particular, domestication, as a special selection, has played important role in crop cultivation with divergence of many genes controlling important agronomic traits. Recent studies have indicated that a pair of duplicate genes was often sub-functionalized from their ancestral functions held by the parental genes. We previously demonstrated that the rice cell-wall invertase (CWI) gene GIF1 that plays an important role in the grain-filling process was most likely subjected to domestication selection in the promoter region. Here, we report that GIF1 and another CWI gene OsCIN1 constitute a pair of duplicate genes with differentiated expression and function through independent selection.
Results
Through synteny analysis, we show that GIF1 and another cell-wall invertase gene OsCIN1 were paralogues derived from a segmental duplication originated during genome duplication of grasses. Results based on analyses of population genetics and gene phylogenetic tree of 25 cultivars and 25 wild rice sequences demonstrated that OsCIN1 was also artificially selected during rice domestication with a fixed mutation in the coding region, in contrast to GIF1 that was selected in the promoter region. GIF1 and OsCIN1 have evolved into different expression patterns and probable different kinetics parameters of enzymatic activity with the latter displaying less enzymatic activity. Overexpression of GIF1 and OsCIN1 also resulted in different phenotypes, suggesting that OsCIN1 might regulate other unrecognized biological process.
Conclusion
How gene duplication and divergence contribute to genetic novelty and morphological adaptation has been an interesting issue to geneticists and biologists. Our discovery that the duplicated pair of GIF1 and OsCIN1 has experienced sub-functionalization implies that selection could act independently on each duplicate towards different functional specificity, which provides a vivid example for evolution of genetic novelties in a model crop. Our results also further support the established hypothesis that gene duplication with sub-functionalization could be one solution for genetic adaptive conflict.
Background
Gene duplication has long been recognized to be an important way to provide a substrate on which evolution acts. The classical models that predict the most possible fate of one of the duplicate genes is to degenerate to a pseudogene or get lost from the genome due to vagaries of chromosomal remodeling, locus deletion or point mutation [1-5]. A less frequent fate of the duplicate genes is to gain a new function (neo-functionalization) when the other copy still maintains its original function. However, recent studies have indicated that the newly duplicated genes are often sub-functionalized from their ancestral functions held by the parental genes [6-8]. The sub-functionalization model (also referred to as duplication-degeneration-complementation model) explains that the duplicate genes are maintained in the genome relying on complementary degenerative changes in a pair of duplicate genes, such that the duplicate genes together retain the original functions of their single ancestor [1-5,9]. During this process, the expression domain shifting is the most common character of duplicate genes. As a consequence, the duplicates acquired sub-functionalization and then were less constrained by selection than the single ancestor, which had to maintain the capacity to fulfill all functions. Therefore, selection could act independently on each duplicate and increase the gene function specificity [10].
Sequence variation plays an essential role in functional renovation of genes, however, the relationship between DNA variation and functional consequence has been enigmatic for the vast majority of genes in plant and animal kingdoms, despite an increasing number of studies have been reported. Crop species and their wild relatives with available genome information are becoming fascinating subjects for study of correlation between cryptic genetic variation and functional evolution, because they have undergone rapid diversification under intense artificial selection [11-14]. Therefore, investigating crop domestication genes will shed meaningful light on genetic variation that drives cultivation adaptation [15]. Rice was used by human about 11,000 years ago [16,17]. It has been indicated that the divergence of indica and japonica predated rice domestication, suggesting that at least two genetically distinct gene pools of O. rufipogon were cultivated and subsequently domesticated [18-20]. During the long-term cultivation and domestication, tremendous diversity in rice has been selected by human, adapting to various ecosystems and agricultural management, in addition to high yielding characteristics, such as grain number and weight [12,17].
Various evolutionary models have been proposed to interpret the fate of paralogous duplicates, but little is known about the mechanisms of evolutionary change in duplicate genes leading to functional novelty. Rice has been recognized as a cereal model for such a study, and recent studies have discovered that some rice genes have undergone adaptive evolution under domestication selection [17,21]. We previously reported that the rice grain-filling gene GIF1 (OsCIN2) encoding a cell wall invertase (CWI) was most likely subjected to domestication selection [22]. Here, we report that GIF1 and another CWI gene OsCIN1 constitute a pair of duplicate genes with differentiated expression and function. Population genetic analysis showed that the two genes have experienced strong domestication selection, and interestingly, the target of selection in the GIF1 gene is the promoter region and that in OsCIN1 is the coding region.
Results
Evolution of GIF1 and OsCIN1 by gene duplication
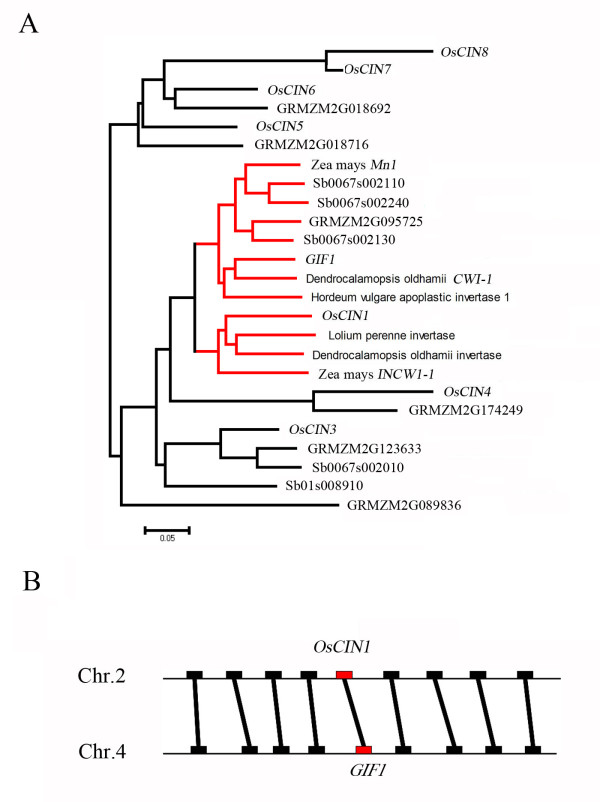
Rice genome has a CWI family consisting of eight members [23,24]. Our previous study has demonstrated that GIF1 is a member of the gene family and required for assimilated carbon partitioning during early grain-filling [22]. A phylogenetic analysis of the known plant CWI genes and predicted CWI genes from the recently released maize and sorghum genomes showed that OsCIN1, located on chromosome 2, is highly similar to GIF1 located on chromosome 4 (Figure 1A). Genetic distance based on amino acid substitutions also indicated that OsCIN1 is most closely related to GIF1 (Additional file 1). To gain insight into their evolutionary relationship, the 500-kb flanking sequences of the GIF1 and OsCIN1 regions were compared. The other eight expressed genes flanking the GIF1 gene on chromosome 4 show good colinearity to the eight counterparts of the OsCIN1 region on chromosome 2 (Figure 1B and Additional file 2). The result indicated that GIF1 and OsCIN1 rose via duplication of a genomic block, which could be as large as 15 Mb (data not shown). As shown in Figure 1A, phylogenetic analysis including cell-wall invertases of Zea mays, Sorghum bicolor, Lolium perenne, Hordeum vulgare, Dendrocalamopsis oldhamii and Oryza sativa showed that GIF1 was closer to cell-wall invertases of Zea mays, Hordeum vulgare and Dendrocalamopsis oldhamii, suggesting that this duplication might occur during the genome duplication of grasses [25]. By directly using synonymous substitution rate between the two paralogs (Ks = 0.57), and assuming the neutral evolutionary rate of rice genes (~6.5 × 10-9substitutions per silent site per year) [26,27], we estimated the time of duplication between GIF1 and OsCIN1 about 44 million years ago (MYA), a time much earlier than the genus Oryza diversified from a common ancestor about 15 MYA [28]. However, this estimated duplication age could be invalid because the regions were likely selected during rice domestication (see below).
Figure 1.
Phylogenetic relationship of cell wall invertases and synteny of the GIF1 and OsCIN1 loci. (A) The N-J phylogenetic tree constructed by MEGA program based on alignment of the DNA sequences of the 8 CWI genes of rice and CWI genes in other species, Lolium perenne, Hordeum vulgare, Dendrocalamopsis oldhamii and the recently released maize and sorghum genomes. Note that the rice GIF1 and OsCIN1 genes were paralogous within two subgroups. (B) Synteny between the GIF1 and OsCIN1 genome regions is illustrated schematically with homologous genes, indicating their duplication event.
To investigate the evidence for functional constraint on both copies at the DNA sequence level, we calculated Ka (non-synonymous substitution rate)/Ks ratios between GIF1, OsCIN1 and their homologs in maize, respectively [27]. The respective Ka/Ks value of GIF1 and OsCIN1 are 0.275 and 0.168 (p = 4.13E-24, p = 1.92E-50) (Table 1), suggesting strong purifying selection.
Table 1.
Summary of nonsynonymous (Ka) and synonymous (Ks) substitutions in OsCIN1 and GIF1
| Ka | Ks | Ka/Ks | p-value (fister) | |
|---|---|---|---|---|
| GIF1 | 0.120098 | 0.436208 | 0.275322 | 4.13E-24 |
| OsCIN1 | 0.124126 | 0.738313 | 0.168121 | 1.92E-50 |
Sub-functionalization of GIF1 and OsCIN1 by expression differentiation
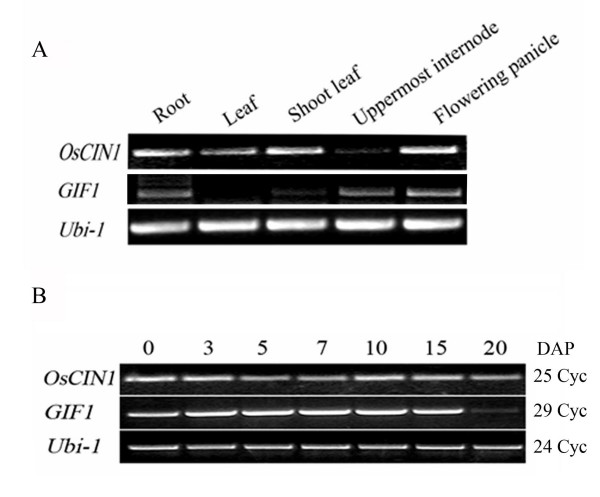
Duplicate genes can be maintained by sub-functionalization (the duplicate genes perform different aspects of the original gene's function), or neo-functionalization (one of the genes acquires a novel function), and may facilitate adaptation to environmental change [6-9]. Our previous research has indicated that other CINs, including OsCIN1, are not functionally redundant to GIF1 [22]. Here we further compared the expression patterns of GIF1 and OsCIN1 in different tissues and grain-filling stages. GIF1 transcripts were detected in roots, elongating internodes, shoots and panicles, but not in leaves. In contrast, OsCIN1 was expressed strongly in leaves, but weakly in elongating internodes (Figure 2A). During the early grain-filling stage, OsCIN1 transcript levels remained high while GIF1 transcript levels decreased after 15 days post-pollination (DAP) (Figure 2B). In situ hybridization experiments further showed that the GIF1 transcript was only detected in the ovular vascular tissue but not in the pericarp and endosperm [22]; in contrast, the OsCIN1 transcript was detected in both the pericarp and endosperm [29]. Consistent with the difference in their expression pattern, GIF1 was induced in the caryopses supplied with sugars, but OsCIN1 was inducible in the leaves treated with sucrose and pathogen [23]. These results evidently showed that GIF1 and OsCIN1 have differentiated in expression pattern after duplication through altering expression patterns in development and response to environment cues.
Figure 2.
Different GIF1 and OsCIN1 expression patterns. (A) Levels of the GIF1 and OsCIN1 transcripts were detected by RT-PCR in different tissues. (B) Levels of the GIF1 and OsCIN1 transcripts during grain filling. Note that OsCIN1 was constitutively expressed in developing grains. The experiments were repeated twice with similar results, Ubi-1 was used as a loading control for RT-PCR. DAP, day after pollination; Cyc, PCR cycles.
Sub-functionalization of GIF1 and OsCIN1 enzymes
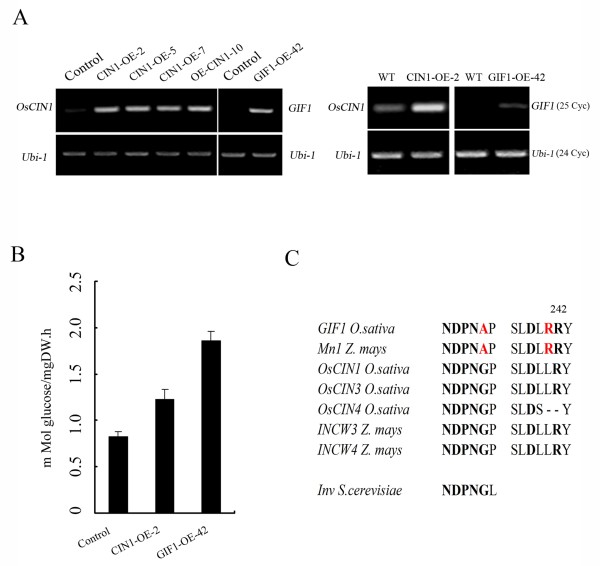
Total activity of cell-wall invertases was reduced to 17% of the wild-type in the gif1 mutant [22], indicating that GIF1 contributes to the majority of cell-wall invertase activity in early developing grains, although OsCIN1 was also expressed at a higher level in developing grains (Figure 2B). In support of this observation, the OsCIN1 T-DNA 'knockout' mutant did not show significant defect in grain filling and weight (J. -S. JEON, personal communication). To further determine the functional differentiation of the GIF1 and OsCIN1enzymes, we developed transgenic plants GIF1-OE [22] and CIN1-OE constitutively expressing GIF1 and OsCIN1 driven by the 35S promoter (Figure 3A). GIF1-OE plants exhibited significantly higher CWI activity than that of CIN1-OE plants although the latter accumulated higher OsCIN1 levels (Figure 3A and 3B). These results suggested that GIF1 and OsCIN1 could have different kinetics parameters such as Km and Vmax.
Figure 3.
Enzymatic activity of GIF1 and OsCIN1. (A) The GIF1 and OsCIN1 transcript levels in leaves detected by RT-PCR showing GIF1 and OsCIN1 overexpression in transgenic lines (left). Each one line of GIF1-OE and OsCIN1-OE was analyzed for GIF1 and OsCIN1 transcript levels respectively, with 25 PCR cycles (right). Ubi-1, a loading control for RT-PCR with 24 cycles. Note that line OsCIN1-OE-2 accumulated higher level of OsCIN1 transcripts than the level of GIF1 transcripts in line GIF1-OE-42. (B) CWI activity in leaves of the OsCIN1-OE and GIF1-OE plants and the empty vector control. C. Multiple sequence alignment of two conserved regions of cell wall invertases in maize, rice and yeast. The NDPNG domain and Asp-239/Lys-242 are shown in bold. The amino acid difference between GIF1 and OsCIN1 are shown in red color.
The difference in the kinetics parameters of enzymatic activity might result from the amino acid substitutions, in particular the GIF1 and OsCIN1 proteins contain Ala and Gly residues in the NDPNG domain (motif), respectively (Figure 3C). Phylogenetic reconstruction (Figure 1A and 3C) revealed that the Gly-26-Ala substitution occurred after the GIF1 and OsCIN1 duplication. The crystal structure of the Arabidopsis CWI indicated that the conserved NDPNG domain is critical for CWI activity [30-32]. However, GIF1 and the homologous maize Mn1 contain a NDPNA motif instead of the NDPNG motif that presents in OsCIN1 and other CWIs, suggesting that the segmental duplication predates maize and rice differentiation. Furthermore, the crystal structure showed that Asp-239 interacted with Lys-242 and both the two amino acids played a crucial role in the transfructosylation process and interacted via H-bonds with the bound substrate [32]. A Thr-241-Arg substitution in the Asp-239/Lys-242 region occurred in GIF1 as well as in Mn1. It is noteworthy that the mutation in the Mn1 gene also caused shrunk grains [33]. The synteny between the GIF1 and Mn1 genome regions suggested that they could be orthologues (Additional file 3). These structure differences might contribute to different enzymatic kinetics of the GIF1 and OsCIN1 proteins. Together, these results suggested that GIF1 and OsCIN1 were subjected to sub-functionalization, or that GIF1 (probable Mn1 too) might have neofunctionalized, albeit we do not know the ancestral function of the CWIs.
Different phenotypes of GIF1-OE and CIN1-OE plants support sub-functionalization of GIF1 and OsCIN1
To further confirm functional divergence of GIF1 and OsCIN1, we determined the phenotypes of GIF1-OE and CIN1-OE plants. In addition to producing shrunken grains (Figure 4A and 4C), GIF1-OE plants were also dwarfed in comparison with wild-type plants (Figure 4E). By contrast, CIN1-OE plants did not exhibit any obvious phenotype in grain-filling and plant development (Figure 4B and 4F). Instead, the CIN1-OE seeds exhibited marked preharvest sprouting in 6 of 10 transgenic lines tested, which expressed high OsCIN1 levels (Figure 4D and data not shown), a phenomenon never occurring to the wild-type japonica control. These observations suggest that OsCIN1 might indirectly modulating hormone signaling pathways through interfering sugar metabolism in seed germination, leading to preharvest sprouting, since the sugar regulates rice alpha amylase (34). Together, our results demonstrate that GIF1 and OsCIN1 have evolved differentially or most likely sub-functionalized after duplication.
Figure 4.
Phenotypes of OsCIN1-OE and GIF1-OE plants. (A) The GIF1-OE plants produced badly-filled grains (right), compared to the empty vector control (left). (B) The OsCIN1-OE plants produced grains completely filled (right). (C) Smaller seeds of the GIF1-OE plants (right), compared to the empty vector control (left). (D) The OsCIN1-OE seeds exhibited preharvest sprouting, while the control seeds did not germinate on panicle at the same time. (E) The GIF1-OE plants grew dwarfing in comparison with the control. (F) The OsCIN1-OE plants were morphologically similar to the control.
Evidence of OsCIN1 domestication-selection
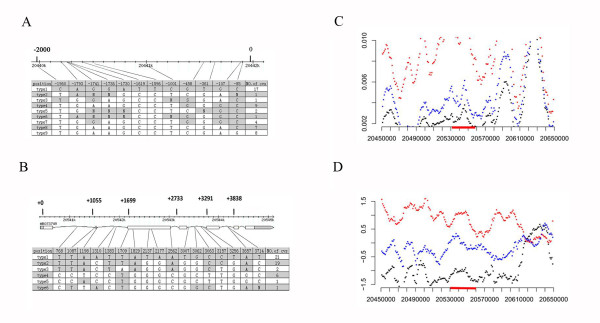
In the previous study, we analyzed artificial selection using the segment sequences of GIF1. Here we further analyzed the 2-kb promoter region of GIF1 in 25 cultivars and 25 wild rice germplasm (AA genome) (Table 2). We identified nine types of promoter sequences (Figure 5A). According to the promoter sequences, nearly all cultivated rice was classified into type 1, further supporting that the GIF1 promoter was artificially selected during rice domestication.
Table 2.
Cultivars and wild rice germplasm used in this study
| Sample name/IRGC no. | Variety name | Origin | Group |
|---|---|---|---|
| 8555 | DZ78 | Bangladesh | indica |
| 12883 | Mehr | Iran | indica |
| 45975 | Kalamkati | India | indica |
| 32399 | Phudugey | Bhutan | indica |
| 6307 | Jhona 349 | India | indica |
| 2540 | Haginomae Mochi | Japan | indica |
| 30416 | - | Brazil | indica |
| 9177 | JC91 | India | indica |
| 8231 | Gie 57 | Vietnam | indica |
| 9148 | TD2 | Thailand | indica |
| 9060 | JC101 | India | japonica |
| 9062 | JC111 | India | japonica |
| 38994 | Bico Branco | Brazil | japonica |
| 12793 | Kitrana 508 | Madagascar | japonica |
| RA4952 | Firooz | Iran | japonica |
| 66756 | Lemont | TX, USA | japonica |
| 50448 | Canella De Ferro | Brazil | japonica |
| 11010 | Maintmolotsy 1226 | Madagascar | japonica |
| 38698 | NPE 844 | Pakistan | japonica |
| 55471 | Chodongji | South Korea | japonica |
| 27630 | Darmali | Nepal | japonica |
| 27762 | Leung Pratew | Thailand | japonica |
| 6513 | - | Bangladesh | Southern Asian indica |
| 60542 | - | Bangladesh | Southern Asian indica |
| 31856 | - | Bangladesh | Southern Asian indica |
| Dongxiang | - | Dongxiang, China | O. rufipogon |
| Yuan3-9 | - | Yunnan, China | O. rufipogon |
| P25 | - | Guangdong, China | O. rufipogon |
| P46 | - | Hainan, China | O. rufipogon |
| P61 | - | Guangxi, China | O. rufipogon |
| 80506 | - | India | O. rufipogon |
| 106505 | - | Papua New Guinea | O. rufipogon |
| 105426 | - | Sri Lanka | O. rufipogon |
| 81982 | - | India | O. rufipogon |
| 81991 | - | Myanmar | O. rufipogon |
| 105912 | - | Thailand | O. rufipogon |
| 105958 | - | Indonesia | O. rufipogon |
| 105960 | - | Bangladesh | O. rufipogon |
| 106161 | - | Laos | O. rufipogon |
| Nepal | - | Nepal | O. rufipogon |
| 80470 | - | India | O. nivara |
| 105705 | - | Nepal | O. nivara |
| 106345 | - | Myanmar | O. nivara |
| 105879 | - | Bangladesh | O. nivara |
| 89215 | - | Cambodia | O. nivara |
| 106154 | - | Laos | O. nivara |
| 105784 | - | Thailand | O. nivara |
| 103407 | - | Sri Lanka | O. nivara |
| 106105 | - | India | O. nivara |
| 105327 | - | India | O. nivara |
Figure 5.
Nucleotide polymorphisms in GIF1 promoters and OsCIN1 gene regions. (A) Nucleotide polymorphisms detected in the 2-kb GIF1 promoter regions, which are classified into 9 types. The location of GIF1 on the chromosome was indicated. (B) Nucleotide polymorphisms in the OsCIN1 gene regions, which are divided into 6 types. The location of the OsCIN1 on the chromosome was indicated. (C) The molecular signature of domestication selection of OsCIN1. The OsCIN1 regions of 25 rice cultivars and 25 wild rice germplasm (Table 2) were sequenced. Haplotype diversity was calculated for nucleotide diversity (π) analysis. (D) Tajima's D-statistics were calculated with DnaSP version 4.0 for the OsCIN1 regions. Sequence positions were indicated with the OsCIN1 loci marked red. Red, wild rice; Blue, indica; black, japonica.
We further sequenced two BACs containing respective OsCIN1 and GIF1 of the BB-genome of wild rice (O. punctata), and found that the coding region of OsCIN1 contains more variation than the coding region of GIF1 in comparison with the reference AA genome sequences (Additional file 4) [28,35], probably suggesting that OsCIN1 and GIF1 might have experienced different selection during Oryza evolution. To carefully investigate the evolution pattern of the OsCIN1 gene, we sequenced the OsCIN1 genome regions of the same set of 25 cultivars and 25 wild rice germplasm (Table 2). Results showed that the silent-site nucleotide, θπ, of OsCIN1 in japonica and indica were 0.0025 and 0.0038, respectively, lower than θπ (0.0097) in wild rice; and also much lower than the genome average θπ (0.0052 and 0.0073) in japonica and indica, respectively (Figure 5C and Additional file 5). Furthermore, the Hudson-Kreitman-Aguade (HKA) test detected a highly significant deviation of OsCIN1 from neutrality for cultivated rice compared with the ADH1 gene (p = 5.89871E-11) [36], using O. punctata as an outgroup (Table 3). The negative deviation in Tajima's D was also consistent with a selective sweep at the OsCIN1 locus in both japonica and indica, but no such a pattern was observed in wild rice (Figure 5D and Additional file 5). These results suggest the OsCIN1 gene might also have been artificially selected. We also estimated genetic variation in upstream and downstream regions of OsCIN1 in the cultivars and wild rice genomes, and found that the region under selective sweep may extend as long as ~100-Kb. We further constructed a gene tree using 4.6-kb gene regions of OsCIN1 from the cultivars and wild rice (Figure 6A). Consequently, all the japonica and indica accessions formed a cluster in the gene tree, in considerable contrast to a genome tree (Figure 6B) established based on SNP data [37,38], suggesting OsCIN1 introgression from one subspecies into another subspecies after domestication-selection, although our data could not rule out the possibility that OsCIN1 was extensively selected during rice domestication independently in the two subspecies. All the results strongly support the hypothesis that OsCIN1 was selected during rice domestication. However, how the OsCIN1 gene has played a role in domestication is still unknown.
Table 3.
HKA tests of the OsCIN1 and GIF1 locia
| pair | polymorphism site number | sequence length | p value |
|---|---|---|---|
| Outgroup/Cultivar_OsCIN1 | 454 | 4636 | 5.90E-11 |
| Cultivar/Cultivar_OsCIN1 | 44 | 4772 | |
| Outgroup/Cultivar_GIF1 | 441 | 5980 | 0.00108 |
| Cultivar/Cultivar_GIF1 | 67 | 6149 | |
| Outgroup/Cultivar_ADH1 | 170 | 2573 | |
| Cultivar/Cultivar_ADH1 | 45 | 2573 |
aThe up-/downstream 2-kb genome regions of OsCIN1 and GIF1 were analyzed for HKA.
Figure 6.
Phylogenetic trees of rice germplasm. (A) Unrooted neighbor-joining phylogenetic tree of the OsCIN1 sequences was established according to sequence divergence of the rice germplasm set (Table 2). (B) Unrooted neighbor-joining phylogenetic tree was created according to whole genome SNP in the rice germplasm set (Table 2).
To narrow down the selection target in the OsCIN1 gene, we examined all the variations in the OsCIN1 genome regions, and found that an amino acid substitution (Arg-212-Leu) almost fixed in the rice cultivars, indicating that, unlike GIF1 which was selected in the promoter region, this site in the coding region could be the target of artificial selection in the OsCIN1 gene (Figure 5B). Further functional characterization of this site will provide more evidence to address how this site has contributed to OsCIN1 function in cultivated rice.
Discussion
Gene duplication and adaptive conflict
Gene duplication plays a fundamental role in organism evolution by providing genetic materials from which novel functions can arise. Large numbers of duplicate genes were found in genomes, which contributed greatly to the genome structure and function evolution [1-5,39]. In general, the duplicate genes have two fates: first, the duplicate gene lost its function due to chromosome remodeling, deletion, and point mutation (known as non-functionalization); second, the duplicate gene retained for the maintenance of ancestral functions [1-5]. According to adaptive conflict model, adaptive mutations could be prohibited in the case of multifunctional genes, or one mutation that can optimize one function, may compromise the other functions, this mutation will be prohibited [6,40]. The adaptive conflict could be solved by sub-functionalization of duplicate genes. In this case, the duplicate genes would be less constrained and be able to evolve new functions under selection [41,42]. With this scenario, it is common that one gene could have multifunction in nature [40,43].
Independent selection of GIF1 and OsCIN1 mutations
The sequenced genomes of Oryza sativa, Arabidopsis thaliana and Populus trichocarpa all contain a family of cell wall invertases [44-46], some members of these gene families were reported to be involved in growth and development, disease resistance, stress responses and cell death, suggesting that the CWI gene families might have undergone sub- or neo-functionalized in these species.
Through genomic synteny analysis, we showed that GIF1 and OsCIN1 derived from a segmental duplication from an ancestor, most likely during genome duplication in grass species. After duplication, GIF1 and OsCIN1 have evolved to gain divergent functions with different expression patterns and enzymatic kinetics parameters through accumulating mutations in cultivated rice. In contrast to GIF1 on which domestication selection mainly occurred in the cis-regulatory region (Figure 5A), the artificial selection occurred mainly in the coding region of OsCIN1 (Figure 5B). Therefore, both GIF1 and OsCIN1 were most likely subjected to domestication selection, resulting in a cultivated GIF1 locus for better harvest, although the biological importance of OsCIN1 in domestication remains enigmatic. With this scenario, GIF1 and OsCIN1 may provide a good genetic model to demonstrate how duplicate genes could evolve and be artificially selected independently during crop domestication with divergent functions derived from accumulation of mutations in the regulatory and coding regions respectively, adding to those systems reported [47,48].
Differential biological functions of GIF1 and OsCIN1
GIF1 is mainly expressed in seed vascular tissues and controls sucrose unloading for starch synthesis at the early grain-filling stage [22]. Overexpression of the GIF1 gene produced plants with marked defects both in grain-filling and development, indicating that over-activity of the GIF1 enzyme disrupts sugar homeostasis, a process important to normal grain and plant development. In contrast, OsCIN1 has lower CWI activity compared to GIF1 in the transgenic plants (Figure 3). Consistent with this, no obvious phenotype was observed in CIN1-OE plants except pre-harvest sprouting (Figure 4). Interestingly, OsCIN1 might be involved in pathogen defense and stress response [23]. It has been reported that sugars interact with signaling pathways mediated by phytohormones such as GA and ABA during seed germination and seedling development [34,49], which are also involved in stress responses. Preharvest sprouting of the CIN1-OE seeds may implicate a role for OsCIN1 in sugar-mediated alpha amylases activation [34]. However, detailed experiments are needed to dissect the OsCIN1 function.
Conclusion
Gene duplication and functional divergence contribute greatly to genetic novelty and adaptive evolution. However, molecular basis of selection and functionalization of duplicate genes remains largely unknown. Based on a set of data including population genetic analysis, fine sequencing of wild rice BACs, phenotyping of transgenic plants and analysis of gene expression and enzymatic activity, we provide a line of evidence that the two rice CWI genes GIF1 and OsCIN1 are a pair of duplicate genes and have been subjected to sub-functionalization during evolution or domestication selection. Therefore, duplicate genes could be independently selected towards different functional specificity, either on promoter for different expression pattern or on coding region for different protein function/activity. Our study provides a vivid example for evolution of genetic novelties in a model crop. The interesting phenotype of preharvest sprouting OsCIN1-OE plants suggests that OsCIN1 overaccumulation might disturb sugar balance during seed germination.
Methods
Duplication and synteny analysis
The 500-kb radiuses of the GIF1 and OsCIN1 regions were scanned for homologous pairs. A homolog pair was defined as a single nr-KOME cDNA and its blastn homolog. A total of 18 homologous genes in both sides of the GIF1 and OsCIN1 loci were compared to establish linearity.
Sequencing and evolution analysis
To investigate the selective forces acting on GIF1 and OsCIN1 on the molecular evolution scale, we estimated the statistic Ka/Ks using the re-sequencing data (see below) and the maize Mn1 and Incw1-1 as the outgroup sequence, where Ka was the number of nonsynonymous substitutions per nonsynonymous site and Ks was the number of synonymous substitutions per synonymous site [27]. Ka/Ks values significantly less than 1 were often taken as evidence of constraint. The mean Ks of nine pair homolog genes, including GIF1 and other eight genes (Additional file 2), in the GIF1 and OsCIN1 regions were used to estimate the duplication time. Two BAC clones of O. punctata (BB genome) from the OMAP project http://www.omap.org/ containing GIF1 and OsCIN1 respectively, were sequenced.
Analysis of OsCIN1 and GIF1 domestication
We deeply analyzed the OsCIN1 and GIF1 sequences from the re-sequenced genomes of 25 rice cultivars and 25 wild rice germplasm (Table 2), which has been done in Dr. Wen Wang's group, using Solexa technology (data not shown). Haplotype diversity was calculated for nucleotide diversity (π), and Tajima's D- statistics were calculated with DnaSP version 4.0. The gene tree was created using MEGA software [50]. The sequences then were aligned. The 2-kb up-/downstream genome sequences and the GIF1, OsCIN1 coding sequences were used for HKA test as described [36]. Sequences from wild rice O. punctata (BB genome) from the OMAP project http://www.omap.org/ were used as outgroups for the HKA test. The DNA phylogenetic tree was constructed by neighbor-joining method using MEGA. The known or predicted CWI genes with high sequence similarity to GIF1 from Oryza sativa, Lolium perenne, Hordeum vulgare, Dendrocalamopsis oldhamii and the recently released Zea mays and Sorghum bicolor genomes were used in this study.
Development and growth of OsCIN1-OE transgenic plants
The full-length OsCIN1 coding sequence was PCR-amplified from ZH11 cDNA by using the primers 5'-TCTAGTACAAAACAATGGGGACTC-3' and 5'-CGGAAAACCTCTTTATTATCTGTA-3'. The amplified fragment was subsequently cloned into the vector 35S-C1301 and transformed into ZH11 to generate 25 independent ectopic expression lines as described [22]. All transgenic materials were assayed in the second (T1) or third (T2) generations with 10-24 sibling plants grown in the paddy field to ensure agronomic traits.
Invertase activity assay
The caryopses were ground in the extraction buffer, and the extraction was centrifuged at 12,000 g for 10 min. The pellet was washed twice then re-suspended in the extraction buffer. Insoluble invertase activity was assayed as described [22].
RNA preparation and analysis
Total RNA was prepared from rice tissues using TRIzol reagent according to the manufacture's protocol (GIBCO BRL). For RT-PCR, 1-5 ug total RNA was used for the first-strand cDNA synthesis with the SuperScript III System (Invitrogen). RT-PCR analysis of GIF1 and OsCIN1 was performed with the primers [22,23].
Accession numbers
All sequences have been deposited in GenBank under accession numbers GU797900-GU798049.
Authors' contributions
EW, SG, B-RL, WW and ZH designed research. EW, LZ, HZ, LL, QW and QL performed research. EW, XX, SG, B-RL, WW and ZH analyzed data. EW, SG, B-RL, WW and ZH wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Summary of the number of amino acid substitutions per site of eight cell wall invertases.
Table S2. Genes and positions on the GIF1 and OsCIN1 chromosome regions.
Figures S1. The synteny between the rice GIF1 genome regions (chromosome 4) and the maize Mn1 genome regions (chromosome 10). High linearity indicates their duplication from the same ancestor fragment(s).
Figure S2. Sequence comparison of the GIF1 and OsCIN1 coding regions in japonica (O. sativa) and O. punctata (BB genome). Two BAC clones containing respective OsCIN1 and GIF1 of the BB-genome wild rice (O. punctata) were sequenced. A. Sequence alignment of the GIF1 coding regions in japonica (O. sativa) and O. punctata. B. Sequence alignment of the OsCIN1 coding regions in japonica (O. sativa) and O. punctata. Note that the OsCIN1 sequence has more divergence than GIF1.
Table S3. Nucleotide polymorphisms and neutrality test for domestication signature of OsCIN1.
Contributor Information
Ertao Wang, Email: etwang@sibs.ac.cn.
Xun Xu, Email: xuxun@genomics.org.cn.
Lin Zhang, Email: zhanglin@sippe.ac.cn.
Hong Zhang, Email: zhanghong@sippe.ac.cn.
Lin Lin, Email: linlin@sibs.ac.cn.
Qin Wang, Email: wangqin0313@163.com.
Qun Li, Email: qli@sippe.ac.cn.
Song Ge, Email: gesong@ibcas.ac.cn.
Bao-Rong Lu, Email: brlu@fudan.edu.cn.
Wen Wang, Email: wwang@mail.kiz.ac.cn.
Zuhua He, Email: zhhe@sibs.ac.cn.
Acknowledgements
We thank M. Long and W. Capoen for critical reading of the manuscript and helpful suggestions; R. A. Wing for O. punctata BAC clones. X. Zhang, L. Zeng and S. Ye for rice growth. This work was supported by grants from the Ministry of Science and Technology of China (2007AA02Z162, 2007AA10Z187), grants from the National Natural Science Foundation of China (30721061) to Z. He
References
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–53. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Bolot S, Abrouk M, Masood-Quraishi U, Stein N, Messing J, Feuillet C, Salse J. The 'inner circle' of the cereal genomes. Curr Opin Plant Biol. 2009;12:119–25. doi: 10.1016/j.pbi.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Gene Sharing and Evolution: The Diversity of Protein Functions. Massachusetts: Harvard Univ Press; 2007. [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploidy yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett B, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager K. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Roos-Ibarra J, Morell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA. 2007;104:8641–8648. doi: 10.1073/pnas.0700643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity: a key factor in the success of polyploidy wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- Mannion AM. Domestication and the origins of agriculture: an appraisal. Prog Phys Geogr. 1999;23:37–56. [Google Scholar]
- Sweeney M, McCouch S. The complex history of the domestication of rice. Ann Bot. 2007;100:951–957. doi: 10.1093/aob/mcm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen J. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Ishii T, Lamy F, Brar D, Panaud O. Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.) Mol Genet Genomics. 2004;272:504–511. doi: 10.1007/s00438-004-1069-6. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- Second G, Rouhan G. Human-mediated emergence as a weed and invasive radiation in the wild of the CD genome allotetraploid rice species (Oryza, Poaceae) in the Neotropics. PLoS One. 2008;3:e2613. doi: 10.1371/journal.pone.0002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Wang JJ, Zhu XD, Hao W, Wang LY, Li Q, Zhang LX, He W, Lu BR, Lin HX, Ma H, Zhang GQ, He ZH. Control of rice grain-filling and yield by a gene with potential signature of domestication. Nat Genet. 2008;40:1270–1274. doi: 10.1038/ng1108-1270. [DOI] [PubMed] [Google Scholar]
- Cho JI, Lee SK, Ko S, Kim HK, Jun SH, Lee YH, Bhoo SH, Lee KW, An G, Hahn TR, Jeon JS. Molecular cloning and expression analysis of the cell wall invertase gene family in rice (Oryza sativa L) Plant Cell Rep. 2005;24:225–236. doi: 10.1007/s00299-004-0910-z. [DOI] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. doi: 10.1016/S1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li S, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Wang J, Deng Y, Ran L, Shi X. The Genomes of Oryza sativa : a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene adh parallel rate differences at the plastid gene rbcL . Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, Talag J, Fan C, Goicoechea JL, Zuccolo A, Chen J, Bennetzen JL, Chen M, Jackson S, Wing RA. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20:3191–3209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T. Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 2002;43:452–459. doi: 10.1093/pcp/pcf055. [DOI] [PubMed] [Google Scholar]
- Lammens W, Le Roy K, Van Laere A, Rabijns A, Ende W Van den. Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose. J Mol Biol. 2008;377:378–385. doi: 10.1016/j.jmb.2007.12.074. [DOI] [PubMed] [Google Scholar]
- Verhaest M, Lammens W, Le Roy K, De Coninck B, De Ranter CJ, Van Laere A, Ende W Van den, Rabijns A. X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr. 2006;62:1555–1563. doi: 10.1107/S0907444906044489. [DOI] [PubMed] [Google Scholar]
- Le Roy K, Lammens W, Verhaest M, De Coninck B, Rabijns A, Van Laere A, Ende W Van den. Unraveling the difference between invertases and fructan exohydrolases: a single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiol. 2007;145:616–625. doi: 10.1104/pp.107.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Taliercio EW, Chourey PS. The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-A, Ho T-HD, Ho S-L, Yu S-M. Three novel Myb proteins with DNA-binding repeat mediate sugar and hormonal regulation of α-amylase gene expression. Plant Cell. 2002;14:1963–1980. doi: 10.1105/tpc.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Ammiraju JS, Sanyal A, Zhang S, Song R, Chen J, Li G, Sui Y, Song X, Cheng Z, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen M. Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci USA. 2009;106:2071–2076. doi: 10.1073/pnas.0812798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J, Wistow G. The recruitment of crystallins: new functions precede gene duplication. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Ji X, Ende W Van Den, Van Laere A, Cheng S, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Sherson SM, Alford HL, Forbes SM, Wallace EG, Smith SM. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J Exp Bot. 2003;54:525–31. doi: 10.1093/jxb/erg055. [DOI] [PubMed] [Google Scholar]
- Bocock PN, Morse AM, Dervinis C, Davis JM. Evolution and diversity of invertase genes in Populus trichocarpa . Planta. 2008;227:565–576. doi: 10.1007/s00425-007-0639-3. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Curr Biol. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Yuan K, Wysocka-Diller J. Phytohormone signaling pathways interact with sugars during seed germination and seedling development. Exp Bot. 2006;57:3359–3367. doi: 10.1093/jxb/erl096. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;4:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the number of amino acid substitutions per site of eight cell wall invertases.
Table S2. Genes and positions on the GIF1 and OsCIN1 chromosome regions.
Figures S1. The synteny between the rice GIF1 genome regions (chromosome 4) and the maize Mn1 genome regions (chromosome 10). High linearity indicates their duplication from the same ancestor fragment(s).
Figure S2. Sequence comparison of the GIF1 and OsCIN1 coding regions in japonica (O. sativa) and O. punctata (BB genome). Two BAC clones containing respective OsCIN1 and GIF1 of the BB-genome wild rice (O. punctata) were sequenced. A. Sequence alignment of the GIF1 coding regions in japonica (O. sativa) and O. punctata. B. Sequence alignment of the OsCIN1 coding regions in japonica (O. sativa) and O. punctata. Note that the OsCIN1 sequence has more divergence than GIF1.
Table S3. Nucleotide polymorphisms and neutrality test for domestication signature of OsCIN1.